Abstract
Transcriptional responses of the yeast Saccharomyces cerevisiae to Zn availability were investigated at a fixed specific growth rate under limiting and abundant Zn concentrations in chemostat culture. To investigate the context dependency of this transcriptional response and eliminate growth rate-dependent variations in transcription, yeast was grown under several chemostat regimens, resulting in various carbon (glucose), nitrogen (ammonium), zinc, and oxygen supplies. A robust set of genes that responded consistently to Zn limitation was identified, and the set enabled the definition of the Zn-specific Zap1p regulon, comprised of 26 genes and characterized by a broader zinc-responsive element consensus (MHHAACCBYNMRGGT) than so far described. Most surprising was the Zn-dependent regulation of genes involved in storage carbohydrate metabolism. Their concerted down-regulation was physiologically relevant as revealed by a substantial decrease in glycogen and trehalose cellular content under Zn limitation. An unexpectedly large number of genes were synergistically or antagonistically regulated by oxygen and Zn availability. This combinatorial regulation suggested a more prominent involvement of Zn in mitochondrial biogenesis and function than hitherto identified.
Zinc is a cofactor of many proteins and is indispensable for their catalytic activity and/or structural stability. Zn is also a ubiquitous component of enzymes involved in transcription and of the Zn finger proteins that regulate gene expression (11). In the yeast Saccharomyces cerevisiae, zinc is estimated to be required for the function of nearly 3% of the proteome (11). Besides its involvement in protein structure and function (53, 73), the interaction of zinc with lipids contributes to the regulation of membrane fluidity (7) and its interaction with nucleic acids helps to prevent deleterious radical reactions (5). The deficiency of this essential trace element can have severe consequences. For example, in beer fermentation, zinc depletion in wort leads to “sluggish” fermentation and, thus, to the deterioration of beer quality (34). While accurate monitoring of the zinc concentration in such industrial fermentations is important, the formation of complexes with polyphenols, proteins, and other compounds (41) implies that the concentration of zinc per se does not always accurately predict its bioavailability to yeast.
Excess zinc is toxic. It can compete with other metal ions for the active sites of enzymes or intracellular transport proteins (26, 37, 54, 57, 65). For this reason, organisms have evolved mechanisms that tightly control intracellular zinc levels. Zinc homeostasis in yeast can be mediated via (i) control of zinc uptake, (ii) storage of zinc in vacuoles, (iii) intracellular binding of zinc by metallothioneins, and (iv) efflux of zinc from the cells. In S. cerevisiae, various proteins involved in zinc uptake and storage have been identified in the last decade. Zinc uptake across the plasma membrane occurs mainly via the transporters Zrt1p and Zrt2p (83, 84). Fet4p and Pho84p, low-affinity and broad-substrate-range transporters of heavy metals, can also transport zinc (79). Zinc storage occurs in the vacuole, and the transport of zinc into this compartment is mediated by Cot1p and Zrc1p (46, 58), while the release of zinc from vacuolar storages is mediated by Zrt3p (51, 52). Msc2p (46) and Yke4p (43) are implicated in the transport of Zn into the lumen of the endoplasmic reticulum and perhaps into an additional organelle involved in the secretory pathway. The genes encoding these transporters are transcriptionally induced by Zap1p (zinc-activated protein) under conditions of zinc limitation or deficiency (85). Contrary to the situation in mammalian cells, no plasma membrane transporter dedicated to zinc export from yeast cells has been identified so far (63). Two cytosolic metallothioneins (Cup1-1p and Cup1-2p) involved in copper chelation can also bind zinc (80). However, the expression of these proteins is not zinc dependent, and their involvement in zinc detoxification has not yet been demonstrated (80).
In order to better define the Zap1p regulon, Lyons and coworkers analyzed the genome-wide transcriptional response of a S. cerevisiae zap1 mutant strain and a control strain to zinc abundance or depletion (50). A combinatorial analysis identified a subset of 46 zinc-responsive genes whose expression was reduced in the zap1 mutant and that possessed a zinc-responsive element (ZRE) (5′-ACCYYNAAGGT-3′). Among the members of this updated defined Zap1p regulon were the well-characterized plasma membrane, vacuole, and endoplasmic reticulum zinc transporters. However, the involvement of many of the proposed Zap1p targets in zinc homeostasis was difficult to interpret and, as suggested by the authors, may be due to the contribution of factors other than zinc depletion. Indeed, these experiments were performed in a shake flask in which the growth conditions could not be strictly monitored and maintained at a constant level, as the pH, the dissolved oxygen, and nutrient concentrations change during growth. Furthermore, zinc depletion and ZAP1 deletions are bound to reduce the specific growth rate compared to zinc-sufficient cultures of a wild-type strain. The regulation of gene expression is therefore affected not only by the difference in growth conditions but also by the specific growth rate (66). This variation in gene regulation can obscure the interpretation of results.
The goal of the present study was to investigate physiological and transcriptional responses of S. cerevisiae to zinc limitation, while minimizing the impact of secondary effects of zinc limitation. To this end, S. cerevisiae was grown at a fixed specific growth rate, oxygen availability, temperature, and pH under zinc limitation in chemostat cultures. By comparing the transcriptome of zinc-limited cultures to those of carbon- and nitrogen-limited cultures, we identified sets of genes that responded uniquely to zinc limitation. Furthermore, these cultures were grown both in the presence and in the complete absence of oxygen in order to identify genes that are subjected to combinatorial control by oxygen and zinc availability.
MATERIALS AND METHODS
Yeast strain and maintenance.
The haploid prototrophic S. cerevisiae strain CEN.PK113-7D (MATa) was obtained from P. Kötter, Frankfurt, Germany. Zinc-depleted cells were obtained by four serial transfers of yeast cells in shake flasks containing synthetic medium (77) from which zinc was omitted, and subsequently, the cells were mixed with glycerol (final concentration of 20%), aliquoted, and stored at −80°C.
Minimizing Zn contamination of culture vessels.
To minimize zinc contamination, all glassware (including shake flasks for precultivation), tubing, and fermenters were subjected to an overnight soak in 2% nitric acid, followed by two washes with deionized water, one wash with 0.1 M EDTA, and four washes with deionized water.
Media for chemostat cultivation.
The synthetic medium composition was based on that described by Verduyn et al. (77). The modifications introduced for carbon-, nitrogen-, and zinc-limited growth are listed in Table 1. In all chemostats except for those limited by carbon, the residual glucose concentration was targeted to 17 g/liter (95 mM) in order to have the same degree of glucose repression (Table 2). Under anaerobic glucose-limited conditions, the glucose concentration was increased to compensate for a low biomass yield. The decreased sulfate concentration [resulting from the reduced (NH4)2SO4 concentration under nitrogen limitation] was compensated by addition K2SO4. The zinc replete cultures (carbon and nitrogen limited) contained excess zinc concentrations, but at subtoxic levels (36). In anaerobic zinc-limited cultures, a minute zinc contamination (probably leaking from the metal fermenter parts) was enough to sustain growth. Conversely, aerobic zinc-limited cultures could not grow at a dilution rate of 0.10 h−1 without the addition of zinc as 0.05 μM zinc sulfate. For anaerobic cultivations, the reservoir medium was supplemented with the anaerobic growth factors Tween 80 and ergosterol (76).
TABLE 1.
Composition of the media used to perform carbon, nitrogen, and zinc limitation under aerobic and anaerobic environments
| Environment | Limiting nutrient | Amt of medium componenta
|
|||
|---|---|---|---|---|---|
| Glucose (g/liter) | (NH4)2SO4 (g/liter) | K2SO4 (g/liter) | ZnSO4·7H2O (mg/liter) | ||
| Aerobic | Carbon | 7.5 | 5 | 4.5 | |
| Nitrogen | 59 | 1 | 5.3 | 4.5 | |
| Zinc | 66 | 5 | 0.014 | ||
| Anaerobic | Carbon | 25 | 5 | 4.5 | |
| Nitrogen | 46 | 1 | 5.3 | 4.5 | |
| Zinc | 58 | 5 | 0 | ||
Numbers in bold indicate the modifications introduced to the synthetic medium described by Verduyn et al. (77) in order to obtain the relevant nutrient limitations.
TABLE 2.
Physiological characteristics of CEN.PK113-7D grown in aerobic and anaerobic carbon-, nitrogen-, or zinc-limited chemostat culturesa
| Growth condition | Residual glucose (mM) | Zn in biomassb | YSXc | Specific consumption and production ratesd
|
RQe | % Carbon recovery | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| qGlucose | qEthanol | qGlycerol | qAcetate | qO2 | qCO2 | ||||||
| Aerobic | |||||||||||
| C | BD | 2.36 ± 0.4 | 0.49 ± 0.00 | −1.1 ± 0.0 | 0.0 ± 0.0 | BD | BD | −2.8 ± 0.3 | 2.8 ± 0.3 | 1.0 ± 0.0 | 98 ± 3 |
| N | 92.7 ± 5.5 | 2.74 ± 1.3 | 0.09 ± 0.00 | −5.8 ± 0.1 | 8.0 ± 0.1 | 0.08 ± 0.01 | 0.03 ± 0.01 | −2.7 ± 0.1 | 12.1 ± 0.2 | 4.5 ± 0.2 | 96 ± 1 |
| Zn | 102.4 ± 6.4 | 0.90 ± 0.2 | 0.10 ± 0.00 | −5.3 ± 0.0 | 8.1 ± 0.2 | 0.08 ± 0.01 | 0.07 ± 0.01 | −2.8 ± 0.0 | 12.3 ± 0.2 | 4.5 ± 0.1 | 105 ± 2 |
| Anaerobic | |||||||||||
| C | BD | 2.10 ± 0.4 | 0.09 ± 0.00 | −6.0 ± 0.0 | 9.6 ± 0.1 | 0.81 ± 0.06 | 0.01 ± 0.00 | NA | 10.3 ± 0.4 | NA | 101 ± 2 |
| N | 100.8 ± 8.6 | 2.74 ± 1.3 | 0.07 ± 0.00 | −8.4 ± 0.0 | 13.5 ± 0.6 | 0.76 ± 0.04 | 0.06 ± 0.05 | NA | 14.8 ± 0.3 | NA | 101 ± 2 |
| Zn | 110.4 ± 3.9 | 0.52 ± 0.03 | 0.07 ± 0.00 | −8.4 ± 0.0 | 13.7 ± 0.2 | 1.09 ± 0.01 | 0.16 ± 0.02 | NA | 15.5 ± 0.5 | NA | 100 ± 1 |
The dilution rate for the cultures was 0.1 h−1. Values are means ± standard deviations. NA, not applicable; ND, not determined; BD, below detection.
Zn in biomass is expressed as μmol·g dry weight−1.
The biomass yield on glucose (YSX) is expressed as g dry weight·g glucose−1.
Specific consumption rates (expressed as mmol·g dry weight −1·h −1) of glucose and oxygen and specific production rates of ethanol, glycerol, acetate, and carbon dioxide.
The respiratory quotient (RQ) was qCO2/qO2.
Chemostat cultivation.
Zinc-depleted precultures were obtained by inoculating shake flasks that contained 100 ml zinc-free synthetic medium with zinc-depleted cells (obtained as described above). After overnight cultivation, these zinc-depleted precultures were inoculated in 2-liter fermenters (Applikon) with a working volume of 1 liter (74). Chemostat cultures were fed with synthetic medium (as described in the previous paragraph) that limited growth by carbon, nitrogen, or zinc, with all other growth requirements in excess and at constant residual concentration (9). The dilution rate was set at 0.10 h−1. Cultures were assumed to be in steady state when, after at least five volume changes, culture dry weight, glucose concentration, carbon dioxide production rate, and oxygen consumption rate varied by less than 2% during one additional volume change (21). Steady-state samples were taken between 10 and 12 generations to avoid strain adaptation due to long-term cultivation (35). Each cultivation condition was performed in triplicate.
The pH was measured online and kept constant at 5.0 by the automatic addition of 2 M KOH by using an Applikon ADI 1030 biocontroller. The stirrer speed was set at 800 rpm. Anaerobic conditions were maintained by sparging the medium reservoir (0.05 liter·min−1) and the fermenter (0.5 liter·min−1) with pure nitrogen gas. Norprene tubing and butyl rubber septa were used to minimize oxygen diffusion into the anaerobic cultures (78). The off-gas was cooled by a condenser connected to a cryostat set at 2°C. Oxygen and carbon dioxide were measured off-line with an NGA 2000 Rosemount gas analyzer.
Analytical methods.
Culture supernatants were obtained after the centrifugation of samples from the chemostats. For the purposes of glucose determination and carbon recovery, culture supernatant and medium were analyzed by high-performance liquid chromatography on an Aminex HPX-87H ion exchange column using 5 mM H2SO4 as the mobile phase. Culture dry weights were determined via filtration as described by Postma et al. (64). Trehalose and glycogen measurements were adapted according to the measurements of François and Parrou (22). Trehalose was determined by triplicate measurements for each chemostat. Glycogen was determined by duplicate measurements for each chemostat. Zn content in the biomass was measured by inductively coupled plasma optical emission spectrometry, using a Varian Vista axial plasma emission spectrometer, according to the procedure developed by Energieonderzoek Centrum Nederland (Petten, The Netherlands).
Microarray analysis.
The sampling of cells from chemostats and total RNA extraction was performed as previously described (1). Probe preparation and hybridization to Affymetrix GeneChip microarrays were performed following Affymetrix instructions. The one-cycle eukaryotic target labeling assay was used, starting with 15 μg of total RNA. The quality of total RNA, cDNA, cRNA, and fragmented cRNA was checked using the Agilent Bioanalyzer 2100 (Agilent Technologies). Results for each growth condition were derived from three independent culture replicates.
Transcriptomics data acquisition and statistical analysis.
Acquisition and quantification of array images and data filtering were performed using Affymetrix GeneChip operating software, version 1.2. Before comparison, all arrays were globally scaled to a target value of 150 by using the average signal from all gene features using GeneChip operating software, version 1.2. To eliminate insignificant variations, genes with expression values below 12 were set to 12 as previously described (9).
To detect genes that exhibited differential expression in at least one of the experimental conditions, an in-house version of significance analysis of microarrays (72) was employed using the multiclass setting. Genes with a q value below the median false discovery rate of 1.5·10−4 were considered differentially expressed.
Transcript data can be downloaded from the Gene Expression Omnibus database under the following series accession numbers: GSE8035 for zinc-limited chemostats, GSE8088 and GSE5926 for carbon-limited chemostats, and GSE8089 for nitrogen-limited chemostats.
Grouping of genes into modules.
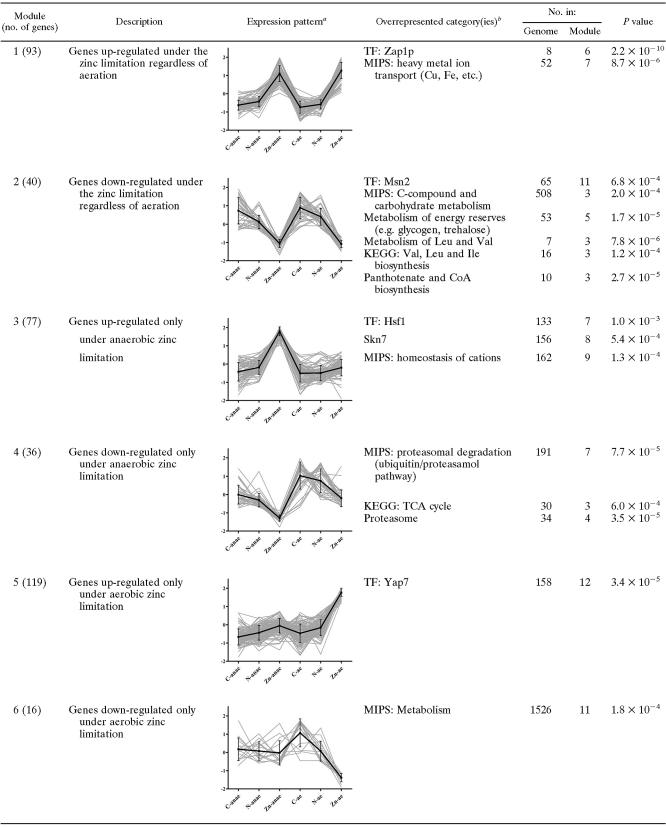
The continuous expression levels of all (1,500) differentially expressed genes were discretized, as described previously by Knijnenburg et al. (39). Resultantly, each gene is represented by a discretized expression pattern of length six, indicating whether the gene is not differentially expressed (0), up-regulated (1), or down-regulated (−1) under each of the six cultivation conditions. For example, a gene that has the discretized expression pattern C-Ana, 0; N-Ana, 1; Zn-Ana, 0; C-Aer, 0; N-Aer, 0; Zn-Aer, −1, where “Ana” means anaerobic and “Aer” means aerobic, is up-regulated when grown anaerobically under a nitrogen limitation (N-Ana) and down-regulated when grown aerobically under a zinc limitation (Zn-Aer), while the four other conditions do not exhibit differential expression. Genes are grouped into modules based on this discretized representation by imposing certain constraints on the discretized expression pattern of a gene in order for it to be part of a particular module. For example, a module could be formed by grouping all genes that have higher discretized expression levels under the zinc limitation than under the other two limitations, both for aerobic and anaerobic growth. This approach provides a coherent and meaningful way to create modules of genes, since the expression behavior of the genes in a module is directly related to the cultivation conditions, allowing for a straightforward interpretation. In our study, six modules were created. The exact constraints on the discretized expression pattern of a gene to be included in one of the six modules are found in the Appendix. Table 3 gives a short verbal description for each of the modules.
TABLE 3.
Clustering of the zinc-responsive genes
Expression pattern of genes of each module along with their averaged expression and standard deviation. Here, the expression levels of each gene are normalized to have zero mean and unit variance (y axis, normalized expression; x axis, culture condition; from left to right: anaerobic carbon, nitrogen, zinc limitation, and aerobic counterparts).
Each data set was analyzed individually for the enrichment of TF binding and functional categories as described in the Materials and Methods. TCA, tricarboxylic acid.
Hypergeometric tests.
The six modules were consulted for enrichment in functional annotation and significant transcription factor (TF) binding. To test for significant relationships, the hypergeometric test was employed. In the case of the TF binding data, the largest available TF binding data set for yeast in its most conservative setting (highest binding confidence) was used (29). This data set, which originally indicates the number of binding sites for each of 102 TFs in the promoter region of each gene, was binarized such that the data indicate whether a TF can bind a gene (upstream). Then, the hypergeometric test assesses whether a TF (or a TF pair) can bind the promoter region of the genes in a module much more frequently than in a randomly selected set of genes. In case of the employed gene annotation information (MIPS [Munich Information Center for Protein Sequences; database for protein sequences, homology data, and yeast genome information] [56] and KEGG [Kyoto encyclopedia of genes and genomes] [38]), it assesses whether the number of genes in a module that belongs to a particular functional category is much larger than would be expected by chance. The P cutoff value to decide whether a relationship is significant is ≤[1/(ncnx)], where nc is the number of modules and nx is the number of TFs (or TF pairs) or the number of MIPS or KEGG annotation categories. This adjustment for multiple testing corresponds with a per-comparison error rate of 1 (24).
Motif discovery.
The promoters (from −800 to −1) of the genes in each module were analyzed for overrepresented regulatory motifs by using the web-based software MEME (2). The P cutoff value to consider a motif significant was 10−4. Other parameter settings included a motif width from 6 to 15 nucleotides, which could be repeated any number of times.
Comparison with the transcriptome study from Lyons and coworkers.
The data from Lyons and coworkers (50) were downloaded from http://genome-www.stanford.edu/zinc/rawdata.html. As this website provides only raw data, the array data were processed following the instructions described in the researchers' publication and 496 genes that were up-regulated in response to zinc depletion were thus isolated. The slightly larger size of the gene set here compared to the one isolated by Lyons et al. (458 genes) probably results from a few differences in data handling.
RESULTS
Establishing Zn-limited chemostat cultures of S. cerevisiae.
While macronutrient limitation in chemostats can be achieved in a straightforward manner, establishing micronutrient limitation still presents an experimental challenge. This holds especially for metals (Zn, Fe, and Cu) that are present in laboratory equipment and that can sustain growth at extremely low concentrations (typically in the micromolar range). Despite thorough and repeated washing steps and use of high-grade medium components, we did not achieve completely Zn-free cultivation conditions, presumably due to Zn leakage from the metal parts (fermenter lid, pipes, and connections). This contamination was sufficient to allow for anaerobic Zn-limited growth at a steady-state biomass concentration of 2.5 g·liter−1. However, 0.05 μM ZnSO4 had to be added to the Zn-deficient medium to enable aerobic Zn-limited growth (steady-state biomass concentration 4.2 g·liter−1). The addition of 15 μM Zn to anaerobic and aerobic Zn-limited cultures resulted in a large increase of the biomass concentration, thus confirming that growth was solely limited by Zn availability (data not shown). The values for Zn content of biomass from Zn-limited cultures were up to fivefold lower than those from carbon- and nitrogen-limited cultures (Table 2). Consistent with a higher Zn requirement for aerobic cultivation, the values for Zn content of biomass from aerobic Zn-limited cultures were twofold higher than those from anaerobic Zn-limited cultures (Table 2). Since genes encoding Zn transporters were not differentially transcribed in the presence and in the absence of oxygen, this difference is unlikely to be due to a different affinity for Zn uptake.
Physiology of Zn-, glucose-, and ammonia-limited chemostat cultures.
Zn-limited cultures were grown at high residual glucose concentrations. A comparison of their physiologies and transcriptomes with those of glucose-limited cultures will therefore also identify changes caused by the different glucose concentrations in the cultures. Therefore, nitrogen-limited cultures, grown at the same residual glucose concentration as the Zn-limited cultures, were included as an additional reference situation. The combination of three nutrient limitations under aerobic and anaerobic conditions resulted in six unique physiological situations (Table 2).
Only in the carbon-limited aerobic cultures was a completely respiratory sugar metabolism observed, resulting in a high biomass yield on glucose (Table 2). In the anaerobic cultures, glucose metabolism was fully fermentative, the main products of glucose dissimilation being ethanol and carbon dioxide. Finally, in glucose-sufficient (i.e., N- or Zn-limited) aerobic cultures, a mixed respirofermentative metabolism was observed. The Zn-limited cultures strongly resembled the nitrogen-limited cultures with respect to biomass yields and rates of product formation. Even under anaerobic conditions, the biomass yield on glucose of these glucose-sufficient cultures was lower than that of glucose-limited cultures, indicating a partial uncoupling of dissimilation and biomass formation under these “energy excess” conditions. The only notable difference was a slightly higher specific rate of acetate and glycerol production in the Zn-limited cultures, which may be related to a reduced in vivo activity of Zn-dependent alcohol dehydrogenases.
Overall transcriptional responses to Zn limitation.
For all six culture conditions described above, microarray analysis was performed on three independent replicate cultures. Statistical analysis (see Materials and Methods) identified 1,500 genes that were differentially transcribed in at least one cultivation condition. A total of 381 of these genes responded specifically to Zn-limited growth. Of these Zn-responsive genes, 81 proteins do not yet have an assigned cellular function. The 381 Zn-responsive genes were subjected to further analysis to identify combinatorial effects of Zn and oxygen availability (Table 3). The majority of the genes that showed a transcriptional response to Zn limitation (248 genes, modules 3 to 6 in Table 3) did so in an oxygen-dependent manner. The remainder (133 genes, modules 1 and 2 in Table 3) of the Zn-responsive genes showed a consistent response to Zn limitation that was independent of oxygen availability. For the identities and transcript levels of the genes contained in the six modules, see Table S1 in the supplemental material. Below, we will analyze these sets of Zn-responsive genes for overrepresentation of genes involved in specific functional categories and/or controlled by specific TFs (see Materials and Methods).
Zinc homeostasis and the Zap1p regulon.
The MIPS functional category “heavy metal transport” was overrepresented among the 93 genes that were transcriptionally up-regulated in response to Zn limitation irrespective of oxygen availability (Table 3, module 1). Of the seven genes belonging to this category found in module 1, six are directly involved in Zn homeostasis. ZRT1, encoding the plasma membrane, high-affinity Zn transporter, was strongly induced (average change of 13-fold) (Table 4). Transcript levels of ZRT2, ZRT3, ZRC1, and ZRG17, involved in Zn transport and homeostasis, were also increased but to a lesser extent than those of ZRT1 (changes ranging from two- to sevenfold). FET4 (up-regulated 3- to 43-fold under Zn limitation) encodes a protein involved in iron transport that has been demonstrated to also be a physiologically relevant Zn carrier (79). The comparison of aerobic and anaerobic cultures confirmed the previously described combinatorial regulation of FET4 by Zn and oxygen availability (79). In addition, a clear hierarchy was observed: while FET4 was strongly regulated by oxygen availability under Zn-sufficient conditions (79), its transcript level in Zn-limited cultures was consistently high regardless of oxygen supply (see Fig. 2). The transcriptional regulation of these six genes was in agreement with previous studies (30, 50) and so was the up-regulation of ZAP1, the transcriptional activator of these six transporters (8- to 20-fold increase relative to Zn-sufficient cultures). FRE1, which also belongs to the “heavy metal transport” category, encodes a protein specifically involved in ferric iron transport (25, 82). FRE1 does not contain a ZRE and its increased transcript levels under Zn-limited conditions suggest an indirect effect.
TABLE 4.
Identity and expression levels of the genes from module 1 consistently up-regulated in response to zinc limitation and containing ZRE sequencesa
| Gene | Description | No. of ZREs | Transcript level
|
|||||
|---|---|---|---|---|---|---|---|---|
| Anaerobic
|
Aerobic
|
|||||||
| C | N | Zn | C | N | Zn | |||
| ZAP1 | Zn-responsive TF | 1 | 23 | 42 | 480 | 34 | 44 | 357 |
| ZRT1 | High-affinity Zn transporter | 3 | 176 | 286 | 2,004 | 68 | 241 | 1,868 |
| ZRT2 | Low-affinity Zn transporter | 2 | 109 | 124 | 739 | 103 | 163 | 552 |
| ZRT3 | Vacuolar Zn efflux | 1 | 237 | 257 | 1,666 | 313 | 283 | 1,709 |
| ZRC1 | Vacuolar Zn influx | 1 | 238 | 367 | 712 | 261 | 338 | 812 |
| ZRG17 | Putative Zn transporter | 1 | 85 | 92 | 446 | 143 | 98 | 527 |
| FET4 | Low-affinity Fe transporter | 1 | 235 | 224 | 743 | 12 | 90 | 444 |
| ADH4 | Alcohol dehydrogenase | 3 | 153 | 206 | 2,890 | 77 | 118 | 2,872 |
| HOR2 | Glycerol-P phosphatase | 1 | 45 | 63 | 141 | 97 | 85 | 199 |
| DPP1 | DAGPP phosphatase | 1 | 307 | 518 | 1,019 | 294 | 637 | 1,233 |
| URA10 | Pyrimidine biosynthesis | 1 | 19 | 25 | 81 | 30 | 17 | 51 |
| FLO11 | Cell surface flocculin | 1 | 1,151 | 1,293 | 1,935 | 42 | 60 | 2,105 |
| ZPS1 | Cell surface mannoprotein | 2 | 75 | 226 | 3,385 | 140 | 119 | 3,349 |
| MNT2 | Mannosyl transferase | 3 | 18 | 12 | 52 | 12 | 12 | 41 |
| KTR6 | Mannosyl transferase | 1 | 450 | 388 | 538 | 314 | 254 | 620 |
| MCD4 | Transferase required for glycosylphosphatidylinositol anchor synthesis | 1 | 337 | 323 | 997 | 232 | 279 | 1,190 |
| ZIP1 | Synaptonemal complex | 1 | 12 | 12 | 46 | 13 | 12 | 25 |
| KTI12 | tRNA modification | 1 | 158 | 134 | 218 | 120 | 122 | 278 |
| VTC3 | Vacuolar transporter chaperone | 1 | 66 | 96 | 175 | 51 | 86 | 273 |
| TEX1 | TREX complex | 1 | 29 | 30 | 92 | 25 | 24 | 178 |
| MUP1 | Methionine transporter | 1 | 70 | 39 | 637 | 129 | 212 | 913 |
| YNL254C | Unknown | 1 | 22 | 28 | 342 | 17 | 32 | 355 |
| YER130C | Unknown | 1 | 32 | 35 | 179 | 31 | 28 | 67 |
| ICY2 | Unknown | 2 | 291 | 204 | 1,939 | 568 | 144 | 1,436 |
| VEL1 | Unknown, similar to YOR387C | 3 | 12 | 12 | 1,047 | 12 | 12 | 859 |
| YOR387C | Unknown, similar to VEL1 | 3 | 12 | 12 | 2,803 | 12 | 12 | 2,612 |
Genes indicated in bold were also part of the regulon defined by Lyons and coworkers (50).
FIG. 2.
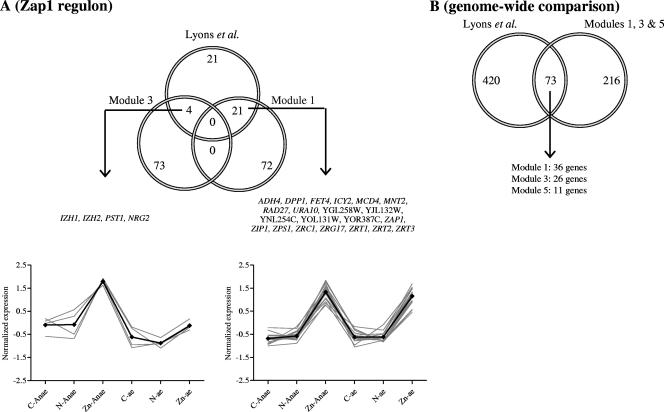
Venn diagram of chemostat-based transcriptome data in comparison with data obtained by Lyons and coworkers (50). (A) Zap1p regulon (modules 1 and 3 in comparison with 46 genes from Lyons and coworkers [50]). (B) Genome-wide comparison (modules 1, 3, and 5 in comparison with all up-regulated genes from Lyons and coworkers [50]). anae, anaerobic; ae, aerobic.
Previous reports have investigated the role of MSC2 in Zn transport into the endoplasmic reticulum (19, 46) and have found that mutations in the latter affect the cellular distribution of zinc (46). In our study, MSC2 was not found among the genes that were transcriptionally induced under Zn limitation. Instead, its transcript levels remained low under the conditions tested. Consistent with this observation, the transcription of MSC2 was not affected in a zap1 mutant (50). ZRG17 encodes a protein that has been proposed to act as a complex with Msc2p (18, 46). The promoter of ZRG17 does contain a ZRE, and its transcript levels were increased in Zn-limited cultures, suggesting that this protein could be the regulatory subunit of the complex.
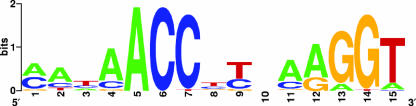
In an attempt to further define the Zap1p regulon, the promoter regions of the 93 genes that showed a robust, oxygen-independent response to Zn limitation (module 1, Table 3) were searched for overrepresented motifs. The web-based software MEME (2), which enables unbiased probability-based motif discovery, identified 26 genes with a 15-nucleotide motif that strongly resembled the previously published ZRE Zap1p-binding consensus sequence (Fig. 1; Table 4). In agreement with previous reports of Zap1p regulation, all six Zn transporters in module 1 as well as ZAP1 itself harbored this element. Twelve additional genes (Table 4) have previously been proven or proposed to be Zap1p targets. An additional seven genes that harbored the 15-nucleotide motif had not previously been implicated as Zap1p targets (50) (Table 4). For a detailed list of the ZRE sequences and positions, see Table S2 in the supplemental material.
FIG. 1.
Consensus ZRE sequence identified by MEME using module 1 as input.
Comparison with previous Zn-related transcriptome studies.
Two previous transcriptome studies investigated yeast adaptation to Zn depletion in batch cultures of an industrial (30) and a laboratory strain of S. cerevisiae (50). Using maltose-grown cultures, Higgins et al. observed a down-regulation of maltose permease and maltase genes (MAL12, MAL32, and MAL31) in Zn-depleted cultures (30). In the present study, growth on glucose resulted in the absence of MAL gene transcripts, thus masking transcriptional responses of these genes to Zn availability. Lyons et al. identified a Zap1p regulon consisting of 46 genes by comparing the transcriptional responses to Zn depletion of a zap1Δ mutant and its parental strain. Three of these 46 genes (COS2, COS4, and COS6) were not represented on the microarrays used in our study. Of the remaining 43 genes, 25 showed increased transcript levels in Zn-limited chemostat cultures (Fig. 2A). The large majority of these (21 genes) were consistently induced in response to Zn limitation irrespective of oxygen availability (module 1, Table 3; Fig. 2A). MEME failed to identify a ZRE sequence in 3 of these 21 genes (RAD27, YJL132W, and YOL131W), which are therefore absent from Table 4. Four genes from the Zap1p regulon defined by Lyons and coworkers (IZH1, IZH2, NRG2, and PST1) were found in module 3 (Table 3), indicating that their transcription was induced under Zn limitation, but only when oxygen was absent. Their identification by Lyons et al. may have been caused by the poor oxygen transfer characteristics of shake-flask cultures (28, 55, 68). Two additional genes (ADE17 and GPG1) identified as Zap1p targets by Lyons et al. were up-regulated in Zn-limited chemostat cultures; however, their expression resulted from intricate regulation by Zn, glucose, and oxygen availability. Both genes responded to zinc limitation under aerobic and anaerobic conditions. They also responded to limiting glucose supply, but this response was oxygen specific; while ADE17 was up-regulated under glucose limitation in the presence of oxygen, GPG1 expression increased under glucose-limited anaerobic growth. The remaining 16 of the 43 genes that were identified as Zap1p targets by Lyons et al. and included on our microarrays did not respond to Zn availability in our chemostat study.
Eight potential Zap1p targets identified in the present study (Table 4) were not found in the study of Lyons et al. However, of these eight genes, HOR2 and TEX1 were found to be transcriptionally induced by Zn depletion in their study. Furthermore, Zap1p was shown to bind TEX1 in chromatin-immunoprecipitation-on-chip experiments (29). Seven genes (HOR2, FLO11, KTR6, KTI12, VTC3, MUP1, and YER130C [Table 4]) are here for the first time proposed to be Zap1p targets. HOR2 encodes a glycerol-3-phosphate phosphatase involved in glycerol biosynthesis (62), which may account for the slightly, but significantly (Student's t test, P value < 0.05), elevated glycerol production observed under zinc-limited growth. VTC3 encodes a vacuolar transport chaperone involved in inorganic ion transport (13). Although it has been shown to be involved in polyphosphate transport, it may also participate in vacuolar Zn transport (61). Alternatively, Zn may be involved in polyphosphate accumulation or may react with polyphosphates. Like the previously identified Zap1p target MNT2 (50) (Table 4), KTR6 encodes a mannosyl transferase involved in the glycosylation of cell wall proteins (49). It can be speculated that Mnt2p and Ktr6p play a role in mannosylation of Zn-scavenging cell wall proteins. For instance ZPS1, a Zap1p target also up-regulated under Zn limitation (Table 4), encodes a cell wall mannoprotein with high similarity to Zn metalloproteinases from filamentous fungi (44, 50). The yeast cell wall and, more specifically, mannoproteins have been shown to fix a substantial fraction of the cellular zinc (15). Zinc fixation by mannoproteins may represent an efficient mechanism to scavenge low zinc concentrations (59). The up-regulation of mannoproteins such as ZPS1 under zinc limitation supports this zinc scavenging function of the cell wall. The consistent up-regulation of FLO11, KTI12, MUP1, and YER130C in Zn-limited cultures and the presence of a ZRE-like motif in their promoters suggest that the encoded proteins have some as-yet-unknown role under Zn-limited conditions, too. For example, Flo11p is known to play an essential role in biofilm formation, filamentation, and invasive growth (47). Studies of Candida albicans have demonstrated that dimorphic switching from budding growth to mycelium formation is regulated by zinc (4, 69). However, in the present study, we did not observe any difference in morphology between the different culture conditions.
When the 289 genes in modules 1, 3, and 5 induced under Zn limitation in chemostat cultures (either in an oxygen-dependent or in an oxygen-independent manner) were compared to the 493 genes that were induced upon Zn depletion in shake flasks (50), 73 genes overlapped between the two studies. For the most part, these were clustered in modules 1 and 3 (Fig. 2B; see Table S3 in the supplemental material). Only a small overlap was observed with module 5 (representing only 9% of the genes in this module), which includes genes that are induced only by Zn limitation under aerobic conditions. As mentioned above, this small overlap may reflect a limiting oxygen supply in the shake-flask studies.
Transcriptional regulation of structural genes for zinc-dependent proteins.
S. cerevisiae contains multiple alcohol dehydrogenases. While the enzymes encoded by ADH1, -2, -3, and -5 all require Zn as a cofactor, Adh4p uses Mg. ADH4 has been shown to be regulated by Zap1p, while expression of the Zn-requiring isoenzymes has been reported to be decreased upon Zn depletion (presumably via Rap1p) (8). In agreement with earlier findings, ADH4 was strongly up-regulated in response to Zn limitation irrespective of the aeration conditions. Transcript levels of other Zn-dependent alcohol dehydrogenase genes were either unchanged or reduced. In addition to alcohol dehydrogenases, many other yeast proteins use Zn as a structural component or cofactor. Regalla and Lyons (54, 65) separated the Zn-dependent protein into two distinct classes, (i) the proteins that use zinc in a catalytic capacity (105 genes) and (ii) the proteins with a structural Zn binding domain (360 genes). Of 105 S. cerevisiae proteins that use Zn as a cofactor (54, 65), none of the structural genes were found to be transcriptionally regulated in response to Zn availability in chemostat cultures (with the clear exception of alcohol dehydrogenases). On the other hand, of the 360 S. cerevisiae proteins that contain a structural Zn binding domain, 16 genes were up-regulated in response to Zn limitation (modules 1, 2, and 5), while 7 were down-regulated (modules 2, 4, and 6). Most of these Zn-responsive genes encoded proteins that have a function in nucleic acid binding (TFs, chromatin reorganizing activity, or mRNA binding). The two homologous TFs Met31p and Met32p that induce the expression of genes involved in methionine biosynthesis were affected only by Zn availability in the presence of oxygen. While MET32 expression increased twofold, MET31 expression decreased twofold. These changes in gene expression probably resulted in modifications of the transcriptional regulation of these transcriptional activators as their target genes displayed a slightly higher expression under conditions of aerobic Zn limitation. This antagonistic regulation of MET31 and MET32 remains difficult to relate to Zn supply as both proteins contain two Zn finger domains and do not have different Zn contents.
In agreement with previous reports (81), SOD1, which encodes the cytosolic Zn-Cu superoxide dismutase, showed a twofold reduction of its transcript level under conditions of low Zn supply. However, SOD2, which encodes mitochondrial manganese-containing superoxide dismutase, did not show an increased transcript level in Zn-limited cultures. In fact, SOD2, which was transcribed only in aerobic cultures, was also down-regulated by ca. twofold under Zn limitation. As proposed previously (81), reduced expression of superoxide dismutase may affect resistance to oxidative stress. A more direct involvement of zinc in oxidative stress resistance was previously suggested via the transcriptional regulation of TSA1, encoding a Zn-dependent peroxiredoxin, by Zap1p (81). Unfortunately, in our experiments TSA1 expression was independent of zinc and oxygen availability. This difference with earlier work may be attributed either to the difference between complete Zn depletion (81) and Zn-limited growth (this study) or to a different strain background. However, close scrutiny of the transcript levels revealed no oxidative stress response (AAD3, AAD6, AAD10, AAD14, AAD15, ATR1, CCP1, GTT2, GRE2, LYS20, OYE2, OYE3, TRR1, TRX2, YDR453C, YLR460C, YNL134C, YMR318C, and YML131W) (40). Although the transcript levels of both SOD1 and SOD2 were reduced, their levels (748 and 295, respectively, under aerobic zinc limitation) may still be high enough to enable efficient processing of ROS and thereby to prevent oxidative stress.
Finally, while we cannot exclude the possibility that Zn sparing and/or mobilization mechanisms occur at a posttranscriptional level(s), these results indicate that a general “Zn sparing” regulation at the transcriptional level is most probably absent in S. cerevisiae. The exceptions of alcohol dehydrogenase and superoxide dismutase may be related to the relative abundance of these proteins and their pivotal role in fermentative and respiratory metabolism, respectively.
Combinatorial response of mitochondrial function to oxygen and zinc availability.
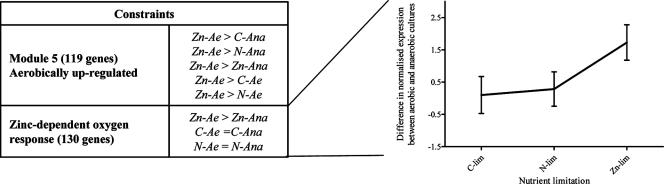
Aerobic Zn limitation of S. cerevisiae resulted in the up-regulation of 119 genes and the down-regulation of 16 genes (Table 3, modules 5 and 6). However, hypergeometric distribution analysis did not reveal clear trends in the identity and function of these oxygen-responsive proteins. In order to better investigate the potential synergetic effects between oxygen and zinc availability, different discretized patterns were considered. As shown in Fig. 3 for the aerobically up-regulated genes, the applied constraints selected genes for which the expression under carbon limitation was unaffected by oxygen and the expression under nitrogen limitation was also oxygen insensitive, but for which the response to zinc limitation was oxygen dependent. A total of 196 genes respecting these constraints were identified, 130 being up-regulated in the presence of oxygen in a Zn-dependent manner and 66 being down-regulated (see Table S4 in the supplemental material). Fisher's exact statistics were then applied to search for overrepresentation of genes involved in specific functional categories and/or controlled by specific TFs. While no enrichment was found within the genes that were down-regulated, the module containing the up-regulated genes showed interesting trends. This module was characterized by enrichment for two functional categories: “respiration” (10 genes) and “mitochondrial biogenesis” (14 genes). The category of “respiration” comprised genes encoding various subunits of the Fo (ATP4, ATP14, ATP18, and ATP20) and F1 (ATP3 and ATP15) domains of mitochondrial ATP synthase (16) but also COX23, COX14, MAM33, and MBA1, involved in the assembly of respiratory complexes in mitochondria (3, 27, 60, 67). The relationship between Zn availability and these proteins remains unclear, although cytochrome c oxidase activity has been shown to be inhibited by Zn. Most of the genes in the “mitochondrial biogenesis” category encoded mitochondrial ribosomal proteins (MRPL10, MRPL11, MRPL37, MNP1, RSM19, and MRPS16), but this category also included MSS116, a gene involved in the splicing of mitochondrial group I and II introns (33). Finally, TIM10/MRS11 also responded synergistically to Zn and oxygen availability. TIM10 encodes a protein involved in the translocation of mitochondrial proteins from the cytoplasm to the mitochondria. For instance, Aac1p and Aac2p, subunits of an ADP/ATP mitochondrial carrier, cannot be translocated in a tim10 mutant (75). This translocation process, also identified in plant (6), requires Zn (48). The present study reveals a more important role for Zn in mitochondrial function and biogenesis than so far assumed. Although still not clearly understood, this role could, at least in part, explain the higher Zn requirement for cells grown in the presence of oxygen, a condition where mitochondria are essential for respiration.
FIG. 3.
Combinatorial regulation of gene expression by Zn and oxygen availability: constraints for the selection of the discretized patterns and average expression profile of these selected patterns. Only the oxygen-induced genes are represented. Error bars indicate standard deviations. lim, limited.
Zn limitation and storage carbohydrate metabolism.
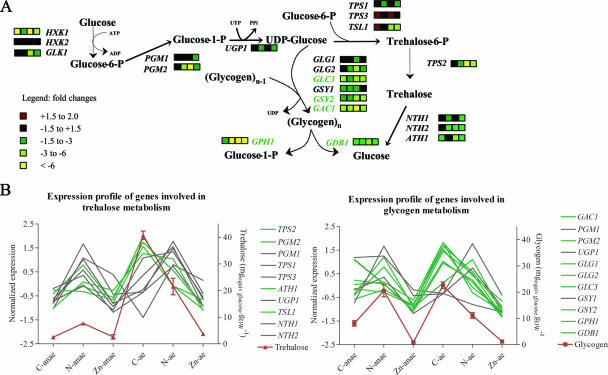
Genes from both glycogen biosynthesis (GSY2, GAC1, and GLC3) and degradation pathways (GDB1 and GPH1) were down-regulated by up to 22-fold in Zn-limited chemostat cultures, regardless of oxygen availability (Fig. 4A). Several additional genes involved in glycogen metabolism that did not pass the very stringent statistical test used in genome-wide analysis displayed a decreased expression upon closer inspection (Fig. 4A). To investigate whether these transcriptional modifications resulted in phenotypic differences, glycogen contents were analyzed in the chemostat cultures on which the transcriptome analyses had been performed (Fig. 4B). Indeed, glycogen accumulation was strongly (10- to 20-fold) reduced in Zn-limited cultures. Genes involved in glycogen metabolism are known to be transcriptionally regulated in response to a wide variety of environmental conditions and signaling pathways (20) (temperature, nutrient supply, and oxidative stress). This regulation is mediated by the general environmental stress response and high-osmolarity glycerol response pathways (22, 23). However, no other target genes of these signaling pathways were found to be differentially transcribed in response to Zn limitation. This suggests that the regulation of glycogen metabolism by Zn occurs via another hitherto unknown signal transduction mechanism.
FIG. 4.
(A) Glycogen and trehalose metabolism in S. cerevisiae. Genes indicated in green are clustered in module 2. The four boxes indicate the following n-fold changes from left to right: zinc versus carbon anaerobic, zinc versus nitrogen anaerobic, zinc versus carbon aerobic, and zinc versus nitrogen aerobic. Intensities of n-fold changes are indicated by the color key. (B) Normalized expression profile of genes involved in glycogen and trehalose metabolism and intracellular glycogen and trehalose concentrations. Error bars indicate standard deviations. anae, anaerobic; ae, aerobic.
Several genes involved in trehalose metabolism were also significantly down-regulated in Zn-limited cultures (PGM1, PGM2, TPS1, TPS2, and TPS3) (Fig. 4A). These down-regulations coincided with substantially lower trehalose biomass contents, an effect that was most pronounced in aerobic cultures (Fig. 4B). These results clearly demonstrated, for the first time, the impact of Zn availability on reserve carbohydrate accumulation. As the genes involved in glycogen and trehalose metabolism do not contain ZREs, their transcriptional regulation is unlikely to be directly mediated by Zap1p. In addition, their down-regulation probably occurs via a stress response element (STRE)-independent mechanism (we did not find overrepresented STRE in the promoters of down-regulated genes). Among the above-mentioned Zap1p regulon, YER130C, encoding a protein of unknown function containing two tandem Zn-finger domains, was up-regulated under zinc limitation. This putative TF may be involved in a Zap1p-dependent regulation of genes involved in trehalose and glycogen and is an interesting candidate for further functional analysis.
DISCUSSION
Analysis of Zn limitation in chemostat cultures.
The unique option of chemostat cultures to control specific growth rate prevented the occurrence of specific-growth-rate-related responses. For example, in a previous study of batch cultures of S. cerevisiae (30), the observed down-regulation of ribosomal proteins in low-Zn cultures is likely to have been caused by a decrease in specific growth rate rather than directly by Zn depletion.
The use of different aeration regimens showed that yeast responses to Zn limitation are strongly context dependent. This result notwithstanding, a set of genes was identified whose specific transcriptional regulation by Zn availability was independent of the oxygen supply. The identification of this gene set enabled us to propose a more precise definition of the Zap1p regulon. Most of these 26 potential Zap1p targets overlapped with those proposed in a previous batch-cultivation study (50). The present study demonstrated that responses of several of the previously identified putative Zap1p targets were not Zn specific. Instead, they were synergistically or antagonistically regulated by carbon, nitrogen, and/or oxygen supply.
Compared to the transcriptional responses observed in chemostat cultures under other nutrient limitations (9, 10, 14, 70), transcriptional responses to Zn limitation were strikingly pleiotropic. Genes involved in a large variety of cellular functions, apparently unrelated to Zn availability, showed marked differences to Zn limitation. Statistical analysis of coregulated genes identified only a very limited number of overrepresented functional categories or DNA binding proteins, with the clear exception of the Zap1p regulon. These observations suggest that the only direct effect of Zn limitation on transcriptional regulation is mediated by Zap1p. Although no concerted transcriptional regulation was observed for genes encoding proteins that contain Zn as a catalytic or structural component, Zn availability is likely to influence the in vivo activity of such proteins, many of which are TFs. The apparently “scattered” transcriptional responses to Zn limitation may further be due to the fact that new roles of Zn in yeast physiology continue to be discovered. For instance, the involvement of Zn in protein translocation by the Tim10p/Tim9p complex has only recently been revealed (48).
Effects of Zn limitation on storage carbohydrate accumulation: a possible cause for stuck fermentations in beer fermentation?
Zn used by yeast during the beer fermentation process comes from barley malt and is extracted during the mashing procedure (starch conversion and extraction). However, Zn content varies largely between fermentations, as its concentration is dependent on the crop quality (32) and is partly removed from wort during lautering (41) or wort separation. Insufficient Zn supply during brewing results in “sluggish” fermentations characterized by low fermentation rates (12). The metabolic and/or regulatory processes in yeast cells that underlie such retarded fermentations are incompletely understood. Yeast crops are commonly reused 4 to 10 times for inoculating succeeding brews and are generally stored around 2°C under starvation (54). Under such conditions, high reserve carbohydrate contents have been shown to be critical for the survival and recovery of metabolic activity of yeast (54). To our knowledge, no published study has investigated how storage carbohydrate metabolism might be affected by Zn deficiency. The present study demonstrates, for the first time, that Zn limitation causes a strong transcriptional down-regulation of genes involved in reserve carbohydrate accumulation. The physiological relevance of this response was verified by an analysis of intracellular glycogen and trehalose contents, which were strongly reduced in Zn-limited cultures. Comparative studies with nitrogen-limited cultures showed that the decreased accumulation of storage carbohydrates was specific for Zn limitation and not merely a consequence of glucose-excess conditions. Furthermore, this effect was independent of the aeration of the cultures and the expression profiles of several genes involved in reserve carbohydrate metabolism perfectly matched the profile of trehalose and glycogen accumulation (Fig. 4B).
While our hypothesis remains to be tested under brewing conditions and with brewing strains of S. cerevisiae, it seems highly probable that the fermentation performance of Zn-limited brewer's yeast will be strongly compromised. Additionally, follow-up research should focus on the molecular mechanisms that link reserve carbohydrate metabolism and Zn availability.
Potential implication of Zn limitation for flavor formation.
Another consequence of limiting Zn supply during the course of beer fermentation might be related to flavor formation. Indeed, three genes involved in the biosynthesis of the branched-chain amino acids leucine, valine, and isoleucine (ILV2, ILV3, and BAT2) were consistently down-regulated under Zn-limited growth, both in the presence and in the absence of oxygen (Table 3). The flux through the branched chain amino acid synthetic pathways has been shown to have a positive impact on desirable flavor compound production, such as isoamyl acetate and isobutyl acetate (45), and Zn supplementation to wort results in increased production of the acetate esters of higher alcohols (31). The present data suggest that this effect of zinc availability on flavor formation may be mediated by the transcriptional regulation of ILV2, ILV3, and BAT2. Maintaining a sufficiently high zinc level during beer fermentation is clearly critical to maintaining the desired balance between several flavor compounds.
Signature transcripts for diagnosing Zn bioavailability in industrial media.
In complex industrial fermentation media, such as wort or other plant biomass hydrolysates, Zn can form complexes with several medium components, thereby reducing its bioavailability for yeast (34, 41, 42). This limits the relevance of chemical analyses of the Zn content to test the bioavailability of zinc in wort and other industrial media. The addition of Zn in the form of salt or trub is a common practice to prevent Zn depletion during the brewing process (71). Especially in beer brewing, this is not risk free, as excess Zn leads to the modification of flavor compound formation (17). Molecular markers can be used to monitor fermentation processes through transcript profiling (30). For such diagnostic purposes, it is preferable to construct small, cost-effective microarrays that contain a limited number of “signature transcripts.” A prerequisite of these signature transcripts is that they are specific to one environmental parameter and show a robust response in various environmental (process) contexts. A comparison of multiple chemostat regimens enabled the identification of such Zn-specific signature transcripts. For instance, ZAP1 and ZRT1 would be very good signature transcripts. Also YOR387C and YGL258W, encoding proteins that have not been characterized yet and that have been previously proposed as potential signature transcripts for Zn depletion (30, 50), were specifically and consistently induced under Zn limitation in chemostat cultures. Conversely, NRG2 and PST1, potential Zap1p targets (50), were here shown to be also regulated by oxygen availability and are therefore not recommended for diagnostic purposes.
APPENDIX
Constraints imposed to group zinc-responsive genes into modules.
Let the discretized expression pattern of a gene be denoted by vector x of length 6. The values of the elements of x can be 0 (no differential expression), 1 (up-regulated), or −1 (down-regulated). The elements of x correspond to the cultivation conditions as follows: C-Ana for x(1), N-Ana for x(2), Zn-Ana for x(3), C-Aer for x(4), N-Aer for x(5), and Zn-Aer for x(6).
In Table A1, we state the constraints on x that must be satisfied in order for a gene to be part of a particular module. Note that all constraints must be met to suffice.
TABLE A1.
Constraints on the discretized expression pattern of a gene to be included in one of the six modules
| Module | Constraints |
|---|---|
| 1 (up-regulated regardless of aeration) | x(3)>x(1), x(3)>x(2), x(3)>x(4), x(3)>x(5), x(6)>x(1), x(6)>x(2), x(6)>x(4), and x(6)>x(5) |
| 2 (down-regulated regardless of aeration) | x(3)<x(1), x(3)<x(2), x(3)<x(4), x(3)<x(5), x(6)<x(1), x(6)<x(2), x(6)<x(4), and x(6)<x(5) |
| 3 (anaerobically up-regulated) | x(3)>x(1), x(3)>x(2), x(3)>x(4), x(3)>x(5), and x(3)>x(6) |
| 4 (anaerobically down-regulated) | x(3)<x(1), x(3)<x(2), x(3)<x(4), x(3)<x(5), and x(3)<x(6) |
| 5 (aerobically up-regulated) | x(6)>x(1), x(6)>x(2), x(6)>x(3), x(6)>x(4), and x(6)>x(5) |
| 6 (aerobically down-regulated) | x(6)<x(1), x(6)<x(2), x(6)<x(3), x(6)<x(4), and x(6)<x(5) |
Supplementary Material
Acknowledgments
The research group of J. T. Pronk is part of the Kluyver Centre for Genomics of Industrial Fermentation, which is supported by The Netherlands Genomics Initiative. Raffaele De Nicola thanks FEMS for the scholarship that allowed him to support his stay in Delft during the execution of this work.
Footnotes
Published ahead of print on 12 October 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abbott, D. A., T. A. Knijnenburg, L. M. de Poorter, M. J. Reinders, J. T. Pronk, and A. J. van Maris. 2007. Generic and specific transcriptional responses to different weak organic acids in anaerobic chemostat cultures of Saccharomyces cerevisiae. FEMS Yeast Res. 7:819-833. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, T. L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28-36. [PubMed] [Google Scholar]
- 3.Barros, M. H., A. Johnson, and A. Tzagoloff. 2004. COX23, a homologue of COX17, is required for cytochrome oxidase assembly. J. Biol. Chem. 279:31943-31947. [DOI] [PubMed] [Google Scholar]
- 4.Bedell, G. W., and D. R. Soll. 1979. Effects of low concentrations of zinc on the growth and dimorphism of Candida albicans: evidence for zinc-resistant and -sensitive pathways for mycelium formation. Infect. Immun. 26:348-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berg, J. M., and Y. Shi. 1996. The galvanization of biology: a growing appreciation for the roles of zinc. Science 271:1081-1085. [DOI] [PubMed] [Google Scholar]
- 6.Bhushan, S., B. Lefebvre, A. Stahl, S. J. Wright, B. D. Bruce, M. Boutry, and E. Glaser. 2003. Dual targeting and function of a protease in mitochondria and chloroplasts. EMBO Rep. 4:1073-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Binder, H., K. Arnold, A. S. Ulrich, and O. Zschornig. 2001. Interaction of Zn2+ with phospholipid membranes. Biophys. Chem. 90:57-74. [DOI] [PubMed] [Google Scholar]
- 8.Bird, A. J., M. Gordon, D. J. Eide, and D. R. Winge. 2006. Repression of ADH1 and ADH3 during zinc deficiency by Zap1-induced intergenic RNA transcripts. EMBO J. 25:5726-5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boer, V. M., J. H. de Winde, J. T. Pronk, and M. D. Piper. 2003. The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus, or sulfur. J. Biol. Chem. 278:3265-3274. [DOI] [PubMed] [Google Scholar]
- 10.Boer, V. M., S. L. Tai, Z. Vuralhan, Y. Arifin, M. C. Walsh, M. D. Piper, J. H. de Winde, J. T. Pronk, and J. M. Daran. 2007. Transcriptional responses of Saccharomyces cerevisiae to preferred and nonpreferred nitrogen sources in glucose-limited chemostat cultures. FEMS Yeast Res. 7:604-620. [DOI] [PubMed] [Google Scholar]
- 11.Böhm, S., D. Frishman, and H. W. Mewes. 1997. Variations of the C2H2 zinc finger motif in the yeast genome and classification of yeast zinc finger proteins. Nucleic Acids Res. 25:2464-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bromberg, S. K., P. A. Bower, G. R. Duncombe, J. Fehring, L. Gerber, V. K. Lau, and M. Tata. 1997. Requirements for zinc, manganese, calcium, and magnesium in wort. J. Am. Soc. Brew. Chem. 55:123-128. [Google Scholar]
- 13.Cohen, A., N. Perzov, H. Nelson, and N. Nelson. 1999. A novel family of yeast chaperons involved in the distribution of V-ATPase and other membrane proteins. J. Biol. Chem. 274:26885-26893. [DOI] [PubMed] [Google Scholar]
- 14.Daran-Lapujade, P., J. M. Daran, P. Kötter, T. Petit, M. D. Piper, and J. T. Pronk. 2003. Comparative genotyping of the Saccharomyces cerevisiae laboratory strains S288C and CEN.PK113-7D using oligonucleotide microarrays. FEMS Yeast Res. 4:259-269. [DOI] [PubMed] [Google Scholar]
- 15.De Nicola, R. 2006. Ph.D. thesis. University of Abertay, Dundee, Scotland.
- 16.Devenish, R. J., M. Prescott, X. Roucou, and P. Nagley. 2000. Insights into ATP synthase assembly and function through the molecular genetic manipulation of subunits of the yeast mitochondrial enzyme complex. Biochim. Biophys. Acta 1458:428-442. [DOI] [PubMed] [Google Scholar]
- 17.Dufour, J. P., P. Malcorps, and P. Silcock. 2003. Control of ester synthesis during brewery fermentation, p. 213-233. In K. A. Smart (ed.), Brewing yeast fermentation performance. Blackwell Science, Oxford, United Kingdom.
- 18.Ellis, C. D., C. W. MacDiarmid, and D. J. Eide. 2005. Heteromeric protein complexes mediate zinc transport into the secretory pathway of eukaryotic cells. J. Biol. Chem. 280:28811-28818. [DOI] [PubMed] [Google Scholar]
- 19.Ellis, C. D., F. Wang, C. W. MacDiarmid, S. Clark, T. Lyons, and D. J. Eide. 2004. Zinc and the Msc2 zinc transporter protein are required for endoplasmic reticulum function. J. Cell Biol. 166:325-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Enjalbert, B., J. L. Parrou, M. A. Teste, and J. Francois. 2004. Combinatorial control by the protein kinases PKA, PHO85 and SNF1 of transcriptional induction of the Saccharomyces cerevisiae GSY2 gene at the diauxic shift. Mol. Genet. Genomics 271:697-708. [DOI] [PubMed] [Google Scholar]
- 21.Ferea, T. L., D. Botstein, P. O. Brown, and R. F. Rosenzweig. 1999. Systematic changes in gene expression patterns following adaptive evolution in yeast. Proc. Natl. Acad. Sci. USA 96:9721-9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.François, J., and J. L. Parrou. 2001. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 25:125-145. [DOI] [PubMed] [Google Scholar]
- 23.Gasch, A. P., P. T. Spellman, C. M. Kao, O. Carmel-Harel, M. B. Eisen, G. Storz, D. Botstein, and P. O. Brown. 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11:4241-4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ge, Y., S. Dudoit, and T. P. Speed. 2003. Resampling-based multiple testing for microarray data analysis. Test 12:1-44. [Google Scholar]
- 25.Georgatsou, E., and D. Alexandraki. 1999. Regulated expression of the Saccharomyces cerevisiae Fre1p/Fre2p Fe/Cu reductase related genes. Yeast 15:573-584. [DOI] [PubMed] [Google Scholar]
- 26.Gitan, R. S., H. Luo, J. Rodgers, M. Broderius, and D. Eide. 1998. Zinc-induced inactivation of the yeast ZRT1 zinc transporter occurs through endocytosis and vacuolar degradation. J. Biol. Chem. 273:28617-28624. [DOI] [PubMed] [Google Scholar]
- 27.Glerum, D. M., T. J. Koerner, and A. Tzagoloff. 1995. Cloning and characterization of COX14, whose product is required for assembly of yeast cytochrome oxidase. J. Biol. Chem. 270:15585-15590. [DOI] [PubMed] [Google Scholar]
- 28.Gupta, A., and G. Rao. 2003. A study of oxygen transfer in shake flasks using a non-invasive oxygen sensor. Biotechnol. Bioeng. 84:351-358. [DOI] [PubMed] [Google Scholar]
- 29.Harbison, C. T., D. B. Gordon, T. I. Lee, N. J. Rinaldi, K. D. Macisaac, T. W. Danford, N. M. Hannett, J. B. Tagne, D. B. Reynolds, J. Yoo, E. G. Jennings, J. Zeitlinger, D. K. Pokholok, M. Kellis, P. A. Rolfe, K. T. Takusagawa, E. S. Lander, D. K. Gifford, E. Fraenkel, and R. A. Young. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431:99-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Higgins, V. J., P. J. Rogers, and I. W. Dawes. 2003. Application of genome-wide expression analysis to identify molecular markers useful in monitoring industrial fermentations. Appl. Environ. Microbiol. 69:7535-7540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hodgson, J. A., and M. Moir. 1990. Control of esters in brewing, p. 266-269. In I. Campbell (ed.), Proceedings of the 3rd Aviemore Conference on Malting, Brewing and Distilling. Institute of Brewing, London, United Kingdom.
- 32.Hough, J. S., D. E. Briggs, R. Stevens, and T. M. Young. 1982. Malting and brewing science, vol. 2, 2nd ed. Chapman and Hall, London, United Kingdom.
- 33.Huang, H. R., C. E. Rowe, S. Mohr, Y. Jiang, A. M. Lambowitz, and P. S. Perlman. 2005. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc. Natl. Acad. Sci. USA 102:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobsen, T., R. Volden, S. Engan, and O. Aubert. 1979. A chemometric study of some beer flavour components. J. Inst. Brew. 89:265-270. [Google Scholar]
- 35.Jansen, M. L., P. Daran-Lapujade, J. H. de Winde, M. D. Piper, and J. T. Pronk. 2004. Prolonged maltose-limited cultivation of Saccharomyces cerevisiae selects for cells with improved maltose affinity and hypersensitivity. Appl. Environ. Microbiol. 70:1956-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones, R. P., and P. F. Greenfield. 1984. A review of yeast ionic nutrition. Process Biochem. 19:48-59. [Google Scholar]
- 37.Kamizono, A., M. Nishizawa, Y. Teranishi, K. Murata, and A. Kimura. 1989. Identification of a gene conferring resistance to zinc and cadmium ions in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 219:161-167. [DOI] [PubMed] [Google Scholar]
- 38.Kanehisa, M., and S. Goto. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28:27-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Knijnenburg, T. A., J. H. de Winde, J. M. Daran, P. Daran-Lapujade, J. T. Pronk, M. J. Reinders, and L. F. Wessels. 2007. Exploiting combinatorial cultivation conditions to infer transcriptional regulation. BMC Genomics 8:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koerkamp, M. G., M. Rep, H. J. Bussemaker, G. P. Hardy, A. Mul, K. Piekarska, C. A. Szigyarto, J. M. De Mattos, and H. F. Tabak. 2002. Dissection of transient oxidative stress response in Saccharomyces cerevisiae by using DNA microarrays. Mol. Biol. Cell 13:2783-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kreder, G. C. 1999. Yeast assimilation of trub-bound zinc. J. Am. Soc. Brew. Chem. 57:129-132. [Google Scholar]
- 42.Kuhbeck, F., W. Back, and M. Krottenthaler. 2006. Influence of lauter turbidity on wort composition, fermentation performance and beer quality—a review. J. Inst. Brew. 112:215-221. [Google Scholar]
- 43.Kumánovics, A., K. E. Poruk, K. A. Osborn, D. M. Ward, and J. Kaplan. 2006. YKE4 (YIL023C) encodes a bidirectional zinc transporter in the endoplasmic reticulum of Saccharomyces cerevisiae. J. Biol. Chem. 281:22566-22574. [DOI] [PubMed] [Google Scholar]
- 44.Lamb, T. M., and A. P. Mitchell. 2003. The transcription factor Rim101p governs ion tolerance and cell differentiation by direct repression of the regulatory genes NRG1 and SMP1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 23:677-686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee, S., K. Villa, and H. Patino. 1995. Yeast strain development for enhanced production of desirable alcohols/esters in beer. J. Am. Soc. Brew. Chem. 53:153-156. [Google Scholar]
- 46.Li, L., and J. Kaplan. 2001. The yeast gene MSC2, a member of the cation diffusion facilitator family, affects the cellular distribution of zinc. J. Biol. Chem. 276:5036-5043. [DOI] [PubMed] [Google Scholar]
- 47.Lo, W. S., and A. M. Dranginis. 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9:161-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu, H., and J. Woodburn. 2005. Zinc binding stabilizes mitochondrial Tim10 in a reduced and import-competent state kinetically. J. Mol. Biol. 353:897-910. [DOI] [PubMed] [Google Scholar]
- 49.Lussier, M., A. M. Sdicu, E. Winnett, D. H. Vo, J. Sheraton, A. Dusterhoft, R. K. Storms, and H. Bussey. 1997. Completion of the Saccharomyces cerevisiae genome sequence allows identification of KTR5, KTR6 and KTR7 and definition of the nine-membered KRE2/MNT1 mannosyltransferase gene family in this organism. Yeast 13:267-274. [DOI] [PubMed] [Google Scholar]
- 50.Lyons, T. J., A. P. Gasch, L. A. Gaither, D. Botstein, P. O. Brown, and D. J. Eide. 2000. Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc. Natl. Acad. Sci. USA 97:7957-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.MacDiarmid, C. W., L. A. Gaither, and D. Eide. 2000. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 19:2845-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.MacDiarmid, C. W., M. A. Milanick, and D. J. Eide. 2002. Biochemical properties of vacuolar zinc transport systems of Saccharomyces cerevisiae. J. Biol. Chem. 277:39187-39194. [DOI] [PubMed] [Google Scholar]
- 53.Magonet, E., P. Hayen, D. Delforge, E. Delaive, and J. Remacle. 1992. Importance of the structural zinc atom for the stability of yeast alcohol dehydrogenase. Biochem. J. 287:361-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martin, V., D. E. Quain, and K. A. Smart. 2003. Brewing yeast oxidative stress responses: impact of brewery handling, p. 61-74. In K. A. Smart (ed.), Brewing yeast fermentation performance. Blackwell Science, Oxford, United Kingdom.
- 55.McDaniel, L. E., E. G. Bailey, and A. Zimmerli. 1965. Effect of oxygen supply rates on growth of Escherichia coli. Appl. Microbiol. 13:115-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mewes, H. W., K. Albermann, K. Heumann, S. Liebl, and F. Pfeiffer. 1997. MIPS: a database for protein sequences, homology data and yeast genome information. Nucleic Acids Res. 25:28-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miyabe, S., S. Izawa, and Y. Inoue. 2000. Expression of ZRC1 coding for suppressor of zinc toxicity is induced by zinc-starvation stress in Zap1-dependent fashion in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 276:879-884. [DOI] [PubMed] [Google Scholar]
- 58.Miyabe, S., S. Izawa, and Y. Inoue. 2001. The Zrc1 is involved in zinc transport system between vacuole and cytosol in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 282:79-83. [DOI] [PubMed] [Google Scholar]
- 59.Mochaba, F., and E. O'Connor-Cox. 1996. Metal ion concentration and release by a brewing yeast: characterization and implications. J. Am. Soc. Brew. Chem. 54:155-163. [Google Scholar]
- 60.Muta, T., D. Kang, S. Kitajima, T. Fujiwara, and N. Hamasaki. 1997. p32 protein, a splicing factor 2-associated protein, is localized in mitochondrial matrix and is functionally important in maintaining oxidative phosphorylation. J. Biol. Chem. 272:24363-24370. [DOI] [PubMed] [Google Scholar]
- 61.Ogawa, N., J. DeRisi, and P. O. Brown. 2000. New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol. Biol. Cell 11:4309-4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Påhlman, A. K., K. Granath, R. Ansell, S. Hohmann, and L. Adler. 2001. The yeast glycerol 3-phosphatases Gpp1p and Gpp2p are required for glycerol biosynthesis and differentially involved in the cellular responses to osmotic, anaerobic, and oxidative stress. J. Biol. Chem. 276:3555-3563. [DOI] [PubMed] [Google Scholar]
- 63.Palmiter, R. D., and S. D. Findley. 1995. Cloning and functional characterization of a mammalian zinc transporter that confers resistance to zinc. EMBO J. 14:639-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Postma, E., A. Kuiper, W. F. Tomasouw, W. A. Scheffers, and J. P. van Dijken. 1989. Competition for glucose between the yeasts Saccharomyces cerevisiae and Candida utilis. Appl. Environ. Microbiol. 55:3214-3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Regalla, L. M., and T. J. Lyons. 2006. Zinc in yeast: mechanisms involved in homeostasis, p. 37-54. In M. J. Tamas and E. Martinoia (ed.), Molecular biology of heavy metal homeostasis and detoxification: from microbes to man. Springer, Berlin, Germany.
- 66.Regenberg, B., T. Grotkjaer, O. Winther, A. Fausboll, M. Akesson, C. Bro, L. K. Hansen, S. Brunak, and J. Nielsen. 2006. Growth-rate regulated genes have profound impact on interpretation of transcriptome profiling in Saccharomyces cerevisiae. Genome Biol. 7:R107. http://genomebiology.com/2006/7/11/R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rep, M., and L. A. Grivell. 1996. MBA1 encodes a mitochondrial membrane-associated protein required for biogenesis of the respiratory chain. FEBS Lett. 388:185-188. [DOI] [PubMed] [Google Scholar]
- 68.Schultz, J. S. 1964. Cotton closure as an aeration barrier in shaken flask fermentation. Appl. Microbiol. 12:305-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Soll, D. R., G. W. Bedell, and M. Brummel. 1981. Zinc and regulation of growth and phenotype in the infectious yeast Candida albicans. Infect. Immun. 32:1139-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tai, S. L., V. M. Boer, P. Daran-Lapujade, M. C. Walsh, J. H. de Winde, J. M. Daran, and J. T. Pronk. 2005. Two-dimensional transcriptome analysis in chemostat cultures. Combinatorial effects of oxygen availability and macronutrient limitation in Saccharomyces cerevisiae. J. Biol. Chem. 280:437-447. [DOI] [PubMed] [Google Scholar]
- 71.Taidi, B., B. Hoogenberg, A. I. Kennedy, and J. A. Hodgson. 2000. Pre-treatment of pitching yeast with zinc. MBAA Tech. Q. 37:431-434. [Google Scholar]
- 72.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vallee, B. L., and D. S. Auld. 1990. Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry 29:5647-5659. [DOI] [PubMed] [Google Scholar]
- 74.van den Berg, M. A., P. de Jong-Gubbels, C. J. Kortland, J. P. van Dijken, J. T. Pronk, and H. Y. Steensma. 1996. The two acetyl-coenzyme A synthetases of Saccharomyces cerevisiae differ with respect to kinetic properties and transcriptional regulation. J. Biol. Chem. 271:28953-28959. [DOI] [PubMed] [Google Scholar]
- 75.Vasiljev, A., U. Ahting, F. E. Nargang, N. E. Go, S. J. Habib, C. Kozany, V. Panneels, I. Sinning, H. Prokisch, W. Neupert, S. Nussberger, and D. Rapaport. 2004. Reconstituted TOM core complex and Tim9/Tim10 complex of mitochondria are sufficient for translocation of the ADP/ATP carrier across membranes. Mol. Biol. Cell 15:1445-1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Verduyn, C., E. Postma, W. A. Scheffers, and J. P. van Dijken. 1990. Energetics of Saccharomyces cerevisiae in anaerobic glucose-limited chemostat cultures. J. Gen. Microbiol. 136:405-412. [DOI] [PubMed] [Google Scholar]
- 77.Verduyn, C., E. Postma, W. A. Scheffers, and J. P. van Dijken. 1992. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8:501-517. [DOI] [PubMed] [Google Scholar]
- 78.Visser, W., W. A. Scheffers, W. H. Batenburg-van der Vegte, and J. P. van Dijken. 1990. Oxygen requirements of yeasts. Appl. Environ. Microbiol. 56:3785-3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Waters, B. M., and D. J. Eide. 2002. Combinatorial control of yeast FET4 gene expression by iron, zinc, and oxygen. J. Biol. Chem. 277:33749-33757. [DOI] [PubMed] [Google Scholar]
- 80.Winge, D. R., K. B. Nielson, W. R. Gray, and D. H. Hamer. 1985. Yeast metallothionein. Sequence and metal-binding properties. J. Biol. Chem. 260:14464-14470. [PubMed] [Google Scholar]
- 81.Wu, C. Y., A. J. Bird, D. R. Winge, and D. J. Eide. 2007. Regulation of the yeast TSA1 peroxiredoxin by ZAP1 is an adaptive response to the oxidative stress of zinc deficiency. J. Biol. Chem. 282:2184-2195. [DOI] [PubMed] [Google Scholar]
- 82.Yun, C. W., M. Bauler, R. E. Moore, P. E. Klebba, and C. C. Philpott. 2001. The role of the FRE family of plasma membrane reductases in the uptake of siderophore-iron in Saccharomyces cerevisiae. J. Biol. Chem. 276:10218-10223. [DOI] [PubMed] [Google Scholar]
- 83.Zhao, H., and D. Eide. 1996. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc. Natl. Acad. Sci. USA 93:2454-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao, H., and D. Eide. 1996. The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J. Biol. Chem. 271:23203-23210. [DOI] [PubMed] [Google Scholar]
- 85.Zhao, H., and D. J. Eide. 1997. Zap1p, a metalloregulatory protein involved in zinc-responsive transcriptional regulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 17:5044-5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.