Abstract
Previous studies using traditional biochemical identification methods to study the ecology of commercial sauerkraut fermentations revealed that four species of lactic acid bacteria, Leuconostoc mesenteroides, Lactobacillus plantarum, Pediococcus pentosaceus, and Lactobacillus brevis, were the primary microorganisms in these fermentations. In this study, 686 isolates were collected from four commercial fermentations and analyzed by DNA fingerprinting. The results indicate that the species of lactic acid bacteria present in sauerkraut fermentations are more diverse than previously reported and include Leuconostoc citreum, Leuconostoc argentinum, Lactobacillus paraplantarum, Lactobacillus coryniformis, and Weissella sp. The newly identified species Leuconostoc fallax was also found. Unexpectedly, only two isolates of P. pentosaceus and 15 isolates of L. brevis were recovered during this study. A better understanding of the microbiota may aid in the development of low-salt fermentations, which may have altered microflora and altered sensory characteristics.
Sauerkraut fermentation involves many physical, chemical, and microbiological changes that influence the quality and safety of the product. This fermentation can be broadly categorized as having successive stages, including an initial heterofermentative stage followed by a homofermentative stage (11, 22). Historically, four species of lactic acid bacteria (LAB) have been identified as organisms that are present in sauerkraut fermentations: Leuconostoc mesenteroides, Lactobacillus brevis, Pediococcus pentosaceus, and Lactobacillus plantarum. The identification of these microorganisms has been based on morphological and biochemical criteria (22). Several species of LAB other than the four species mentioned above have been found in cabbage fermentations, including Lactobacillus curvatus, Lactobacillus sakei, Lactococcus lactis subsp. lactis, and Leuconostoc fallax (3, 19, 25). Recently, six L. fallax strains were isolated from brine samples obtained from sauerkraut fermentations (2). The methods used for taxonomic characterization of LAB have been modified, and new species have been identified using molecular techniques (1, 10, 20, 24). Improvements in molecular identification techniques for the study of microbial ecology have created new opportunities for the analysis of food fermentations.
This study was carried out because of the need to reduce sodium chloride (salt) waste from commercial vegetable fermentations. It is well documented that the concentration of salt has a controlling influence on the microbial succession in a typical sauerkraut fermentation (11, 12, 22). It may be possible to reduce salt waste by fermenting cabbage with 1% salt instead of 2% salt, the concentration typically used. The introduction of an L. mesenteroides starter culture to the fermentation could help ensure that the initial stage of the fermentation produces the desirable flavor compounds (11). A method has been developed (23) to determine the ability of an unmarked starter culture to predominate over the indigenous microbiota in sauerkraut fermentations. In that study (23), however, the effect of the starter culture on the indigenous microbiota was not determined.
During the sauerkraut fermentation, there is a rapid turnover of LAB species. The dominant species present in the fermentation shifts within 2 to 3 days from less-acid-tolerant heterolactic LAB species to more-acid-tolerant homolactic fermenting LAB species, with the sequential populations each reaching concentrations of 108 to 109 CFU/g (11). Under normal conditions, the fermentation is essentially complete within 2 weeks, with the most-acid-tolerant species, L. plantarum, predominating. Our objective was to characterize the dominant LAB species in the successive stages of fermentation.
There is strong evidence that the genetic diversity of many ecosystems as assessed by molecular techniques exceeds the microbial diversity determined by traditional culture-based identification methods (26). However, due to the rapid succession of LAB in sauerkraut and the potential impact of large numbers of dead cells on culture-free nucleic acid-based methods, we carried out this study using bacterial isolates. In this study, we characterized isolates from the microbiota of commercial sauerkraut fermentations using an rRNA gene intergenic transcribed spacer (ITS)-PCR method with a database of known ITS-PCR patterns for LAB (4; this study), supplemented with 16S rRNA gene sequence analysis. Several species of LAB not previously found in sauerkraut were observed. Notably, Weissella and Leuconostoc citreum were found in the heterolactic phase of the fermentations, and two of the four LAB species expected to be present (P. pentosaceus and L. brevis) were apparently minor constituents of the microbiota. A better understanding of the microbial ecology of sauerkraut fermentations may aid in the development of low-salt fermentation technology, which may alter the normal microflora and sauerkraut flavor (F. Breidt, v. Plengvidhya, Z. Lu, and H. P. Fleming, unpublished).
MATERIALS AND METHODS
Bacterial strains.
Bacterial strains were obtained from the Food Fermentation Culture Collection, U.S. Department of Agriculture, Agricultural Research Service, Raleigh, NC (see Table S1 in the supplemental material) or were isolated as described below. LAB were maintained at −80°C in De Man-Rogosa-Sharpe (MRS) broth (Difco Laboratories) supplemented with 16% glycerol and were grown in MRS broth or on MRS agar plates (MRS broth supplemented with 1.5% agar; Difco). To inhibit aerobic yeast or mold growth on MRS agar plates with sauerkraut isolates, MRS agar was supplemented with 0.2% sodium azide (Sigma Chemical Co., St. Louis, MO) or plates were incubated in an anaerobic hood (Coy Laboratories, Detroit, MI).
Commercial sauerkraut fermentation and sample collection.
Samples of sauerkraut fermentations from a commercial processing plant in Wisconsin consisting of approximately 100 Mg of cabbage were used for this study; samples were obtained from one tank in the first year of the study (Y1) and from three different tanks in the second year (Y2). Only one sauerkraut production facility in Wisconsin was used in this study because there are currently very few commercial production facilities in the United States. The fermentation tanks were cement tanks, and the approximate dimensions were as follows: length, 5 m; width, 5 m; and depth, 4 m. Each fermentation was carried out with approximately 2.3% (final equilibrated concentration) NaCl, which was added by a dry salting process using shredded cabbage (mixed cultivars). The cabbage was manually spread in the tanks, covered with plastic sheeting, and initially weighted down with water on top of the plastic sheeting. The fermentation temperature was not controlled, but the average temperature in these commercial fermentations typically was 18°C. For analysis of cabbage prior to fermentation, shredded cabbage samples (approximately 500 g) were collected in sterile plastic bags prior to salting. Brine samples (100 ml each) from the fermentations were obtained for microbial and biochemical analysis, with a 1-cm-diameter, perforated stainless steel tube that was sealed at the bottom, from a depth of approximately 60 cm from the top of each fermentation tank and about 60 cm from the sides. The sampling apparatus was sanitized with a dilute (10%) Clorox solution and rinsed with sterile water prior to use. Brine and cabbage samples were obtained on days 1, 3, 7, and 14 after the start of fermentation and were placed in two 50-ml sterile plastic tubes (catalog no. 430829; Corning, Inc., Corning, NY). The cabbage and brine samples were transported to the laboratory by overnight mail in insulated boxes containing wet ice packs to maintain the temperature between 0 and 5°C. Samples were processed immediately upon arrival.
Chemical and microbial analysis of cabbage samples.
For chemical and microbiological analysis, 100 g of the shredded cabbage was blended in sterile glass blender jars with 200 g of water for 3 min using a Waring blender (Waring Products, Torrington, CT) and then homogenized with a stomacher (Stomacher 400; Tekmar, Cincinnati, OH) for 3 min at the maximum speed using bags containing an internal filter. Filtrate from the extract (approximately 30 ml) was transferred to a 50-ml sterile plastic tube and frozen at −20°C for subsequent chemical analysis. Prior to freezing, 1 ml of the cabbage extract was removed for microbiological analysis (see below). A chemical analysis was carried out using high-performance liquid chromatography. Organic acids and ethanol were analyzed using an anion exchange column (Aminex HPX-87H; Bio-Rad Laboratories) and 0.03 N H2SO4 at a flow rate of 0.8 ml/min at 75°C. A UV detector (UV-6000; Thermo Separation Products, Inc., San Jose, CA) and a differential refractometer (Waters 410; Waters, Milford, MA) were connected in series for detection of organic acids (at 210 nm) and ethanol. Sugars and mannitol were separated by using a Carbopac PA1 column (Dionex Corp., Sunnyvale, CA) and 0.12 N NaOH at a flow rate of 0.8 ml/min at room temperature and were detected with a pulsed amperometric detector (model PAD-2; Dionex). The salt (NaCl) content in the brine was determined by titration with an AgNO3 solution using 4′5′-dichlorofluorescein as the indicator (13).
For microbial analysis, samples were diluted in sterile saline (0.85% NaCl) and plated on plate count agar (Difco Laboratories, Detroit, MI), modified violet-red bile agar (violet-red bile agar [Difco] supplemented with 1% glucose), modified MRS agar (MRS agar [Difco] supplemented with 0.2% sodium azide), and yeast extract malt agar (Difco) containing 250 mg/liter chlortetracycline and 250 mg/liter chloramphenicol (Sigma Aldrich, St. Louis, MO) to enumerate the total aerobic microbiota, Enterobacteriaceae, LAB, and yeasts and molds, respectively. In addition, each brine sample was plated on unmodified MRS agar (without sodium azide) for collection of LAB isolates. Plating and plate counting were performed using a spiral plater (model 4000; Spiral Biotech, Norwood, MA) with an automated colony counter (Protos Plus; Microbiology International, Frederick, MD). For each of the five sampling times (days 1, 3, 5, 7, and 14 after the start of fermentation) in Y1, 96 isolated colonies were randomly selected from MRS agar and isolated on MRS agar, and then cells were frozen in MRS broth containing 16% glycerol at −80°C. For the four sampling times in Y2 (days 1, 3, 7, and 14), 20 isolates were obtained from each of three fermentation tanks. This resulted in a combined total for both years of the study of 720 possible isolates, although only 686 isolates were recovered. The isolates were screened for gas production using Durham tubes (6 by 50 mm; Kimble) inverted in 5 ml MRS broth. In addition, cells were cultured on homolactic-heterolactic differentiation medium (17).
DNA extraction and PCR amplification.
MRS broth cultures of each fermentation isolate were incubated at 30°C for 12 to 16 h and then subjected to DNA extraction. Genomic DNA was isolated using a Wizard genomic DNA purification kit (Promega Corporation, Madison, WI) in accordance with the manufacturer's instructions, with minor modifications. Twenty microliters of mutanolysin (2.4 mg/ml; Sigma-Aldrich) was substituted for lysostaphin. The method of Breidt and Fleming (4) was used to amplify the ITS region between the 16S and 23S rRNA genes. Each 100 μl of reaction mixture consisted of 10 μl of 10× PCR buffer (500 mM KCl, 100 mM Tris-Cl, pH 8.0), 10 μl of 25 mM MgCl2, 1 μl of a deoxynucleoside triphosphate mixture (25 mM of each deoxynucleoside triphosphate; Stratagene), 4 μl of a DNA preparation as described above, 70 μl of water, 1 μl of Taq DNA polymerase (5 U/μl), and 2 μl of each primer. The primers used for PCR amplification (4) were GAAGTCGTAACAAGG and GGGTTTCCCCATTCGGA. All primers in this study were obtained from Sigma-Genosys (Sigma-Aldrich, St. Louis, MO). PCRs were carried out using a model GTC-2 genetic thermal cycler with a model LTM-2 refrigeration unit (Precision Scientific, Inc., Chicago, IL). The temperature program consisted of an initial heat denaturation step of 94°C for 5 min and then 25 cycles of 1 min at 94°C, 5 min at 55°C, and 2 min at 72°C, followed by 5 min at 72°C. DNA products from the PCR were treated (without purification) with 1 μl of an RsaI enzyme solution (16 U/μl) (catalog no. 500890; Stratagene, La Jolla, CA) for 1 h at 37°C. The restriction digest samples were analyzed by electrophoresis in 5% nondenaturing polyacrylamide gels using a vertical gel electrophoresis apparatus (BRL model V16; Invitrogen, Carlsbad, CA). The DNA banding profiles were identified by ethidium bromide staining and were subsequently analyzed using GelCompar II software (Applied Maths, Inc., Austin, TX). For sequencing of 16S rRNA gene variable regions V1 and V2 (21), the primers (2) used for amplification of the 5′ end (approximately 300 bases) of the 16S rRNA gene were 5′-AGAGTTTGATCCTGGCTCAG-3′ and 5′-GTCTCAGTCCCAATGTGGCC-3′. The PCR program consisted of 10 min at 94°C, followed by 25 cycles of 1 min at 94°C, 2 min at 61°C, and 2 min at 72°C and then 5 min at 72°C. Alternatively, primers 5′-AGTTTGATCMTGGCTCAG-3′ (M = A or C) and 5′-AGGAGGTGATCCARCCGCA-3′ (R = A or G) were used to amplify the entire 16S rRNA gene (7) using an annealing temperature of 55°C. PCR products were purified using a QIAquick PCR purification kit (Qiagen, Inc., Valencia, CA), and the DNA fragments were sequenced commercially (Davis Sequencing, Davis, CA). Sequences were analyzed by performing a BLAST (Basic Local Alignment Search Tool) search of the National Center for Biotechnology Information nonredundant DNA sequence database (http://www.ncbi.nlm.nih.gov/).
RESULTS
Fermentation microbiology and chemistry.
The shredded cabbage used for all fermentations in this study contained between 4 × 106 and 6 × 106 CFU/g total aerobes, 2 × 106 to 3 × 106 CFU/g total Enterobacteriaceae, and less than 102 CFU/g yeasts and molds. However, the sizes of the initial LAB populations in the shredded cabbage varied from 104 CFU/g in the Y1 cabbage to 106 CFU/g in two of the Y2 cabbage samples. It is possible that some growth of LAB occurred during transport prior to analysis. As previously reported (11, 22), during the first week of each fermentation rapid increases in the numbers of total bacteria and LAB occurred, as did a rapid decrease in the number of Enterobacteriaceae (data not shown).
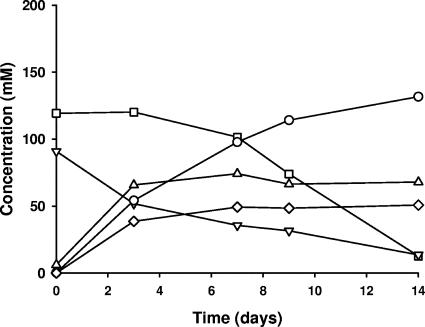
Glucose and fructose were the primary fermentable sugars in the cabbage (the concentrations were between 1.5 and 2.2%, respectively) (Table 1). Sucrose accounted for only a small amount of the fermentable sugars (less than 0.2% of the cabbage by weight) and was not detectable in Y2 samples. Overall, the cabbage used in Y1 contained more sugar than the Y2 samples. However, the sugar utilization, acid production, and pH profiles were similar in the commercial tanks in Y1 and Y2. The results of the chemical analysis indicated that the fermentations in the four commercial sauerkraut fermentation tanks in Y1 (Fig. 1) and Y2 (not shown) were normal and consistent with those described by Fleming et al. (14) and Pederson and Albury (22). During the first week of fermentation, lactic acid, acetic acid, and mannitol were produced. The pH on day 14 for all fermentations was in the range from 3.4 to 3.7 (data not shown).
TABLE 1.
Biochemistry of cabbage used in sauerkraut fermentations
| Yr | Tank | Glucose concn
|
Fructose concn
|
Sucrose concna
|
Malic acid concn
|
||||
|---|---|---|---|---|---|---|---|---|---|
| mM | % (wt/wt) | mM | % (wt/wt) | mM | % (wt/wt) | mM | % (wt/wt) | ||
| 1 | 1 | 119.2 | 2.15 | 90.8 | 1.64 | 4.8 | 0.17 | 5.6 | 0.08 |
| 2 | 1 | 92.4 | 1.66 | 81.7 | 1.47 | ND | ND | 3.9 | 0.05 |
| 2 | 90.8 | 1.63 | 83.5 | 1.50 | ND | ND | 3.5 | 0.05 | |
| 3 | 81.7 | 1.47 | 73.8 | 1.33 | ND | ND | 4.7 | 0.06 | |
ND, not detected.
FIG. 1.
Biochemistry of the Y1 fermentation. Changes in the concentrations of acids and sugars during the first 14 days after the start of the Y1 fermentation are shown. Symbols: ○, lactic acid; ⋄, acetic acid; ▵, mannitol; □, glucose; ▿, fructose.
ITS-PCR database and 16S rRNA gene sequence analysis.
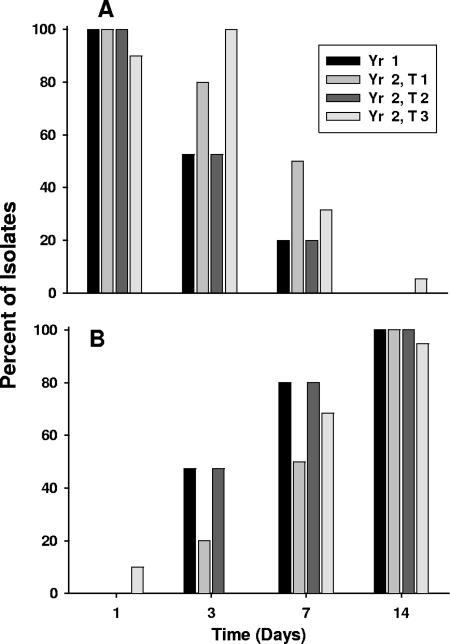
A database of ITS-PCR gel banding patterns was generated using 64 LAB strains belonging to 42 species (see Table S1 and Fig. S1 in the supplemental material), using both the RsaI-digested and undigested PCR products. The 686 fermentation isolates obtained from brine during the 2-year study were grouped by ITS-PCR banding patterns using GenCompar II software for both the RsaI-digested and undigested PCR products. Banding patterns for the isolates were grouped by unique ITS-PCR banding patterns, and 652 isolates were tentatively identified using the ITS-PCR database. One or more representative cultures from each group with a unique banding pattern were then further characterized by sequencing the 5′ end of the 16S rRNA gene. The sequence accession numbers and the corresponding bacterial strain identifications determined by BLAST analysis are shown in Table S2 in the supplemental material. Using this method, L. mesenteroides was identified as the predominant species (179 of 686 isolates) in the early heterofermentative stage of fermentation and L. plantarum was identified as the predominant species involved in the late homofermentative stage (280 of 686 isolates). Together, these two species accounted for two-thirds of all isolates (67%). Weissella sp. and Lactobacillus curvatus were the next most common species among the isolates, with 57 (8.3%) and 40 (5.8%) isolates in the 2-year study. The majority of the LAB isolates (90 to 100%) from Y1 and Y2 samples obtained on days 1 and 3 after the start of fermentation were members of heterofermentative species. Homofermentative LAB accounted for a similar majority of the isolates for day 14 (Fig. 2A and B).
FIG. 2.
Changes in gas production by microorganisms during commercial sauerkraut fermentations. The percentages of total isolates that were heterofermentative (A) or homofermentative (B) on each day (days 1, 3, 7, and 14) for each of the four commercial fermentations sampled are shown. T, tank.
Microbial diversity in commercial sauerkraut fermentations.
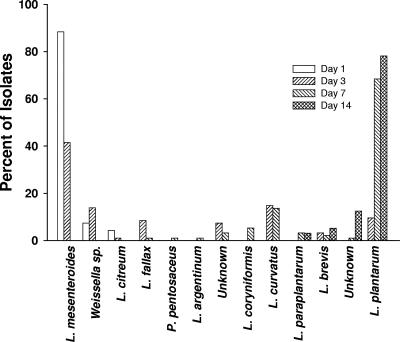
Three different bacterial species were found in the Y1 day 1 samples: L. mesenteroides, Weissella sp., and Leuconostoc citreum (Table 2 and Fig. 3). As expected, L. mesenteroides was the predominant organism associated with the fermentation, comprising 88% (84 of 95) of the isolates in the samples from the first day. L. mesenteroides was the most frequently isolated species in the brine samples from day 3, but it accounted for only 41% (38 of 93) of the total isolates. Other species found on day 3 included L. curvatus (15%), Weissella sp. (14%), L. fallax (9%), and others (Table 2). Homofermentative L. plantarum strains were observed in the fermentation samples from day 3 but accounted for only 9.7% of the isolates. We observed variations in the pattern of hetero- and homofermentative LAB between the Y1 and the three Y2 fermentations. Samples from tank 3 in Y2 were unique compared to the samples from the other tanks because both heterofermentative and homofermentative LAB were present in all samples.
TABLE 2.
Identification of Y1 isolates
| Species | No. of isolates
|
||||
|---|---|---|---|---|---|
| Day 1 | Day 3 | Day 7 | Day 9 | Day 14 | |
| L. mesenteroides | 84 | 38 | |||
| Weissella sp. | 7 | 20 | |||
| L. citreum | 4 | 1 | |||
| L. curvatus | 14 | 14 | 3 | ||
| L. fallax | 8 | 1 | 1 | ||
| L. plantarum | 9 | 64 | 51 | 75 | |
| L. brevis | 3 | 3 | 3 | 5 | |
| L. coryniformis | 5 | 1 | |||
| L. argentinum | 1 | 11 | |||
| P. pentosaceus | 1 | ||||
| L. paraplantarum | 3 | 6 | 3 | ||
| Not identified | 1 | 14 | 13 | ||
| Total analyzed | 95 | 93 | 93 | 90 | 96 |
FIG. 3.
Microbial diversity of the Y1 fermentation as determined by ITS-PCR and 16S rRNA gene sequencing. The percentages of the total isolates (95, 93, 93, and 90 isolates on days 1, 3, 7, and 14, respectively) that were members of different species are indicated by the bars.
As expected, the bacterial isolates from samples obtained in Y1 on days 7, 9, and 14 included no heterofermentative L. mesenteroides or Weissella sp. isolates. Overall, the diversity of species was greatest in the day 9 samples obtained in Y1, when nine different species were isolated out of a total of 90 isolates. The majority of the isolates from the brine samples taken on days 7, 9, and 14 were identified as L. plantarum (68.8, 56.7, and 78.1%, respectively). L. argentinum, which has not previously been found in sauerkraut fermentations, accounted for 10% of the total isolates recovered from samples obtained in Y1 on day 9. Interestingly, only 1 isolate of P. pentosaceus and 14 isolates of L. brevis were found among the 467 Y1 isolates. Of the 279 Y1 isolates from samples obtained on days 7, 9, and 14, a total of 28 were not identified by the ITS-PCR pattern or 16S rRNA gene sequencing analysis.
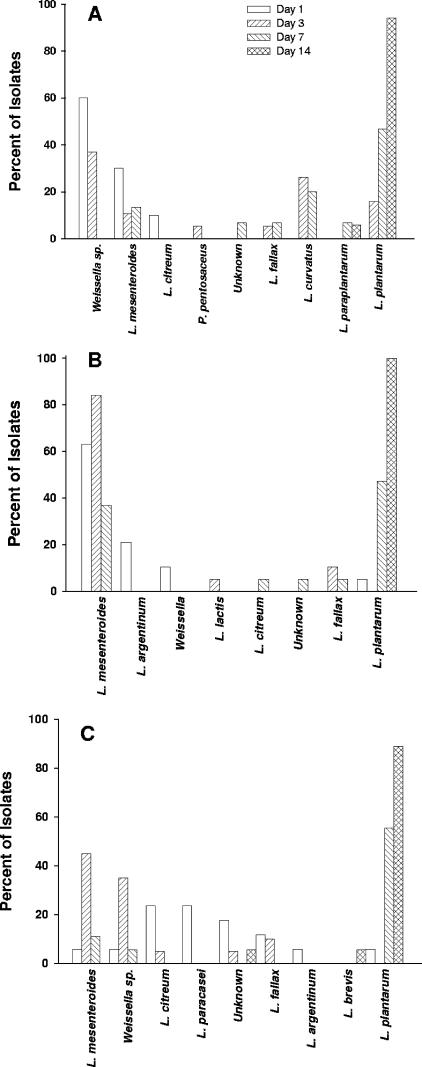
Isolates from the three Y2 fermentation tanks had the bacterial species profiles shown in Fig. 4 and Table 3. For tank 1, three heterofermentative LAB species (L. mesenteroides, Weissella sp., and L. citreum) were recovered from samples obtained on day 1. However, Weissella sp. accounted for the majority of the isolates recovered (12 of 20 isolates for day 1 and 7 of 19 isolates for day 3). L. curvatus, L. fallax, and L. plantarum were also recovered from day 3 brine samples. L. plantarum was the major LAB species isolated from the Y2 tank 1 samples obtained on days 7 and 14 and accounted for over 46 and 94% of the isolates, respectively. The bacterial species obtained from tank 2 were similar to the tank 1 fermentation species (Table 3). For day 14, L. plantarum was the only LAB isolated from the brine sample; it accounted for 100% (18 of 18) of the isolates. In tank 3, however, eight different species were identified for the 17 isolates obtained from the day 1 sample. Only 1 P. pentosaceus isolate and 1 L. brevis isolate were among the 219 Y2 isolates.
FIG. 4.
Microbial diversity of the Y2 fermentations as determined by ITS-PCR and 16S rRNA gene sequencing. The percentages of the total isolates (95, 93, 93, and 90 isolates on days 1, 3, 7, and 14, respectively) that were members of different species are indicated by the bars. The results for tank 1 (A), tank 2 (B), and tank 3 (C) are shown.
TABLE 3.
Identification of Y2 isolates
| Species | No. of isolates
|
|||
|---|---|---|---|---|
| Day 1 | Day 3 | Day 7 | Day 14 | |
| Tank 1 | ||||
| L. mesenteroides | 6 | 2 | 2 | |
| Weissella sp. | 12 | 7 | ||
| L. citreum | 2 | |||
| L. curvatus | 5 | 3 | ||
| L. fallax | 1 | 1 | ||
| L. plantarum | 3 | 7 | 16 | |
| P. pentosaceus | 1 | |||
| L. paraplantarum | 1 | 1 | ||
| Not identified | 1 | |||
| Total analyzed | 20 | 19 | 15 | 17 |
| Tank 2 | ||||
| L. mesenteroides | 12 | 16 | 7 | |
| Weissella sp. | 2 | |||
| L. citreum | 1 | |||
| L. fallax | 2 | 1 | ||
| L. plantarum | 1 | 9 | 18 | |
| L. lactis | 1 | |||
| L. argentinum | 4 | |||
| Lactobacillus mali | 1 | |||
| Total analyzed | 19 | 19 | 19 | 18 |
| Tank 3 | ||||
| L. mesenteroides | 1 | 9 | 2 | |
| Weissella sp. | 1 | 7 | 1 | |
| L. citreum | 4 | 1 | ||
| L. curvatus | 1 | |||
| L. fallax | 2 | 2 | 4 | |
| L. plantarum | 1 | 10 | 16 | |
| L. brevis | 1 | |||
| L. argentinum | 1 | |||
| L. paracasei | 4 | |||
| Not identified | 3 | 1 | 1 | |
| Total analyzed | 17 | 20 | 18 | 18 |
DISCUSSION
We used the ITS-PCR database to identify the major bacterial species present in commercial sauerkraut fermentations. It is possible that different bacterial species that have very similar or indistinguishable ITS patterns were present or that some bacterial species did not grow on MRS medium. Because we sequenced only the 16S RNA gene of isolates with representative ITS-PCR patterns, we may have failed to identify some species that were present among the isolates. The data presented here, therefore, may underreport the microbial diversity in the sauerkraut fermentations. The results of the analysis of the ITS-PCR patterns and 16S rRNA gene sequences of the isolates, however, agreed with the results of the biochemical analysis, indicating that heterofermentative LAB species dominated the first week of the commercial sauerkraut fermentations and that homofermentative species were predominate in the second week. Weissella sp. isolates were recovered together with L. mesenteroides isolates on both day 1 and day 3. We could not identify the species of any of the Weissella isolates by 16S rRNA gene sequence comparisons with the GenBank database because of the sequence similarity of Weissella confusa, Weissella cibaria, and Weissella kimchii. Weissella species have previously been recovered from plant products (5, 8, 18) and have very similar 16S rRNA gene sequences (3, 5). Complete sequencing of the 16S genes from selected Weissella isolates did not allow conclusive identification of known species (data not shown). Weissella spp. were previously characterized as members of the genus Leuconostoc (6). There was variation between the microbiota in the three Y2 fermentations during the heterolactic (days 1 and 3) stage of fermentation, which is believed to be vital to the quality of sauerkraut. This variation may have been due in part to the small sample size (20 isolates from each sampling time and from each fermentation in Y2) but could have represented differences in the microbiota. These data indicate that the use of L. mesenteroides starter cultures may help improve consistency in flavor development in commercial sauerkraut.
During the transition period between the heterofermentation and homofermentation phases (days 3 to 9), a variety of LAB species, including L. curvatus and Leuconostoc argentinum, were isolated. L. argentinum was originally isolated from raw milk in Argentina (9). L. plantarum became the dominant microorganism after 7 days of fermentation, when the pH decreased to 3.9 or less in all four fermentations. However, we recovered relatively few L. brevis isolates (15 of 686 isolates) and only two isolates of P. pentosaceus. This was surprising because previous research showed that both of these species were considered major bacterial species involved in sauerkraut fermentation (22).
This study and a related study of the bacteriophage ecology of commercial sauerkraut fermentations (15) have significantly altered the classical understanding (11, 22) of the microbiota present in sauerkraut fermentations. Several LAB isolated from the 2-year study had never been previously recovered from sauerkraut fermentations, including Weissella sp., L. argentinum, Lactobacillus coryniformis, L. citreum, Lactobacillus paraplantarum, and Lactobacillus paracasei. This study and a recent study of the comparative genomics of lactic acid bacteria, including L. mesenteroides (16), may also aid in the development of starter cultures and increase our understanding of cabbage fermentation biochemistry and ecology.
Supplementary Material
Acknowledgments
This investigation was supported in part by a research grant from Pickle Packers International, Inc. (Washington, DC). We thank Bush Brothers and Company and the Great Lakes Kraut Company for their help and support of this research.
We thank Janet Hayes and Roger McFeeters for aid with experiments and helpful discussions and Sandra L. Parker for excellent secretarial assistance.
Mention of a trademark or proprietary product does not constitute a guarantee or warranty of the product by the U.S. Department of Agriculture or North Carolina Agricultural Research Service, nor does it imply approval to the exclusion of other products that may be suitable.
Footnotes
Published ahead of print on 5 October 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
Paper no. FSR06-22 of the Journal Series of the Department of Food Science, North Carolina State University, Raleigh.
REFERENCES
- 1.Aguirre, M., and M. D. Collins. 1993. Lactic acid bacteria and human clinical infection. J. Appl. Bacteriol. 75:95-107. [DOI] [PubMed] [Google Scholar]
- 2.Barrangou, R., S.-S. Yoon, F. Breidt, H. P. Fleming, and T. R. Klaenhammer. 2002. Characterization of six Leuconostoc fallax bacteriophages isolated from an industrial sauerkraut fermentation. Appl. Environ. Microbiol. 68:5452-5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bjorkroth, K. J., U. Schillinger, R. Geisen, N. Weiss, B. Hoste, W. H. Holzapfel, H. J. Korkeala, and P. Vandamme. 2002. Taxonomic study of Weissella confusa and description of Weissella cibaria sp. nov., detected in food and clinical samples. Int. J. Syst. Evol. Microbiol. 52:141-148. [DOI] [PubMed] [Google Scholar]
- 4.Breidt, F., and H. P. Fleming. 1996. Identification of lactic acid bacteria by ribotyping. J. Rapid Methods Automation Microbiol. 4:219-233. [Google Scholar]
- 5.Choi, H. K., C. C. Ick, K. S. Bo, L. J. Choul, L. D. Woo, C. S. Won, P. J. Min, and P. Y. Ryang. 2002. Weissella kimchii sp. nov., a novel lactic acid bacterium from kimchi. Int. J. Syst. Evol. Microbiol. 52:507-511. [DOI] [PubMed] [Google Scholar]
- 6.Collins, M. D., J. Samelis, J. Metaxopoulos, and S. Wallbanks. 1993. Taxonomic studies on some Leuconostoc-like organisms from fermented sausages: description of a new genus, Weissella, for the Leuconostoc paramesenteroides group of species. J. Appl. Bacteriol. 75:595-603. [DOI] [PubMed] [Google Scholar]
- 7.Dasen, G., J. Smutney, M. Teuber, and L. Meile. 1998. Classification and identification of propionibacteria based on ribosomal RNA genes and PCR. Syst. Appl. Microbiol. 21:251-259. [DOI] [PubMed] [Google Scholar]
- 8.De Vuyst, L., V. Schrijvers, S. Pararmithiotis, B. Hoste, M. Vancanneyt, J. Swings, G. Kalantzopoulos, E. Tsakalidou, and W. Messens. 2002. The biodiversity of lactic acid bacteria in Greek traditional wheat sourdoughs is reflected in both composition and metabolite formation. Appl. Environ. Microbiol. 68:6059-6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dicks, L. M., T. L. Fantuzzi, F. C. Gonzalez, M. Du Toit, and F. Dellaglio. 1993. Leuconostoc argentinum sp. nov., isolated from Argentine raw milk. Int. J. Syst. Bacteriol. 43:347-351. [Google Scholar]
- 10.Du Plessis, E. M., and L. M. T. Dicks. 1995. Evaluation of random amplified polymorphic DNA (RAPD)-PCR as a method to differentiate Lactobacillus acidophilus, Lactobacillus crispatus, Lactobacillus amylovorus, Lactobacillus gallinarum, Lactobacillus gasseri, and Lactobacillus johnsonii. Curr. Microbiol. 31:114-118. [DOI] [PubMed] [Google Scholar]
- 11.Fleming, H. P., K. H. Kyung, and F. Breidt. 1995. Vegetable fermentations, p. 629-661. In H.-J. Rehm and G. Reed (ed.), Biotechnology, 2nd ed. VCH Publishers, Inc., New York, NY.
- 12.Fleming, H. P., R. F. McFeeters, and M. A. Daeschel. 1985. The lactobacilli, pediococci, and leuconostocs: vegetable products. CRC Press, Inc., Boca Raton, FL.
- 13.Fleming, H. P., R. F. McFeeters, and M. A. Daeschel. 1992. Fermented and acidified vegetables, p. 929-952. In C. Vanderzant and D. F. Splittstoesser (ed.), Compendium of methods for the microbiological examination of foods, 3rd ed. American Public Health Association, Washington, DC.
- 14.Fleming, H. P., R. F. McFeeters, J. L. Etchells, and T. A. Bell. 1984. Pickled vegetables, p. 663-681. In M. L. Speck (ed.), Compendium of methods for the microbiological examination of foods, 2nd ed. American Public Health Association, Washington, DC.
- 15.Lu, Z., F. Breidt, Jr., V. Plengvidhya, and H. P. Fleming. 2003. Bacteriophage ecology in commercial sauerkraut fermentations. Appl. Environ. Microbiol. 69:3192-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makarova, K., A. Slesarev, Y. Wolf, A. Sorokin, B. Mirkin, E. Koonin, A. Pavlov, N. Pavlova, V. Karamychev, N. Polouchine, V. Shakhova, I. Grigoriev, Y. Lou, D. Rohksar, S. Lucas, K. Huang, D. M. Goodstein, T. Hawkins, V. Plengvidhya, D. Welker, J. Hughes, Y. Goh, A. Benson, K. Baldwin, J.-H. Lee, I. Díaz-Muñiz, B. Dosti, V. Smeianov, W. Wechter, R. Barabote, G. Lorca, E. Altermann, R. Barrangou, B. Ganesan, Y. Xie, H. Rawsthorne, D. Tamir, C. Parker, F. Breidt, Jr., J. Broadbent, R. Hutkins, D. O' Sullivan, J. Steele, G. Unlu, M. Saier, T. Klaenhammer, P. Richardson, S. Kozyavkin, B. Weimer, and D. Mills. 2006. Comparative genomics of the lactic acid bacteria. Proc. Natl. Acad. Sci. USA 103:15611-15616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDonald, L. C., R. F. McFeeters, M. A. Daeschel, and H. P. Fleming. 1987. A differential medium for the enumeration of homofermentative and heterofermentative lactic acid bacteria. Appl. Environ. Microbiol. 53:1382-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mugula, J. K., S. Nnko, J. A. Narvhus, and T. Sorhaug. 2003. Microbiological and fermentation characteristics of togwa, a Tanzanian fermented food. Int. J. Food Microbiol. 80:187-199. [DOI] [PubMed] [Google Scholar]
- 19.Murcia-Martinez, A. J., and M. D. Collins. 1990. A phylogenetic analysis of the genus Leuconostoc based on reverse transcriptase sequencing of 16S rRNA. FEMS Microbiol. Lett. 58:73-83. [DOI] [PubMed] [Google Scholar]
- 20.Murcia-Martinez, A. J., and M. D. Collins. 1991. A phylogenetic analysis of an atypical leuconostoc: description of Leuoconostoc fallax. FEMS Microbiol. Lett. 82:55-60. [DOI] [PubMed] [Google Scholar]
- 21.Neefs, J. M., Y. Van de Peer, P. De Rijk, S. Chapelle, and R. De Wachter. 1993. Compilation of small ribosomal subunit RNA structures. Nucleic Acids Res. 21:3025-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pederson, C. S., and M. N. Albury. 1969. The sauerkraut fermentation. Technical bulletin 824. New York State Agricultural Experiment Station, Geneva, NY.
- 23.Plengvidhya, V., F. Breidt, and H. P. Fleming. 2004. Use of RAPD-PCR as a method to follow the progress of starter cultures in sauerkraut fermentations. Int. J. Food Microbiol. 93:287-296. [DOI] [PubMed] [Google Scholar]
- 24.Schleifer, K. H., M. Ehrmann, C. Beimfohr, E. Brockmann, W. Ludwig, and R. Amann. 1995. Application of molecular methods for the classification and identification of lactic acid bacteria. Int. Dairy J. 5:1081-1094. [Google Scholar]
- 25.Vogel, R. F., M. Lohmann, A. N. Weller, M. Hugas, and W. P. Hammer. 1991. Structural similarity and distribution of small cryptic plasmids of Lactobacillus curvatus and L. sakei. FEMS Microbiol. Lett. 84:183-190. [DOI] [PubMed] [Google Scholar]
- 26.Ward, D. M., R. Weller, and M. M. Bateson. 1990. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345:63-65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.