Abstract
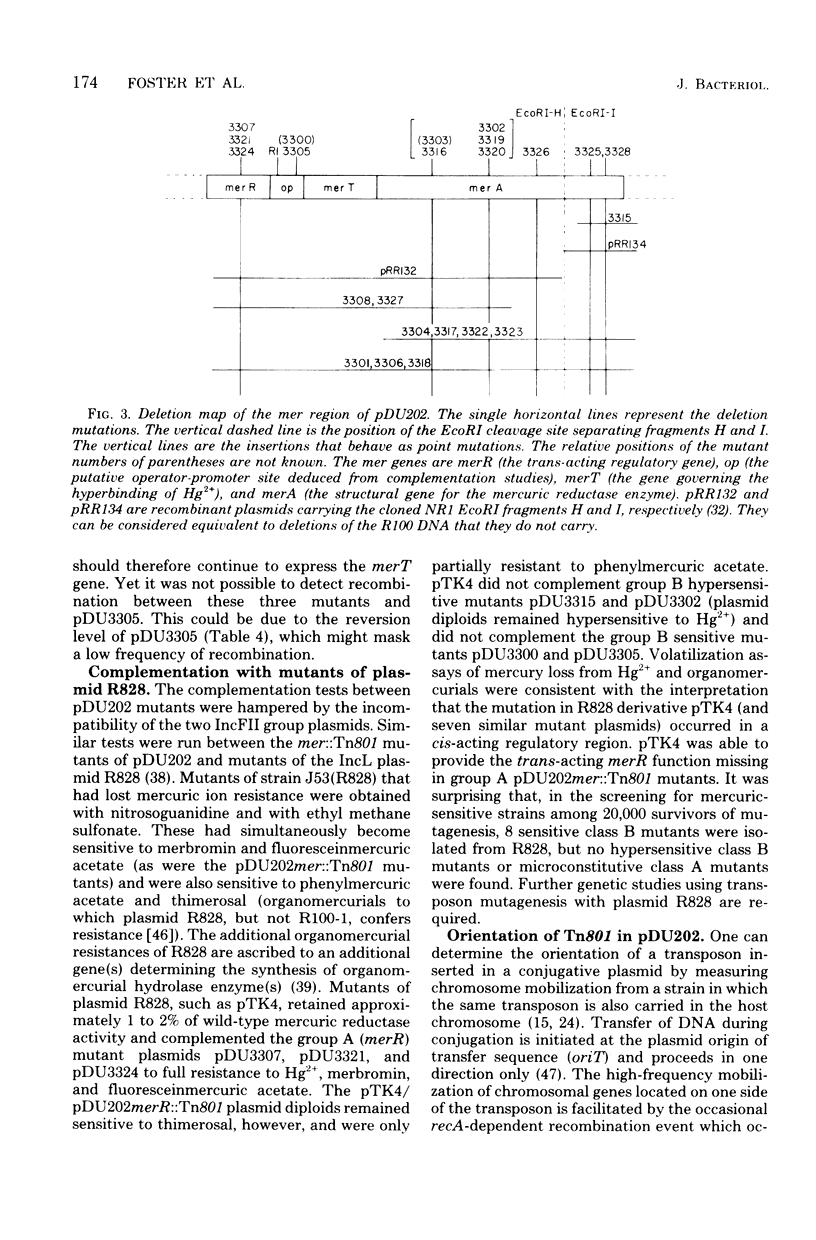
A series of 23 transposon 801(Tn801)-induced mutations of plasmid R100-1 from mercuric salts resistance to sensitivity was studied. Although Tn801 transposed frequently into the mer region of the plasmid, fine structural analysis showed that the site of insertion within mer varied. About one-half of the Tn801 insertion events also caused a deletion of greater than 1 megadalton. Genetic and restriction endonuclease EcoRI and BamHI analysis of the mutant plasmid deoxyribonucleic acid elucidated the organization of the mer operon and suggested the existence of a trans-acting regulatory factor governing resistance to mercuric salts. Tn801 insertions leading to mercuric sensitivity occurred in the restriction endonuclease fragments EcoRI-H and EcoRI-I. Regulatory mutations leading to a 50-fold-reduced synthesis of mercuric reductase enzyme occurred in two complementation classes thought to represent the gene for a trans-acting inducer molecule and a cis-acting operator-promoter sequence. Mutations leading to total loss of the enzyme mercuric reductase occurred on both the EcoRI-H and EcoRI-I fragments, showing that the structural gene for this enzyme (merA) bridges the EcoRI cleavage site separating the segments. Hypersensitivity to mercuric salts resulted when Tn801 insertion occurred in the reductase gene in the operatordistal portion of the operon. Hypersensitive cells inducibly bound three to five times more Hg2+ at low concentrations than did sensitive (plasmidless) cells. This finding led to the proposal that another gene (merT) controls uptake of Hg2+ by the cells. Transcription of the operon was deduced to start in the EcoRI-H fragment and to move into the EcoRI-I fragment of the plasmid genome.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Achtman M., Willetts N., Clark A. J. Beginning a genetic analysis of conjugational transfer determined by the F factor in Escherichia coli by isolation and characterization of transfer-deficient mutants. J Bacteriol. 1971 May;106(2):529–538. doi: 10.1128/jb.106.2.529-538.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J., Low K. B., Taylor A. L. Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev. 1976 Mar;40(1):116–167. doi: 10.1128/br.40.1.116-167.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth P. T., Grinter N. J., Bradley D. E. Conjugal transfer system of plasmid RP4: analysis by transposon 7 insertion. J Bacteriol. 1978 Jan;133(1):43–52. doi: 10.1128/jb.133.1.43-52.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett P. M., Richmond M. H. Translocation of a discrete piece of deoxyribonucleic acid carrying an amp gene between replicons in Eschericha coli. J Bacteriol. 1976 Apr;126(1):1–6. doi: 10.1128/jb.126.1.1-6.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M., Allet B., Gallay E., Boy de La Tour E., Caro L. Involvement of IS1 in the dissociation of the r-determinant and RTF components of the plasmid R100.1. Mol Gen Genet. 1977 Jun 24;153(3):289–295. doi: 10.1007/BF00431594. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Properties of a supercoiled deoxyribonucleic acid-protein relaxation complex and strand specificity of the relaxation event. Biochemistry. 1970 Oct 27;9(22):4428–4440. doi: 10.1021/bi00824a026. [DOI] [PubMed] [Google Scholar]
- Cohen S. N. Transposable genetic elements and plasmid evolution. Nature. 1976 Oct 28;263(5580):731–738. doi: 10.1038/263731a0. [DOI] [PubMed] [Google Scholar]
- Dempsey W. B., McIntire S. A., Willetts N., Schottel J., Kinscherf T. G., Silver S., Shannon W. A., Jr Properties of lambda transducing bacteriophages carrying R100 plasmid DNA: mercury resistance genes. J Bacteriol. 1978 Dec;136(3):1084–1093. doi: 10.1128/jb.136.3.1084-1093.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan G., Sherratt D. The transposon Tn1 as a probe for studying ColE1 structure and function. Mol Gen Genet. 1977 Mar 7;151(2):151–160. doi: 10.1007/BF00338689. [DOI] [PubMed] [Google Scholar]
- Foster T. J. Insertion of the tetracycline resistance translocation unit Tn10 in the lac operon of Escherichia coli K12. Mol Gen Genet. 1977 Sep 9;154(3):305–309. doi: 10.1007/BF00571287. [DOI] [PubMed] [Google Scholar]
- Foster T. J., Nakahara H. Deletions in the r-determinant mer region of plasmid R100-1 selected for loss of mercury hypersensitivy. J Bacteriol. 1979 Oct;140(1):301–305. doi: 10.1128/jb.140.1.301-305.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T. J., Walsh A. Phenotypic characterization of R-factor tetracycline resistance determinants. Genet Res. 1974 Dec;24(3):333–343. doi: 10.1017/s0016672300015330. [DOI] [PubMed] [Google Scholar]
- Foster T. J., Willetts N. S. Genetic analysis of deletions of R100-1 that are both transfer-deficient and tetracycline-sensitive. J Gen Microbiol. 1976 Mar;93(1):133–140. doi: 10.1099/00221287-93-1-133. [DOI] [PubMed] [Google Scholar]
- Grinsted J., Saunders J. R., Ingram L. C., Sykes R. B., Richmond M. H. Properties of a R factor which originated in Pseudomonas aeruginosa 1822. J Bacteriol. 1972 May;110(2):529–537. doi: 10.1128/jb.110.2.529-537.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., Rubens C., Falkow S. Translocation of a plasmid DNA sequence which mediates ampicillin resistance: molecular nature and specificity of insertion. Proc Natl Acad Sci U S A. 1975 Sep;72(9):3623–3627. doi: 10.1073/pnas.72.9.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inselburg J. Isolation, mapping, and examination of effects of TnA insertions in ColE1 plasmids. J Bacteriol. 1977 Jan;129(1):482–491. doi: 10.1128/jb.129.1.482-491.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaki K., Tashiro Y., Funaba T. Mechanism of mercuric chloride resistance in microorganisms. 3. Purification and properties of a mercuric ion reducing enzyme from Escherichia coli bearing R factor. J Biochem. 1974 Mar;75(3):591–599. doi: 10.1093/oxfordjournals.jbchem.a130427. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Roth J., Botstein D. Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J Mol Biol. 1977 Oct 15;116(1):125–159. doi: 10.1016/0022-2836(77)90123-1. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Translocatable elements in procaryotes. Cell. 1977 May;11(1):11–23. doi: 10.1016/0092-8674(77)90313-0. [DOI] [PubMed] [Google Scholar]
- Kopecko D. J., Brevet J., Cohen S. N. Involvement of multiple translocating DNA segments and recombinational hotspots in the structural evolution of bacterial plasmids. J Mol Biol. 1976 Dec;108(2):333–360. doi: 10.1016/s0022-2836(76)80124-6. [DOI] [PubMed] [Google Scholar]
- Kretschmer P. J., Cohen S. N. Selected translocation of plasmid genes: frequency and regional specificity of translocation of the Tn3 element. J Bacteriol. 1977 May;130(2):888–899. doi: 10.1128/jb.130.2.888-899.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D., Chandler M. Mapping of the drug resistance genes carried by the r-determinant of the R100.1 plasmid. Mol Gen Genet. 1977 Nov 29;157(1):17–23. doi: 10.1007/BF00268682. [DOI] [PubMed] [Google Scholar]
- Meynell E., Datta N. The relation of resistance transfer factors to the F-factor (sex-factor) of Escherichia coli K12. Genet Res. 1966 Feb;7(1):134–140. doi: 10.1017/s0016672300009538. [DOI] [PubMed] [Google Scholar]
- Mickel S., Ohtsubo E., Bauer W. Heteroduplex mapping of small plasmids derived from R-factor R12: in vivo recombination occurs at IS1 insertion sequences. Gene. 1977;2(3-4):193–210. doi: 10.1016/0378-1119(77)90017-8. [DOI] [PubMed] [Google Scholar]
- Miki T., Easton A. M., Rownd R. H. Mapping of the resistance genes of the R plasmid NR1. Mol Gen Genet. 1978 Jan 17;158(3):217–224. doi: 10.1007/BF00267192. [DOI] [PubMed] [Google Scholar]
- Nakahara H., Kinscherf T. G., Silver S., Miki T., Easton A. M., Rownd R. H. Gene copy number effects in the mer operon of plasmid NR1. J Bacteriol. 1979 Apr;138(1):284–287. doi: 10.1128/jb.138.1.284-287.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahara H., Silver S., Miki T., Rownd R. H. Hypersensitivity to Hg2+ and hyperbinding activity associated with cloned fragments of the mercurial resistance operon of plasmid NR1. J Bacteriol. 1979 Oct;140(1):161–166. doi: 10.1128/jb.140.1.161-166.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisen P. D., Kopecko D. J., Chou J., Cohen S. N. Site-specific DNA deletions occurring adjacent to the termini of a transposable ampicillin resistance element (Tn3). J Mol Biol. 1977 Dec 25;117(4):975–978. doi: 10.1016/s0022-2836(77)80008-9. [DOI] [PubMed] [Google Scholar]
- Ohtsubo H., Ohtsubo E. Nucleotide sequence of an insertion element, IS1. Proc Natl Acad Sci U S A. 1978 Feb;75(2):615–619. doi: 10.1073/pnas.75.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savageau M. A. Genetic regulatory mechanisms and the ecological niche of Escherichia coli. Proc Natl Acad Sci U S A. 1974 Jun;71(6):2453–2455. doi: 10.1073/pnas.71.6.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schottel J. L. The mercuric and organomercurial detoxifying enzymes from a plasmid-bearing strain of Escherichia coli. J Biol Chem. 1978 Jun 25;253(12):4341–4349. [PubMed] [Google Scholar]
- Schottel J., Mandal A., Clark D., Silver S., Hedges R. W. Volatilisation of mercury and organomercurials determined by inducible R-factor systems in enteric bacteria. Nature. 1974 Sep 27;251(5473):335–337. doi: 10.1038/251335a0. [DOI] [PubMed] [Google Scholar]
- Sharp P. A., Cohen S. N., Davidson N. Electron microscope heteroduplex studies of sequence relations among plasmids of Escherichia coli. II. Structure of drug resistance (R) factors and F factors. J Mol Biol. 1973 Apr 5;75(2):235–255. doi: 10.1016/0022-2836(73)90018-1. [DOI] [PubMed] [Google Scholar]
- Smith B. R., Schleif R. Nucleotide sequence of the L-arabinose regulatory region of Escherichia coli K12. J Biol Chem. 1978 Oct 10;253(19):6931–6933. [PubMed] [Google Scholar]
- So M., Heffron F., Falkow S. Method for the genetic labeling of cryptic plasmids. J Bacteriol. 1978 Mar;133(3):1520–1523. doi: 10.1128/jb.133.3.1520-1523.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers A. O., Silver S. Mercury resistance in a plasmid-bearing strain of Escherichia coli. J Bacteriol. 1972 Dec;112(3):1228–1236. doi: 10.1128/jb.112.3.1228-1236.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers A. O., Silver S. Microbial transformations of metals. Annu Rev Microbiol. 1978;32:637–672. doi: 10.1146/annurev.mi.32.100178.003225. [DOI] [PubMed] [Google Scholar]
- Tanak N., Cramer J. H., Rownd R. H. EcoRI restriction endonuclease map of the composite R plasmid NR1. J Bacteriol. 1976 Jul;127(1):619–636. doi: 10.1128/jb.127.1.619-636.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willetts N. S. Location of the origin of transfer of the sex factor F. J Bacteriol. 1972 Nov;112(2):773–778. doi: 10.1128/jb.112.2.773-778.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]



