Abstract
Growth arrest-specific (gas) genes are expressed preferentially in cells that enter a quiescent state. gas7, which we identified in serum-starved murine fibroblasts, is reported here to be expressed in vivo selectively in neuronal cells of the mature cerebral cortex, hippocampus, and cerebellum. gas7 transcripts encode a 48-kDa protein containing a structural domain that resembles sequences of OCT2, a POU transcription factor implicated in neuronal development, and synapsins, which have a role in modulating neurotransmitter release. Using in situ hybridization and immunocytochemical analysis, we show that GAS7 expression occurs prominently in cerebellar Purkinje cells and that inhibition of production in terminally differentiating cultures of embryonic murine cerebellum impedes neurite outgrowth from maturing Purkinje cells. Conversely, GAS7 overexpression in undifferentiated neuroblastoma cell cultures dramatically promotes neurite-like outgrowth. Collectively, our results provide evidence for an association between expression of this gas gene and neuronal development.
Growth arrest-specific (gas) genes are expressed preferentially in cultured cells that enter a quiescent state following serum deprivation or growth to confluence (1–3). In vivo, gas gene expression has been observed during the growth arrest that accompanies terminal differentiation of cells during development of peripheral nerves (4, 5). Individual gas genes have been implicated in a variety of biological functions, including the control of microfilament organization (6), nerve cell growth or differentiation (1, 4, 7–9), apoptosis (10), tyrosine kinase receptor activity (9), and the negative (11) and positive (3, 9) control of cell cycling. No sequence similarity or common structural features have been found among the gas genes identified thus far.
Earlier work done by using retroviral-based gene search (“gene trap”) vectors that fuse mouse chromosomal DNA sequences to the Escherichia coli lacZ reporter gene identified two chromosomal loci whose expression was activated during growth arrest of cultured NIH 3T3 fibroblasts (2). We describe here the isolation and characterization of gas7, a gas gene present at one of these loci. We show that gas7 expression occurs primarily in vivo in terminally differentiated brain cells and particularly prominently in mature cerebellar Purkinje neurons. Interference with GAS7 production in cultured embryonic cerebellar cells by gas7-specific antisense oligonucleotides inhibits the neurite formation normally observed during development of these neurons. Overexpression of gas7 in mouse neuroblastoma cell culture induces neurite-like outgrowth from these cells. Our results suggest that gas7 expression and the growth arrest associated with it may have a role in the developmental maturation of cerebellar neurons.
MATERIALS AND METHODS
Cloning and Sequencing Procedures.
A lacZ-containing fragment of genomic DNA from 354–7 cells, in which a Moloney murine leukemia virus (Mo-MuLV)lac provirus is integrated downstream of the gas7 genomic DNA (2), was introduced by cloning and subcloning into the pSP72NOT plasmid (Promega), yielding pGas354–7. A series of nested deletions of this fragment was sequenced (12) by using a primer complementary to a region near the 5′ end of the retroviral long terminal repeat. 5′ rapid amplification of cDNA ends (RACE) cloning (13) of cDNA corresponding to the chromosomal sequence fused to the lacZ reporter gene used poly(A) RNA isolated from serum-starved 354–7 cells (48 h in 0.5% calf serum) and a primer complementary to an internal sequence of the lacZ mRNA. A poly(dA) tail was added to the 3′ terminus of the DNA strand by using terminal deoxynucleotidyltransferase (Life Technologies, Grand Island, NY) and the resulting single-strand cDNA was amplified by two rounds of PCR, using initial and nested lacZ-specific and 5′ primers. Primer sequences and additional experimental details are published as supplemental data to this article on the PNAS web site (www.pnas.org). 3′ RACE cloning of gas7 cDNA from uninfected cells was carried out by using a kit (Life Technologies) and poly(A) RNA was isolated from serum-starved NIH 3T3 cells.
Chromosomal Localization of the Mouse gas7 Gene and Detection of gas7 Expression.
Mapping was carried out by using hybrid DNAs prepared from a panel of 21 Chinese hamster/mouse hybrid cell lines (kindly provided by U. Francke and X. Li of Stanford University). These were analyzed by PCR using oligonucleotide primers derived from intronic sequences of the mouse gas7 gene. The forward and reverse primers were 5′ TAG AGT TAG GCA ACT AGC AGG CT 3′ and 5′ CTT CAC ATT ATC TAG CAT CAT GCA 3′, respectively.
RNA in situ hybridization was carried out as described (14), except that the sagittal sections used were 7 μm thick and paraffin was removed by treatment with 100% xylene for 5 min before annealing.
For immunocytological detection of the GAS7 protein, rabbit anti-GAS7 antiserum was generated against histidine-tagged full-length GAS7 protein expressed in E. coli from plasmid pET15b. Fresh brain tissue taken from anesthetized adult male mice (ICR strain) was fixed in paraformaldehyde and washed, and sagittal sections 50 μm thick were prepared. Analysis was performed by using anti-GAS7 polyclonal antibodies (6–10 mg/ml in PBS), an avidin-biotin-peroxidase system (Vectastain, ABC system), and a diaminobenzidine substrate kit, both from Vector Laboratories (Burlingame, CA). For controls, the primary antibodies were replaced by preimmune serum.
Treatment of Primary Cerebellar Cell Cultures with Oligonucleotides.
Dissociated cells were prepared (15) from cerebella obtained from ICR-strain mice at day 18 of embryonic development and were treated with a mixture of two gas7 antisense oligonucleotides, gas7-p1 and gas7-p2 (the nucleotide sequences of gas7-p1 and -p2 are 5′-GGC CAT GAG AAG AGA TGG CAA CGG C and 5′-ATG TCC AAC ATG GAG AAC AGC TTT G, respectively) at a concentration of 10 μM. gas7-p1 was predicted to hybridize to the translation initiation region of gas7 mRNA; gas7-p2 contains a sequence complementary to the 5′-end of an alternatively spliced gas7 transcript shown in Fig. 1b. Control cultures were treated with the corresponding sense oligonucleotides, gas7-p3 and -p4, under the same conditions. The sequences of oligonucleotides selected did not match any known sequences in the GenBank database. Incubation of cell cultures treated with the oligonucleotides was continued for 3 days before immunocytochemical analysis.
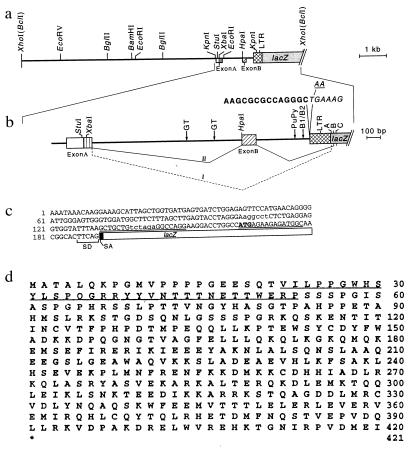
Figure 1.
(a) Map and structure of the chromosomal DNA in the region of integration of Mo-MuLVlac in cell line 354–7. The scale at the right indicates DNA length in kb pairs. The solid horizontal lines represent introns. Exon A is indicated by the open box adjoining the box with vertical lines; exon B is indicated by the box with diagonal lines. Restriction endonuclease cleavage sites are indicated, as are the long terminal repeat (LTR) (crosshatched boxes) and lacZ segments of the retrovirus construct. (b) Enlargement of the region containing the provirus integration site. The symbols are as described for a. The nucleotides shown by bold letters indicate the chromosomal sequence, and the italicized lettering indicates the Mo-MuLVlac/chromosomal DNA junction. The letters A, B, and C above the lacZ gene mark the locations of three splice acceptor sites. The locations of GT-rich repeats in the chromosomal DNA are indicated, as are the sites of polypurine/polypyrimidine (Pu/Py) segments and B1/B2 repeats. I and II represent alternatively spliced transcripts. Exon A is directly linked to lacZ at splice acceptor site A in transcript I. In transcript II, the 3′ region of exon A (box with vertical lines) is linked to exon B, which joins lacZ at splice acceptor site B. (c) The structure and sequence of the lacZ-fusion cDNA obtained by 5′ RACE cloning from cell line 354–7 is shown. The translation start codon (ATG) for the fusion protein is shown in bold letters. The splice donor sequence (SD) on chromosomal DNA and the splice acceptor sequence (SA) (shaded box) on the vector are also shown. The underlined sequences indicate the locations of the primers used for 3′ RACE cloning of gas7 cDNA. Lowercase letters identify the StuI and XbaI sites mapped in a and b. (d) Predicted amino acid sequence encoded by gas7 cDNA. The full-length cDNA sequence (GenBank accession no. U19860) contains a hexanucleotide sequence (AGAAGA) commonly present 5′ to a translation start codon. The translation stop codon is indicated by an asterisk. The underlined amino acid residues indicate the region of the GAS7 protein that resembles synapsin I.
Immunodetection of GAS7 in Cerebellar Primary and Neuroblastoma Cell Cultures.
Oligonucleotide-treated cerebellar cells grown on coverslips were examined as described by Baptista et al. (16). The same immunodetection procedures were used to detect cytoskeleton proteins [MAPII or glial fibrillary acidic protein (GFAP), a marker for nonneuronal glial cells], except that cells were fixed and permeabilized with cold methanol for 10 min at −20°C. Analysis was performed by using a Zeiss microscope (Axioplan). The neuroblastoma cell line, Neuro 2A (ATCC CCL-131), was cultured in DMEM with 10% fetal bovine serum, transfected with pcDNA3 expressing either gas7 or lacZ, and analyzed 48 h after transfection by using anti-GAS7 or anti-LacZ polyclonal antibodies.
RESULTS
Characterization of Genomic DNA Flanking Mo-MuLVlac Retrovirus Gene Search Vector Insert.
The NIH 3T3-derived cell line 354–7 (2) contains a single chromosomal insertion of a Mo-MuLV retrovirus carrying the E. coli lacZ gene, which is expressed in this insert from a chromosomally located promoter induced by serum starvation or by growth of cells to confluence. Genomic DNA from 354–7 was cloned in pGEM11 and fragments containing lacZ were identified by hybridization. A plasmid carrying a 10.5-kb insert whose map (Fig. 1) corresponds to the chromosomal DNA region containing the Mo-MuLVlac retrovirus (2) was designated pGas 354–7.
Sequence analysis of pGas 354–7 showed that insertion of the retrovirus into the mouse chromosome had occurred within a B1/B2 element, a dispersed repetitive sequence common in transcriptionally silent intergenic regions and in untranslated regions of active genes (17). A polypurine/polypyrimidine sequence (18) seen typically in S1 hypersensitive regions 5′ to genes is located near the B1 repeat. The terminal two base pairs of the native long terminal repeat sequence of the retrovirus (Fig. 1b: indicated as AA on the coding strand) were deleted at the chromosome/provirus junction, as is characteristic of retroviral integrations (19). Segments (33- and 28-bp) of GT repeats, which have been implicated in the formation of Z DNA within enhancer elements (20), are approximately 900 bp 5′ to this junction.
Cell Line 354–7 Encodes Two Species of lacZ Fusion Transcripts.
Sequence analysis of eight separate cDNA clones generated from 354–7 mRNA by 5′ RACE cloning showed two distinct species of fusion transcripts (Fig. 1b, transcripts I and II), which were mapped on the corresponding genomic DNA segment. In four clones, the terminus of an exon (designated here as exon A) was joined to a splice acceptor site located upstream of the lacZ gene (2), yielding transcript species I; in the remaining clones, the 3′ half of exon A was spliced to a more distal exon (designated as exon B), which in turn was spliced to the splice acceptor site located 5′ to lacZ to produce transcript species II. The sequences at exon/intron junctions correspond to previously reported consensus sequences at known splice sites (21), implying that accurate splicing had occurred. The chromosomal gene encoding the 5′ portion of these fusion transcripts was designated as gas7. A chromosomally encoded ATG codon in frame with lacZ was observed 27 bp 5′ to the splice junction in transcript I (Fig. 1c); in transcript II, an ATG translational start codon was generated at the splice junction. The presence of translational start codons at these locations is consistent with the finding that the LacZ fusion proteins observed previously in cell line 354–7 are approximately the same size as the native LacZ protein (2).
Cloning and Characterization of gas7 cDNA from Uninfected NIH 3T3 Cells.
Using information obtained from sequence analysis of gas7/lacZ fusion transcripts, we designed primers for 3′ RACE synthesis of cDNA corresponding to native gas7 transcripts; the template was poly(A) mRNA isolated from NIH 3T3 cells that had been cultured in 0.5% calf serum. Eight clones produced a cDNA species derived from transcripts in which exon A had been joined to the splice acceptor site 5′ to lacZ (i.e., transcript I in Fig. 1b). The translational ORF of 421 aa in these transcripts specifies a peptide predicted to have a molecular mass of 48,285 daltons (Fig. 1d; for the full-length cDNA sequence, see GenBank U19860). The hexanucleotide sequence AGAAGA, which commonly is present 5′ to translational start sites of mammalian cell mRNA (22), occurs at the predicted location. At the 3′ terminus of the gas7 transcript is a lengthy untranslated region that contains a poly(A) addition but lacks an obvious polyadenylation signal (Fig. 2a).
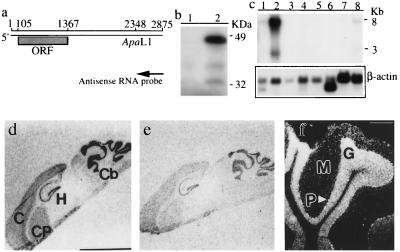
Figure 2.
Protein encoded by the GAS7 ORF and differential expression of gas7 mRNA in mouse tissues. (a) The filled box indicates the location of the computer-predicted GAS7 ORF. The orientation and location of the antisense RNA probe used for in situ hybridization are shown. The ApaLI restriction site was used to linearize plasmid DNA for in vitro synthesis of RNA probes. The nucleotide coordinates correspond to the sequence shown in GenBank accession no. U19860. (b) Autoradiograph of [35S]methionine-labeled GAS7 synthesized in vitro by using rabbit reticulocyte lysate (Promega) and analyzed on a 10% SDS-polyacrylamide gel. Lane 1: protein encoded by cDNA containing a 5′ deletion of approximately one-third of the cDNA sequence. Lane 2: protein synthesized from full-length gas7 cDNA. Molecular size markers in kilodaltons (kDa) are indicated. (c) Radioautograph of multiple-tissue RNA blot probed with full-length gas7 cDNA (Upper) or a fragment of β-actin cDNA (Lower) – both 32P-labeled by random priming (29). The multiple-tissue RNA blot (CLONTECH) contains poly(A) RNA (1 μg per lane) isolated from various tissues. β-Actin DNA (CLONTECH) was used as an internal control for the amount of RNA present in each lane; lane 1, heart; lane 2, brain; lane 3, spleen; lane 4, lung; lane 5, liver; lane 6, muscle; lane 7, kidney; lane 8, testis. (d–f) Differential expression of gas7 mRNA in the adult mouse brain. (d) Autoradiograph of a sagittal section hybridized in situ with the antisense RNA probe. (e) Same as d, except that the in situ hybridization mixture contained an excess amount of unlabeled gas7 antisense RNA. (f) Same as d, but shown in dark-field photomicrograph of emulsion autoradiogram. The antisense RNA probe used is shown in a. The brain regions indicated are cerebral cortex (C), hippocampus (H), caudate putamen (CP), and cerebellum (Cb). The molecular layer (M), Purkinje layer (P) and granule layer (G) are also indicated. (Scale bar: d, 4.5 mm; f, 400 μm.)
A protein of the size predicted from the GAS7 ORF was produced by gas7 cDNA in an in vitro transcription/translation assay (Fig. 2b, lane 2); deletion of the 5′ end of the cDNA prevented its synthesis (Fig. 2b, lane 1). The region encompassing amino acids 3–30 of the predicted GAS7 protein shows 43% identity to the proline-rich C terminus of OCT2, a member of the POU family of transcription factors that is known to have a role in neuronal development (23, 24); the same segment (amino acids 2–29) also has 48% identity to, and an additional 40% conserved amino acid substitutions that match, a region (amino acids 23–47, GenBank accession no. P17600) of synapsins Ia and Ib—proteins involved in modulating neurotransmitter release (25). A motif/pattern search showed a WW (Trp-Trp) module that mediates protein–protein interactions through the linking of proline-rich regions (26) between amino acids 22–60 of GAS7. Also encoded by gas7 mRNA is a short segment that strongly resembles (seven of eight amino acids are identical) a region present in CDC9, a yeast ATP synthetase known to be cell-cycle regulated (27).
Chromosomal Location of the Mouse gas7 Gene.
Intron-based oliogonucleotide primers flanking a 542-bp region of gas7 genomic DNA were tested for the ability to generate a fragment of the expected size by PCR analysis of a panel of mouse/hamster hybrid DNAs. A 542-bp fragment was observed by electrophoresis of DNA amplified from a cell line that contains mouse chromosome 11, but not for DNA from other mouse/hamster hybrids or the parental Chinese hamster cell line (data not shown). A reported human DNA fragment (SHGC-1222) having 85% sequence identity with gas7 maps to the short arm of chromosome 17, which is largely syngeneic with mouse chromosome 11 (28).
Expression of gas7 Occurs in Vivo Primarily in Brain Neurons.
Northern blot analysis of mRNA from a variety of adult mouse tissues showed that 3-kb and 8-kb gas7-hybridizing RNA species are produced abundantly in the brain (Fig. 2c, lane 2) and in much lower amounts in the heart and testes (Fig. 2c, lanes 1 and 8, respectively). In situ hybridization of a gas7 antisense RNA probe to a sagittal section of brain (Fig. 2d) showed gas7 mRNA expression most prominently in the cerebellum, moderately in the hippocampus, and less extensively in the cerebral cortex and caudate putamen. Addition of unlabeled gas7-antisense RNA interfered with this hybridization (Fig. 2e). More precise localization in the mouse cerebellum by dark-field microscopic analysis of emulsion autoradiograms showed that gas7 mRNA is present prominently in the Purkinje layer, but also in the granule and molecular layers (Fig. 2f).
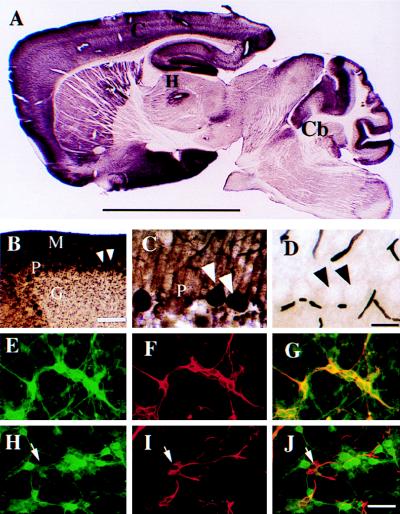
Immunohistochemical analysis of mouse brain slices by using antibody raised against histidine-tagged, full-length GAS7 protein showed high expression in the cerebellum and hippocampus (Fig. 3 A), consistent with the results of RNA in situ hybridization (Fig. 2d). In the cerebellum, GAS7 was detected prominently in the cell body and neurites of Purkinje neurons, and to a lesser extent in granule neurons (Fig. 3 B and C).
Figure 3.
Expression of GAS7 protein in vivo. Brain regions and layer description are the same as in Fig. 2. (A) Low magnification of a sagittal section of whole mouse brain probed with GAS7 antibody shows positive immunoreactivity in the C, H and Cb. (B) Magnified photograph of the layered structure of a cerebellar folia from A showing strong positive staining in the M and P layers and light staining in the G layer. (C) Magnified photograph of B. For a negative control, D, a sagittal section prepared from the same mouse brain was similarly analyzed, except that preimmune serum, instead of the GAS7 antibody, was used. Scale bar: A, 5 mm; B, 120 μm; C and D, 60 μm. (E–J) GAS7 is preferentially detected by confocal microscopy of neurons in primary cell culture of mouse cerebella. E and H were probed with anti-GAS7 antibodies, F and I were probed with anti-MAPII and anti-GFAP, respectively. G and J show overlaid photographs of E and F, H and I, respectively. The arrowheads in H–J show a glial cell that is positively stained with antibody against GFAP but not against GAS7. (Scale bar E–J: 40 μm.)
The identity of GAS7-expressing cells was studied further by using primary cultures of embryonic cerebellar neurons (16, 30, 31) and antibodies against cell-specific markers: MAPII (a microtubule-associated protein and a brain neuron marker), calbindin (a marker for Purkinje neurons), and GFAP. GAS7 expression in these cultures was similar to expression in cerebella of postnatal day 1 mice (Fig. 4 A, lanes 1–4). Confocal microscope analysis of cells doubly immunostained with antibodies showed GAS7 expression in cells that produce MAPII (Fig. 3 E–G) and calbindin (Fig. 4 B, b and c), but not in cells that express GFAP (Fig. 3 H–J). GAS7 was detected prominently in cytoplasm of both cell bodies and neurites of expressing cells (Fig. 3 E and H).
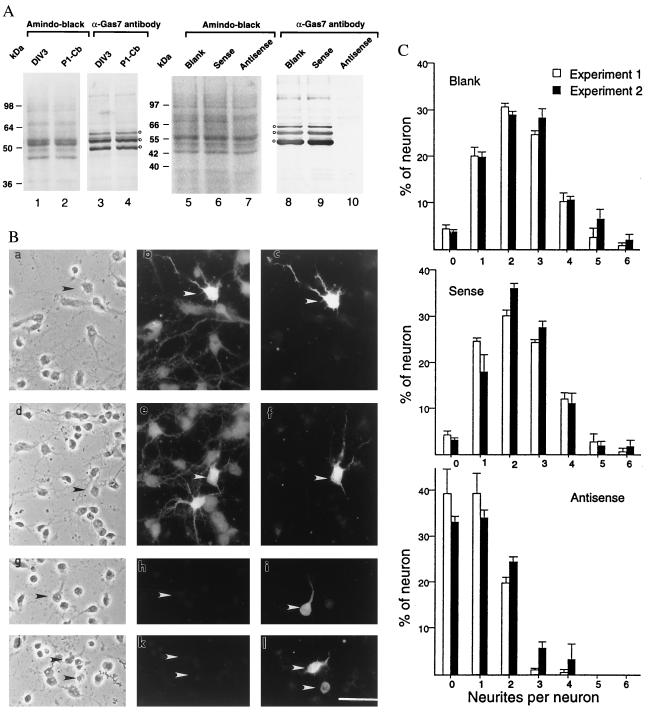
Figure 4.
(A) Specific reduction in the expression of GAS7 in a primary cerebellar cell culture treated with gas7-antisense oligonucleotides. Extracts of the sense-, antisense-, or no (Blank) oligonucleotide-treated cells prepared as described in Materials and Methods were electrophoresed on SDS/PAGE gels (lanes 5–7), and expression of GAS7 was detected by Western blot analysis using anti-GAS7 antiserum (lanes 8–10). Lanes 1–4 and 5–7 show expression of GAS7 in extracts prepared from the cerebella of postnatal day 1 mice (P1-Cb) or from 3-day in vitro primary cerebellar cell cultures (DIV3). Amido-black staining showing approximately equal loading of protein extracts in each lane before probing with GAS7 antibody is also shown (lanes 1, 2, 5, 6, and 7). Molecular mass size markers in kDa are indicated. (B) Photomicrographs of cells treated with gas7-antisense oligonucleotide. Cerebellar cell primary cultures were untreated (a–c), or treated with a gas7-sense (d–f) or with gas7-antisense (g–l) oligonucleotide. Cells were examined by phase-contrast microscopy (a, d, g, and j), or stained with GAS7 antibodies (b, e, h, and k) or calbindin antibodies (c, f, i, and l). Arrowheads indicate Purkinje cells that reacted with antibodies against calbindin. (Bar = 70 μm.) (C) Quantitative comparison of the numbers of neurites formed in the Purkinje cells (followed by the staining of calbindin antibodies) after treatment with gas7 sense or antisense oligonucleotides. A neurite was defined in this analysis as a cellular projection longer than the length of the cell body. The data shown are the cumulative results (mean ± SD) of two independent experiments. In each experiment, at least 300 calbindin-stained neurons were examined.
Western blot analysis of extracts prepared from mouse cerebella or cultured cerebellar neurons by using GAS7 antibodies showed three dominant bands of 48 kDa, 55 kDa, and 63 kDa (Fig. 4A, lanes 3, 4, 8, and 9). The N-terminal amino acid sequence for the 48-kDa protein (which was the most abundant species and was purified by immunoprecipitation) was identical to the GAS7 N-terminal sequence (PPPPG) predicted from analysis of cloned gas7 cDNA (Fig. 1d). We were unable to determine the N-terminal sequence of the other immunoreactive bands.
Reduced gas7 Expression Is Associated with Inhibition of Neurite Formation in Cultured Purkinje Cells.
Our finding that gas7, which was originally identified because of its selective expression in growth-arrested fibroblasts (2), is also expressed abundantly and selectively in mature (i.e., growth-arrested) cerebellar neurons led us to investigate whether gas7 expression has a role in neuronal maturation. Treatment of cultured cerebellar neurons with antisense oligonucleotides complementary to the 5′-end of gas7 transcripts greatly decreased the abundance of all three bands detected by GAS7 antibodies in Western blots (Fig. 4A, lane 10), identifying all of these bands as gas7 gene products. Mock treatment or treatment with sense oligonucleotides had no effect on GAS7 production (Fig. 4A, lanes 8 and 9). Immunostaining of antisense oligonucleotide-treated cerebellar cultured neurons also showed a decrease in gas7 expression, which was associated with an overall reduction in neurite formation in gas7-expressing cells (Fig. 4B, g–l). Staining with the Purkinje marker, calbindin, showed that Purkinje cells were among the specific neurons that showed decreased gas7 expression and inhibited neurite outgrowth during treatment with antisense oligoneucleotides (Fig. 4B, h and k vs. i and l). Whereas untreated calbindin-producing Purkinje neurons and those treated with sense oligonucleotides often had up to five or six neurites, few Purkinje cells treated with the antisense oligonucleotide contained more than two neurites and none showed more than four neurites (Fig. 4B, g–l, and 4C). Cells containing zero neurites increased 8-fold during antisense oligonucleotide treatment (from 4% to 36%); the percentage of cells containing more than one neurite was reduced from 74% for untreated and sense oligonucleotide-treated cells to 27% for antisense oligonucleotide-treated cells. The modal number of neurites was reduced from 2 to 3 in untreated cells or in cells treated with sense oligonucleotide to 1 or 0 in cultures treated with antisense oligonucleotide (Fig. 4B, a, d vs. g, j or c, f vs. i, l, and 4C). Antisense oligonucleotide inhibition of neuritogenesis, but not of calbidin production, during Purkinje cell maturation suggests that gas7 expression may be associated with morphological but not biochemical differentiation.
Overproduction of GAS7 in Neuroblastoma Cell Cultures Promotes Neurite-Like Outgrowth.
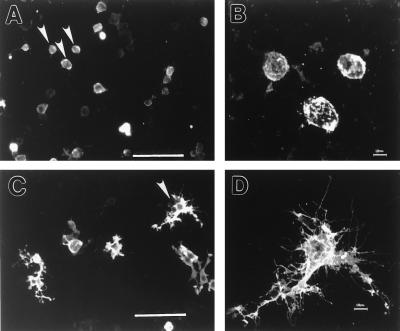
The mouse neuroblastoma cell line Neuro 2A normally grows as round or flat cells that show very limited morphological resemblance to differentiated neurons but have been reported to form short cellular extensions in about 10% of the population (32). In the presence of retinoic acid, Neuro 2A cells are induced to produce extended neurite processes and to express the differentiation marker, MAPII (32). As seen in Fig. 5 C and D, overproduction of gas7 in Neuro 2A cells induced dramatic morphological changes, promoting abundant lengthy neurite-like extensions from 65% of cells. In contrast, less than 17% of control cells overexpressing LacZ showed projections, and these remained short and stubby (Fig. 5 A and B). However, unlike the neurite formation in Neuro 2A, which is induced by retinoic acid, the morphological changes induced by gas7 overexpression in Neuro 2A cells were not accompanied by immunocytologically detectable evidence of MAPII production (data not shown). This observation is consistent with the notion that gas7 expression may lead to morphological, but not biochemical, differentiation.
Figure 5.
Effect of overproduction of gas7 in neuroblastoma cells. (A and B) LacZ-expressing Neuro-2A cells. (C and D) GAS7-expressing Neuro-2A cells. Enlargements of cells designated by arrowheads in A and C are shown in B and D, respectively. [Bars = 100 μm (A and C); 10 μm (B and D).]
DISCUSSION
The results reported here indicate that the growth arrest-specific gene gas7 (2) is expressed preferentially in the brain and particularly in the Purkinje neurons of the cerebellum. Treatment of primary cerebellar cell cultures with antisense oligonucleotides inhibited gas7 expression and interfered with neurite formation in Purkinje cells. Conversely, overproduction of gas7 in mouse neuroblastoma cells promoted neurite-like outgrowth. Together, these results suggest that gas7 may have a role in promoting, and possibly maintaining, maturation and morphological differentiation of these cerebellar neurons. However, in contrast to the morphological consequences of inhibiting gas7 expression or overproducing the gas7 gene product, at least some biochemical concomitants of neuronal differentiation do not appear to be affected by gas7 in either Purkinje cells or the Neuro 2A cell line.
The N-terminal end of GAS7 contains a region (amino acids 2–29) highly similar to a region (amino acids 23–47) of synapsins Ia/b, which tether synaptic vesicles to the cytoskeleton and regulate the function of synapses (25). Overexpression of synapsins in NIH 3T3 (33) or neuroblastoma cells (34) results in the outgrowth of highly elongated neurites and the formation of synapse-like cell–cell contacts. However, we do not know the relevance of the region of gas7/synapsin homology in these effects. The same GAS7 region that shows homology to synapsins (amino acids 3–30) resembles the proline-rich transcription activation domain of OCT2 (35), which is known to regulate targeted gene expression in neuronal cells (23).
A human genomic DNA nucleotide sequence showing 85% identity to mouse gas7 cDNA was identified on human chromosome 17 (National Center for Biotechnology Information no. G13706), which is largely syngeneic to mouse chromosome 11, where we have mapped gas7. Additionally, a second human cDNA clone (AB007854/KIAA0394) recently isolated from the brain has been predicted from its sequence to encode an ORF of 412 amino acids, of which 401 (97%) are identical to the amino acid sequences of mouse GAS7.
The gas7/lacZ fusion transcripts we isolated from growth-arrested NIH 3T3 fibroblasts showed differential splicing. Our Northern blot analysis of mRNA from mouse brain tissue (Fig. 2c) detected two species of gas7 transcripts, one of which is much larger then the gas7 cDNA species we studied in the experiments reported here, suggesting that differential splicing of gas7 mRNA occurs in the normal brain as well as in cultured serum-starved fibroblasts. A human cDNA that appears to be homologous to mouse gas7 (AB007854/KIAA0394) contains 7,979 nucleotides and is about the same size as the larger of the two gas7 transcripts shown in Fig. 2c. This transcript is similar in sequence to the mouse transcript we analyzed, except that it contains a much longer 3′ untranslated region. This finding suggests that differential splicing at a second site in the 3′ untranslated region may also have occurred in the production of the mouse transcripts.
Multiple species of cerebellar GAS7 proteins were detected by Western blot analysis (Fig. 4A). From the size and N-terminal sequence of the 48-kDa protein, we conclude that this protein is encoded by transcripts corresponding to the cDNA we isolated from uninfected NIH 3T3 cells. Other proteins may be encoded by alternatively spliced gas7 RNA (i.e., transcript II; Fig. 1b).
Two other gas genes previously isolated from growth-arrested NIH 3T3 cells, gas3 and gas6, have been implicated in the growth or differentiation of nerve cells. GAS3 (later renamed PMP22; ref. 36) has extensive sequence similarity to SR13, a myelin sheath protein of Schwann cells that is strongly down-regulated following sciatic nerve injury (37). GAS3/PMP22 is capable of delaying the transition from G0/G1 to S phase in Schwann cells and increasing the fraction of G1 cells in actively cycling NIH 3T3 cell populations (8). When overexpressed, GAS3/PMP22 produces an apoptosis-like phenotype (10). Furthermore, both hyper-myelination and de-myelinating peripheral neuropathy occur in mice homozygously mutated in GAS3/PMP22 (4). Although GAS3 plays a role in negatively regulating cell growth, GAS6 is a ligand for protein tyrosine kinase receptors of the Axl/Rse and Sky families and promotes the growth of at least Schwann cells (9).
Supplementary Material
Acknowledgments
We thank Drs. N. Heintz, R. Scheller, L. Reichardt, C.-H. Lin, S.-D. Wang, C.-L. Chien, and A. Chien-Chang for helpful comments and suggestions, and G.-S. Wang for technical assistance in determining certain cDNA sequences. This study fulfilled in part the requirements for the Ph.D. thesis of Y.-T.J., National Yang-Ming University, Taiwan, Republic of China. These studies were supported by an intramural fund from Academia Sinica, by grants from the National Science Council of Taiwan, Republic of China, to S.L.-C. (NSC 83-0412-B001-067 M15 and 86-2311-B001-093), M.-L.T. (NSC85-2311-B010-003B16), C.C.-K.C. (NSC84-2331-B182-065M02), and S.N.C. (NSC84-0412/2331-B001-094-Y), and by grant HG00325 to S.N.C. from the National Institutes of Health. B.-R.S. received a postdoctoral fellowship from the National Science Council of Taiwan, Republic of China.
ABBREVIATIONS
- Mo-MuLV
Moloney murine leukemia virus
- RACE
rapid amplification of cDNA ends
- GFAP
glial fibrillary acidic protein
References
- 1.Schneider C, King R M, Philipson L. Cell. 1988;54:787–793. doi: 10.1016/s0092-8674(88)91065-3. [DOI] [PubMed] [Google Scholar]
- 2.Brenner D G, Lin-Chao S, Cohen S N. Proc Natl Acad Sci USA. 1989;86:5517–5521. doi: 10.1073/pnas.86.14.5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lih C-J, Cohen S N, Wang C, Lin-Chao S. Proc Natl Acad Sci USA. 1996;93:4617–4622. doi: 10.1073/pnas.93.10.4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adlkofer K, Martini R, Aguzzi A, Zielasek J, Toyka K V, Suter U. Nat Genet. 1995;11:274–280. doi: 10.1038/ng1195-274. [DOI] [PubMed] [Google Scholar]
- 5.Magyar J P, Martini R, Ruelicke T, Aguzzi A, Adlkofer K, Dembic Z, Zielasek J, Toyka K V, Suter U. J Neurosci. 1996;16:5351–5360. doi: 10.1523/JNEUROSCI.16-17-05351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brancolini C, Bottega S, Schneider C. J Cell Biol. 1992;117:1251–1261. doi: 10.1083/jcb.117.6.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manfioletti G, Ruaro M E, Del Sal G, Philipson L, Schneider C. Mol Cell Biol. 1990;10:2924–2930. doi: 10.1128/mcb.10.6.2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zoidl G, Blass-Kampmann S, D’Urso D, Schmalenbach C, Müller H W. EMBO J. 1995;14:1122–1128. doi: 10.1002/j.1460-2075.1995.tb07095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li R, Chen J, Hammonds G, Phillips H, Armanini M, Wood P, Bunge R, Godowski P J, Sliwkowski M X, Mather J P. J Neurosci. 1996;16:2012–2019. doi: 10.1523/JNEUROSCI.16-06-02012.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fabbretti E, Edomi P, Brancolini C, Schneider C. Genes Dev. 1995;9:1846–1856. doi: 10.1101/gad.9.15.1846. [DOI] [PubMed] [Google Scholar]
- 11.Del Sal G, Collavin L, Ruaro M E, Saccone S, Della Valle G, Schneider C. Proc Natl Acad Sci USA. 1994;91:1848–1852. doi: 10.1073/pnas.91.5.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nancy F, Nolan C, Ferguson M. In: Molecular Cloning: A Laboratory Manual. 2nd Ed. Sambrook J, Fritsch E F, Maniatis T, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. pp. 60–80. [Google Scholar]
- 13.Frohman M A. In: PCR Protocols: A Guide to Methods and Applications. Innis M A, Gelfand D H, Snindky J J, White T J, editors. San Diego: Academic; 1990. pp. 28–38. [Google Scholar]
- 14.Tsaur M-L, Sheng M, Lowenstein D H, Jan Y N, Jan L Y. Neuron. 1992;8:1055–1067. doi: 10.1016/0896-6273(92)90127-y. [DOI] [PubMed] [Google Scholar]
- 15.Brewer G J, Torricelli J R, Evege E K, Price P J. J Neurosci Res. 1993;35:567–576. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- 16.Baptista C A, Hatten M E, Blazeski R, Mason C A. Neuron. 1994;12:243–260. doi: 10.1016/0896-6273(94)90268-2. [DOI] [PubMed] [Google Scholar]
- 17.Ireland R C, Kotarski M A, Johnston L A, Stadler U, Birkenmeier E, Kozak L P. J Biol Chem. 1986;261:11779–11785. [PubMed] [Google Scholar]
- 18.Hoffman-Liebermann B, Liebermann D, Troutt A, Kedes L H, Cohen S N. Mol Cell Biol. 1986;6:3632–3642. doi: 10.1128/mcb.6.11.3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colicelli J, Goff S P. J Mol Biol. 1988;199:47–59. doi: 10.1016/0022-2836(88)90378-6. [DOI] [PubMed] [Google Scholar]
- 20.Thomas M J, Freeland T M, Strobl J S. Mol Cell Biol. 1990;10:5378–5387. doi: 10.1128/mcb.10.10.5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharp P A. Science. 1987;235:766–771. doi: 10.1126/science.3544217. [DOI] [PubMed] [Google Scholar]
- 22.Kozak M. J Biol Chem. 1991;266:19867–19870. [PubMed] [Google Scholar]
- 23.Ensor E, Kendall G, Allchorne A, Woolf C J, Latchman D S. Neurosci Lett. 1996;204:29–32. doi: 10.1016/0304-3940(96)12308-9. [DOI] [PubMed] [Google Scholar]
- 24.Treacy M N, He X, Rosenfeld M G. Nature (London) 1991;350:577–584. doi: 10.1038/350577a0. [DOI] [PubMed] [Google Scholar]
- 25.Südhof T C. Nature (London) 1995;375:645–653. doi: 10.1038/375645a0. [DOI] [PubMed] [Google Scholar]
- 26.Sudol M, Chen H I, Bougeret C, Einbond A, Bork P. FEBS Lett. 1995;369:67–71. doi: 10.1016/0014-5793(95)00550-s. [DOI] [PubMed] [Google Scholar]
- 27.Barker D G, White J H, Johnston L H. Nucleic Acids Res. 1985;13:8323–8337. doi: 10.1093/nar/13.23.8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kurtz A, Zimmer A. Mamm Genome. 1995;6:379–380. doi: 10.1007/BF00364810. [DOI] [PubMed] [Google Scholar]
- 29.Feinberg A P, Vogelstein B. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 30.Mount H T J, Dean D O, Alberch J, Dreyfus C F, Black I B. Proc Natl Acad Sci USA. 1995;92:9092–9096. doi: 10.1073/pnas.92.20.9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caceres A, Kosik K S. Nature (London) 1990;343:461–463. doi: 10.1038/343461a0. [DOI] [PubMed] [Google Scholar]
- 32.Fischer I, Shea T B, Sapirstein V S, Kosik K S. Dev Brain Res. 1986;25:99–109. doi: 10.1016/0165-3806(86)90156-2. [DOI] [PubMed] [Google Scholar]
- 33.Han H-Q, Greengard P. Proc Natl Acad Sci USA. 1994;91:8557–8561. doi: 10.1073/pnas.91.18.8557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han H-Q, Nichols R A, Rubin M R, Bähler M, Greengard P. Nature (London) 1991;349:697–700. doi: 10.1038/349697a0. [DOI] [PubMed] [Google Scholar]
- 35.Tanaka M, Clouston W M, Herr W. Mol Cell Biol. 1994;14:6046–6055. doi: 10.1128/mcb.14.9.6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Snipes G J, Suter U, Welcher A A, Shooter E M. J Cell Biol. 1992;117:225–238. doi: 10.1083/jcb.117.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Welcher A A, Suter U, De Leon M, Snipes G J, Shooter E M. Proc Natl Acad Sci USA. 1991;88:7195–7199. doi: 10.1073/pnas.88.16.7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.