Abstract
Postharvest processing (PHP) is used to reduce levels of Vibrio vulnificus in oysters, but process validation is labor-intensive and expensive. Therefore, quantitative PCR was evaluated as a rapid confirmation method for most-probable-number enumeration (QPCR-MPN) of V. vulnificus bacteria in PHP oysters. QPCR-MPN showed excellent correlation (R2 = 0.97) with standard MPN and increased assay sensitivity and efficiency.
Vibrio vulnificus can cause life-threatening, systemic disease (2, 9, 14) that is associated with the consumption of raw oysters. The bacterium is distributed throughout temperate estuaries worldwide (6, 7, 17, 24, 25), and environmental conditions of warmer water temperature and lower salinity favor its growth in molluscan shellfish (5, 13, 16, 21, 23, 25). Warning labels on oyster products and educational programs have not been effective in reducing disease mortality rates for at-risk individuals with underlying diseases, such as cirrhosis, hemochromatosis (iron overload), diabetes, or immune system dysfunction (8). Therefore, the FDA and the Interstate Shellfish Sanitation Conference (ISSC) have mandated postharvest processing (PHP) of oysters harvested from Gulf Coast states in order to reduce V. vulnificus infections (11). Application of PHP methodology requires validation and verification in order to ensure that the process will substantially reduce numbers of V. vulnificus bacteria to levels that are below the predicted threshold for disease. Validation trials for PHP are labor-intensive and cost-prohibitive (10, 12), and improved protocols for industry compliance and for risk assessment of V. vulnificus in PHP oysters are urgently needed.
The standard method for validating PHP requires that three independent lots of oysters meet the specification of <30 most probable numbers (MPN)/g of V. vulnificus by using the geometric mean of 10 samples/lot. Levels of V. vulnificus bacteria in oysters are enumerated by MPN endpoint titration of replicate samples in enrichment broth cultures (12), and species-specific growth is determined by isolating typical V. vulnificus colonies on selective medium, with subsequent confirmation by DNA probe (26). The present study evaluated a real-time quantitative PCR (QPCR) assay for detection of V. vulnificus growth in MPN enrichment cultures (QPCR-MPN). QPCR increases assay throughput by using automated species-specific confirmation, which is not available with standard PCR. Also, the limit of detection for direct QPCR enumeration of V. vulnificus bacteria in oysters without enrichment is generally 100 CFU/g, but QPCR-MPN assays generally increase the sensitivity of detection to 1 bacterium/g after enrichment (1, 18, 19, 20) and permit detection of V. vulnificus at levels (30 CFU/g) required for validation protocols.
Field trials were conducted to assess application of QPCR-MPN to oyster PHP validation, as application of QPCR-MPN to enumerate V. vulnificus bacteria in PHP oysters has not been examined previously and merits further scrutiny. For example, large numbers of dead bacteria may accumulate in the oyster product as a consequence of PHP and could provide a DNA template for false-positive amplification of nonviable bacteria. The present study used immersion of oysters in liquid nitrogen, followed by extended frozen storage at −20°C as an ultralow-temperature PHP for oysters. Samples were examined before, during, and after exposure to PHP to provide a side-by-side comparison of the standard MPN protocol to QPCR-MPN confirmation. Results support the application of QPCR-MPN for improved assessment of validation and verification protocols related to oyster PHP.
TaqMan versus SYBR green I QPCR assays for V. vulnificus.
We previously reported a TaqMan QPCR assay that targeted the V. vulnificus vvhA gene, which encodes a hemolytic cytolysin. Prior examination of target (n = 28) and nontarget (n = 22) strains showed that V. vulnificus QPCR was 100% specific and detected about 102 CFU/g in oysters (4). The assay was modified in the present study to use boiling lysis rather than Qiagen tissue kits for DNA extraction. SYBR green I dye detection was compared to TaqMan in order to reduce assay cost and simplify the protocol for subsequent MPN applications. The type of thermocycler (Cepheid) and addition of SmartMix beads (Cepheid) also differed from the prior assay, which used an Applied Biosystems thermocycler and reagents. For boiling lysis, cultures (1 ml) were centrifuged (15,000 × g, 10 min), resuspended in phosphate-buffered saline (PBS) (1 ml), boiled for 10 min, and subsequently centrifuged to remove particulates. Supernatants were stored at −20°C. Primers (Geno-mechanix, Gainesville, FL) from the prior study were used for SYBR green I or TaqMan detection at 100 nM or 900 nM and with 1× SYBR green I dye (Cepheid) or 0.25 mM TaqMan probe (Applied Biosystems), respectively. DNA template (2 μl) and water were added to QPCR reactions for a total volume of 25 μl. The TaqMan protocol consisted of incubation at 50°C for 2 min followed by denaturation at 95°C for 10 min and 40 cycles of 15 s at 95°C and 60°C for 1 min. The SYBR green I assay used 2 min at 95°C, followed by 40 cycles of the parameters given above.
QPCR examination of DNA from V. vulnificus (n = 25) and non-V. vulnificus (n = 28) strains (Table 1) showed both TaqMan detection and SYBR green I detection were 100% sensitive and species specific for V. vulnificus. Cycle threshold (CT) values (number of cycles required to reach threshold for detection) for SYBR green I detection of V. vulnificus strains were comparable to those for TaqMan QPCR, with mean CT values (± standard deviations) of 16.48 ± 0.79 and 16.61 ± 0.87, respectively. All V. vulnificus strains were positive by TaqMan assay, while nontarget species were all negative, including false-positive strains (shown in bold in Table 1) described in the prior report (19). Although the SYBR green I assay detected CT values above threshold for nontarget strains, detection occurred only after extended PCR cycling (mean number of cycles, 34.86 ± 2.28) and is likely to be a consequence of artifactual signal (22). First-derivative analysis of melting curves (melting temperature) provides a sensitive discrimination of nucleotide differences in the DNA sequence of amplicons (21, 22), and species-specific detection of PCR product by SYBR green I was confirmed by single melt peaks with consistent values (mean, 88.02 ± 0.26) from V. vulnificus strains (Table 1). In contrast, melt peak values for nontarget species averaged >22 standard deviations apart from the means of positive controls.
TABLE 1.
Specificity and sensitivity of V. vulnificus QPCR detection with SYBR green I and TaqMan detection
| Straina | QPCR resultb
|
||
|---|---|---|---|
| TaqMan (CT) | SYBR green I (CT) | Melt peak (temp [°C]) | |
| Target strains | |||
| Vibrio vulnificus | |||
| 1009 | 16.46 | 16.15 | 88.29 |
| MO6-24/O | 16.14 | 16.17 | 88.10 |
| MLT365 | NDc | 18.29 | 88.15 |
| 6353 | 16.45 | 15.92 | 87.91 |
| MLT367 | 17.21 | 17.6 | 88.42 |
| CVD752 | 15.94 | 14.87 | 88.26 |
| 345/T | 15.60 | 16.36 | 87.77 |
| BO6312 | 17.56 | 16.44 | 88.12 |
| 5C1326 | ND | 16.16 | 88.14 |
| NJMSA | 15.91 | 15.65 | 87.83 |
| UNCC1015 | 15.92 | 15.98 | 87.98 |
| CVD737 | ND | 16.13 | 87.93 |
| LC4 | 15.62 | 16.27 | 87.86 |
| UNCC9 | ND | 16.32 | 88.02 |
| 85A667 | ND | 15.61 | 87.92 |
| 1015 | 16.16 | 15.87 | 88.13 |
| 345/O | 16.56 | 16.64 | 87.91 |
| 80363 | 15.72 | 16.09 | 88.78 |
| LC4/T | 16.95 | 17.29 | 88.13 |
| E4125 | 16.49 | 15.62 | 87.83 |
| 2400112 | 18.2 | 17.34 | 88.08 |
| 52785 | ND | 17.71 | 87.46 |
| EDL174 | ND | 16.5 | 87.91 |
| MLT403 | 17.13 | 17.31 | 87.77 |
| LL728 | 17.69 | 17.29 | 87.81 |
| Nontarget strains | |||
| Aeromonas hydrophila 7965 | 0 | 34.77 | 70.76 |
| Escherichia coli | |||
| JM109 | 0 | 37.42 | 82.68 |
| HB101 | 0 | 35.12 | 79.43 |
| Listeria monocytogenes | 0 | 36.11 | 78.15 |
| Pseudomonas aeruginosa | 0 | 35.15 | 86.4 |
| Plesiomonas shigelloides 14029 | 0 | 35.12 | 76.69 |
| Salmonella enterica serovar Cholerasius 10708 | 0 | 36.04 | 77.97 |
| Salmonella enterica subsp. enterica 10112 | 0 | 37.64 | 62.4 |
| Salmonella enterica serovar Enteritidis 13076 | 0 | 39.38 | 63.37 |
| Salmonella enterica serovar Enteritidis 14050 | 0 | 38.99 | 62.66 |
| V. cholerae | |||
| JVY212 | 0 | 34.7 | 79.47 |
| JVB 52 | 0 | 33.38 | 74.24 |
| JVY210 | 0 | 28.3 | 73.88 |
| JVB 25 | 0 | 30.36 | 74.9 |
| 2076 | 0 | 35.06 | 79.59 |
| A5 | 0 | 35.44 | 79.59 |
| V. alginolyticus | 0 | 33.18 | 77.16 |
| V. fischeri ES114 | 0 | 38.44 | 63.17 |
| V. fluvialis 1959-2 | 0 | 33.14 | 78.04 |
| V. furnissii 1958-83 | 0 | 34.35 | 78.76 |
| V. hollisae 89A7053 | 0 | 31.37 | 78.07 |
| V. parahaemolyticus | |||
| LM 5674 | 0 | 31.93 | 72.51 |
| 10290 | 0 | 34.32 | 72.71 |
| LM 4892 | 0 | 36.31 | 78.76 |
| N4 3483R | 0 | 39.27 | 78.9 |
| NY3547 | 0 | 33.06 | 71.91 |
| NVY3483 | 0 | 33.06 | 86.8 |
| TX2103 | 0 | 33.14 | 86.35 |
Strains in bold were reported to be positive by a prior study (16).
CT values are shown for QPCR as described in the text, with melt peak analysis results for the SYBR green I assay. The average values for the target strains were a CT of 16.61 ± 0.87 for the TaqMan assay, a CT of 16.48 ± 0.79 for the SYBR green I assay, and a melt peak of 88.02°C ± 0.26°C. The average values for the nontarget strains were a CT of 0 for the TaqMan assay, a CT of 34.81 ± 2.66 for the SYBR green I assay, and a melt peak of 75.90°C ± 6.75°C.
ND, not done.
The strains examined in this study included other Vibrio species that have genes encoding hemolysins that are somewhat related to vvhA. Our prior work with the vvhA TaqMan probe did not show any cross-reaction with nontarget species; however, false negatives were reported for some nontarget species in another report but only after extensive PCR cycling (19). These false-negative strains were generously provided by A. K. Bej and were included in the present comparison of TaqMan and SYBR green I QPCR detection (shown in bold in Table 1). Although some species (i.e., Vibrio cholerae, V. fluvialis, and Aeromonas hydrophila) exhibit hemolysins with limited deduced amino acid similarity (about 30%) to VvhA, BLAST two-sequence or genomic comparisons did not indicate significant identity to vvhA probes and primers at the nucleotide level. The present results also did not show false-negative amplification for any of these strains by TaqMan PCR, and SYBR melt curve analysis confirmed that PCR products detected after extensive cycling (>30 cycles) were not related to a V. vulnificus product. Therefore, we conclude that the prior report of a false-negative signal was not based on amplification of homologous DNA.
QPCR-MPN of artificially inoculated PHP oysters.
In order to compare assay sensitivities of QPCR-MPN versus standard MPN, PHP oyster homogenates were seeded with known concentrations of V. vulnificus. Oysters were obtained immediately postharvest from Leavins Seafood, and the PHP protocol was performed on site in Apalachicola, FL. High initial numbers (>104 CFU/g) of V. vulnificus bacteria are required for PHP validation (11); therefore, oysters were heat abused prior to processing by overnight incubation (18 to 20 h) at 26°C in order to elevate V. vulnificus numbers, followed by refrigeration of oysters at 4°C for 5 to 6 h. Ultralow-temperature PHP oysters were briefly (<30 min) exposed to liquid nitrogen aspersion in a freezing tunnel, followed by direct immersion in liquid nitrogen (−87°C) until the meat detached from the shell. Frozen oysters were subsequently kept in extended (≥21 days) dry storage at −20°C in order to obtain V. vulnificus-free oysters for seeding studies. PHP oysters (n = 12) were homogenized (Warring blender) in PBS (1:2, wt/wt), and dilutions of overnight cultures of V. vulnificus M06-24/O in alkaline peptone water (APW) (1 ml) were inoculated into triplicate APW enrichment broths (8 ml) containing either PBS (1 ml) alone or 0.01 or 0.10 g oyster homogenate (1 ml) in PBS. Three-tube replicates of seeded enrichment broth cultures were assessed at 0 and 24 h postincubation at 37°C by SYBR green I or TaqMan QPCR as described above and also by standard MPN, as confirmed by growth of typical colonies isolated on modified colistin polymyxin cellobiose (mCPC) agar (12) incubated at 40°C. The QPCR standard curve for each experiment was based on plate counts of dilutions of V. vulnificus on nonselective T1N1 agar (12) as determined immediately postinoculation. Positive (target DNA) and negative (nontarget DNA and no template) controls were included for all QPCR assays. Media were purchased from Fisher Scientific or Difco.
The standard curve for QPCR of V. vulnificus in APW with or without the addition of oyster homogenate demonstrated a linear range of detection from about 102 to 105 for 0.01 g homogenate, with increasing CT values for decreasing inocula (Fig. 1). However, a significant loss of sensitivity was observed with the addition of 0.10 g oyster tissue at lower inocula. Confirmation of positive samples in seeded homogenates prior to growth in APW was about 100-fold more sensitive by QPCR melt peak than by recovery on mCPC (Table 2). However, after 24 h of enrichment all concentrations of seeded homogenates were positive, as indicated by both growth on mCPC and SYBR QPCR melt peak for both 0.10- and 0.01-g homogenates. SYBR green I and TaqMan QPCR results after 24 h of enrichment were similar, and CT values ranged from 15.55 to 20.72 or 16.74 to 20.27, respectively (Table 2). Duplicate experiments showed identical MPN results and similar CT values at 24 h of incubation (not shown). Results confirmed that approximately one cell in the original inoculum could be detected by QPCR-MPN, in agreement with previous reports (1, 18, 19, 20).
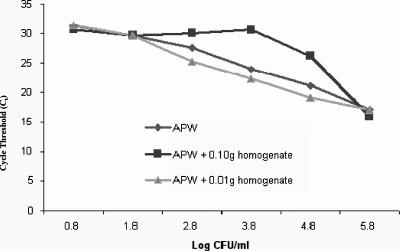
FIG. 1.
QPCR standard curve of V. vulnificus in APW with or without oyster homogenate. QPCR was performed for dilutions of V. vulnificus seeded in triplicate samples of APW with or without addition of oyster homogenate (0.01 g or 0.10 g). CT values for APW alone or with addition of 0.01 g oyster homogenate showed linearity (R2 = 0.99) at concentrations of 1.8 to 5.8 log CFU/ml. Standard deviations of the means ranged from 0.01 to 0.60.
TABLE 2.
Detection of V. vulnificus in artificially inoculated APW enrichmenta
| Inoculum (log CFU/ml)b | % Positive samples before APW enrichment
|
% Positive samples after APW enrichment
|
Postenrichment CT by QPCR
|
|||
|---|---|---|---|---|---|---|
| mCPC | QPCR melt | mCPC | QPCR melt | SYBR green I | TaqMan | |
| 0.01 g oyster homogenate | ||||||
| 5.40 | 100 | 100 | 100 | 100 | 17.94 ± 0.56 | 16.74 ± 0.44 |
| 4.40 | 100 | 100 | 100 | 100 | 18.40 ± 0.40 | 16.97 ± 0.34 |
| 3.40 | 100 | 100 | 100 | 100 | 17.90 ± 0.58 | 17.64 ± 0.20 |
| 2.40 | 33 | 100 | 100 | 100 | 17.90 ± 0.56 | 16.83 ± 0.09 |
| 1.40 | 0 | 100 | 100 | 100 | 17.70 ± 0.60 | 18.56 ± 0.32 |
| 0.40 | 0 | 33 | 100 | 100 | 18.70 ± 0.21 | 17.86 ± 0.30 |
| 0.04 | 0 | 0 | 100 | 100 | 20.01 ± 2.09 | 19.87 ± 2.44 |
| Uninoculated | 0 | 0 | 0 | 0 | 35.38 ± 0.25 | 0.00 |
| 0.10 g oyster homogenate | ||||||
| 5.40 | 100 | 100 | 100 | 100 | 15.84 ± 0.27 | 16.99 ± 0.77 |
| 4.40 | 100 | 100 | 100 | 100 | 16.40 ± 0.07 | 16.76 ± 0.10 |
| 3.40 | 33 | 100 | 100 | 100 | 16.29 ± 0.02 | 17.57 ± 0.21 |
| 2.40 | 0 | 33 | 100 | 100 | 17.01 ± 1.61 | 17.44 ± 1.41 |
| 1.40 | 0 | 0 | 100 | 100 | 17.93 ± 2.89 | 18.35 ± 1.63 |
| 0.40 | 0 | 0 | 100 | 100 | 15.55 ± 0.70 | 16.92 ± 0.23 |
| 0.04 | 0 | 0 | 100 | 100 | 20.72 ± 1.27 | 20.27 ± 1.43 |
| Uninoculated | 0 | 0 | 0 | 0 | 33.18 ± 2.43 | 0.00 |
Detection of V. vulnificus in APW, determined as percent positive samples from three tubes, was based either on observation of V. vulnificus typical colonies on selective agar (mCPC) or on melt peak analysis (QPCR) for pre- and postenrichment. Values are the means (±standard deviations) of duplicate experiments with identical results.
V. vulnificus inocula (log CFU/ml as determined by plate count) for seeding of MPN enrichment of PHP oyster homogenates (0.01 or 0.10 g).
QPCR-MPN validation of oyster PHP.
Validation of ultralow freezing as a PHP method for oysters was conducted using both standard and QPCR methodologies for confirmation of MPN. SYBR green I detection was selected because results with this dye in the seeding studies described above were comparable to results with TaqMan but with lower cost and additional confirmation by melt peak analysis. A comparison of QPCR-based MPN to standard MPN used concurrent examinations of four independent lots (n = 3 samples/lot) of oysters (n = 12 for each sample) that were processed by ultralow freezing as described above. Lots 1 to 3 were evaluated by MPN prior to processing and at days 1 and 21 of frozen storage. An additional lot (lot 4) was evaluated only on day 21. Homogenized oysters were prepared as described above and serially diluted in PBS to yield 1.0 to 0.00001 g for three-tube enrichment cultures (10 ml). V. vulnificus-positive growth was determined by standard MPN on mCPC as described above but with DNA probe confirmation (26) and by SYBR green I QPCR analysis. Positive (target DNA) and negative (nontarget DNA and no template) controls along with standard curve analysis of extracted DNA from dilutions of V. vulnificus cells in APW were included with each sample for quality control.
QPCR-MPN values for all lots were comparable (R2 = 0.97 by Pearson's correlation coefficient) to standard MPN results for confirmation of growth of V. vulnificus following enrichment (Table 3). A total of 1,232 enrichment culture tubes were examined, with 84.9% agreement between both assays: 14.8% of cultures were QPCR positive and probe negative, and only one sample (0.3%) was probe positive and PCR negative. Discrepancies between the two assays were observed only at higher homogenate concentrations (1.0 to 0.1 g enrichment culture), while results with more diluted concentrations (0.01 to 0.0001 g) were consistent for both assays. These results agree with prior observations that MPN values derived from higher concentrations of homogenate plated on CPC may yield false-negative results that are inconsistent with positive results from more dilute concentrations of homogenate (12). Thus, high concentrations of homogenates appear to inhibit either growth of V. vulnificus in APW or recovery on mCPC. In our study, discrepant samples showed positive melt peaks but with high CT values (CT of >30), indicating very little growth in APW.
TABLE 3.
Comparison of standard MPN to QPCR-MPN analysis of PHP oyster samples
| Oyster lot | Treatmenta | Avg (±SD) log MPN/gb
|
|
|---|---|---|---|
| FDA MPN | QPCR MPN | ||
| 1 | Pre-PHP | 2.7 ± 1.5 | 3.2 ± 0.3 |
| 2 | Pre-PHP | 4.4 ± 0.4 | 4.8 ± 0.2 |
| 3 | Pre-PHP | 4.1 ± 1.0 | 4.3 ± 0.5 |
| 1 | PHP 1D | 0.9 ± 0.5 | 1.7 ± 1.1 |
| 2 | PHP 1D | 1.9 ± 0.6 | 2.3 ± 0.3 |
| 3 | PHP 1D | 3.7 ± 0.3 | 3.8 ± 0.2 |
| 1 | PHP 21D | 1.5 ± 0.4 | 2.0 ± 0.1 |
| 2 | PHP 21D | 0.6 ± 0.3 | 0.6 ± 0.3 |
| 3 | PHP 21D | 0.5 ± 0.0 | 0.5 ± 0.0 |
| 4 | PHP 21D | 1.1 ± 0.2 | 0.9 ± 0.3 |
Individual oyster lots (n = 4) were heat abused by incubation at 26°C for 24 h (pre-PHP), followed by processing with ultralow freezing in liquid nitrogen and frozen storage at −10°C for 1 (PHP 1D) or 21 (PHP 21D) days following PHP.
For each lot, oysters (n = 12) were sampled in triplicate, and log MPN/g values were determined by the standard FDA bacteriological analytical manual method (FDA MPN) or by MPN using QCPR confirmation with SYBR green I (QPCR-MPN), as described in the text. Lots 1 to 3 were examined before and after PHP, and lot 4 was examined only at 21 days after PHP.
Both dead cells and viable but not culturable cells are detected by PCR (3) and represent a possible source of contamination for PCR-based MPN. However, the standard MPN assay is dependent on viability of target bacteria, and agreement of QPCR-MPN with this assay demonstrated that residual DNA from nonviable or nonculturable cells did not contribute significantly to QPCR-MPN. Enriched samples that were PCR positive but negative on selective media invariably were derived from higher concentrations of homogenates that were falsely negative on mCPC, as indicated by agreement of positive mCPC and QPCR results in more-diluted inocula of the same sample. DNA from dead cells is likely to be degraded after overnight incubation of oyster homogenates or diluted below the limit of QPCR detection. For example, the addition of 0.10 or 0.01 g of oyster tissue containing 104 MPN/g of V. vulnificus would yield about 101 to 102 CFU/ml, respectively, in 10 ml of APW enrichment. Thus, without bacterial growth, the level of V. vulnificus DNA in broth culture is at or below the limit of detection for QPCR. Bacterial growth during MPN enrichment amplifies the signal for QPCR by increasing cell density and results in CT values equivalent to >104 to 105 CFU/ml based on seeding studies, greatly exceeding possible contamination DNA from dead cells. Additional DNA purification or concentration steps may increase QPCR assay sensitivity but could also increase false-positive detection of DNA from dead cells.
Summary.
These data demonstrated that QPCR provides a sensitive and cost-effective alternative to standard methods for confirmation of MPN. Field trials indicated that QPCR offered an improved confirmatory assay compared to the standard method, as QPCR showed more-sensitive detection at higher concentrations of oyster tissue. Other studies have also reported a lack of growth of some strains of V. vulnificus (serovar E) on CPC or growth of nontarget species (15). Results support adoption of a QPCR confirmation to ensure detection of V. vulnificus at higher concentrations of oyster homogenate. Our data conflicted with a prior report (19) of detection of false positives by the TaqMan assay. Detection of non-Vibrio species (Aeromonas and Pseudomonas species) cannot be attributed to primer hybridization with homologous DNA, as these species do not have sequences with sufficient DNA identity to align with V. vulnificus primers by two-sequence BLAST analysis. We found that discrimination of low-level detection (CT values that were just slightly above threshold after >30 cycles of extended PCR cycling) was provided by the melt peak analysis in the present study. False-positive data may result from numerous differences in methodology (extraction protocol, template concentration, reagents, equipment, etc.) or may reflect contamination, PCR artifact, or excess template. Development of robust, highly reproducible QPCR assays requires optimization, standardization, and interlaboratory verifications that are difficult to achieve but are needed for adoption of these protocols by the industry. The application of internal control standards, as opposed to the external controls and standard curve analysis used herein, would also simplify and enhance this assay. Results show that combining QPCR with MPN increased assay reliability and sensitivity compared to standard methods and support the application of this technology for PHP validation of oysters.
Acknowledgments
This research was funded in part by Sea grants R/LR-Q-26A and R/LF-Q-30 and a USDA special grant.
Technical assistance was provided by Joel Fernandes, Koo-Whang Chung, Charlene Burke, and ABC Research, Inc. Oysters were provided by Leavins Seafood.
Footnotes
Published ahead of print on 28 September 2007.
REFERENCES
- 1.Blackstone, G. M., J. L. Nordstrom, M. C. L. Vickery, M. D. Bowen, R. F. Meyer, and A. Depaola. 2003. Detection of pathogenic Vibrio parahaemolyticus in oyster enrichments by real time PCR. J. Microbiol. Methods 53:149-155. [DOI] [PubMed] [Google Scholar]
- 2.Blake, P. A., M. H. Merson, R. E. Weaver, D. G. Hollis, and P. C. Heulein. 1979. Disease caused by a marine vibrio: clinical characteristics and epidemiology. N. Engl. J. Med. 300:1-5. [DOI] [PubMed] [Google Scholar]
- 3.Brauns, L. A., M. C. Hudson, and J. D. Oliver. 1991. Use of the polymerase chain reaction in detection of culturable and nonculturable Vibrio vulnificus cells. Appl. Environ. Microbiol. 57:2641-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell, M. S., and A. C. Wright. 2003. Real-time PCR analysis of Vibrio vulnificus from oysters. Appl. Environ. Microbiol. 69:7137-7144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cook, D. W., and A. D. Ruple. 1992. Cold storage and mild heat treatment as processing aids to reduce the numbers of Vibrio vulnificus in raw oysters. J. Food. Prot. 55:985-989. [DOI] [PubMed] [Google Scholar]
- 6.Dalsgaard, A., N. Frimodt-Moller, B. Bruun, L. Hoi, and J. L. Larsen. 1996. Clinical manifestations and molecular epidemiology of Vibrio vulnificus infections in Denmark. Eur. J. Clin. Microbiol. Infect. Dis. 15:227-232. [DOI] [PubMed] [Google Scholar]
- 7.DePaola, A., G. M. Capers, and D. Alexander. 1994. Densities of Vibrio vulnificus in the intestines of fish from the U.S. Gulf Coast. Appl. Environ. Microbiol. 60:984-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldhusen, F. 2000. The role of seafood in bacterial foodborne diseases. Microbes Infect. 2:1651-1660. [DOI] [PubMed] [Google Scholar]
- 9.Gulig, P. A., K. L. Bourdage, and A. M. Starks. 2005. Molecular pathogenesis of Vibrio vulnificus. J. Microbiol. 43:118-131. [PubMed] [Google Scholar]
- 10.Harwood, V. J., J. P. Gandhi, and A. C. Wright. 2004. Methods for isolation and confirmation of Vibrio vulnificus from oysters and environmental sources: a review. J. Microbiol. Methods 59:301-316. [DOI] [PubMed] [Google Scholar]
- 11.Interstate Shellfish Sanitation Conference. 2003. Issue relating to a Vibrio vulnificus risk management plan for oysters. ISSC, Columbia, SC.
- 12.Kaysner, C. A., and A. DePaola. 2004. Vibrio cholerae, V. parahaemolyticus, V. vulnificus, and other Vibrio spp. Bacteriological analytical manual online, 8th ed. U.S. FDA, Center for Food Safety and Applied Nutrition. http://www.cfsan.fda.gov/∼ebam/bam-toc.html.
- 13.Kelly, M. T. 1982. Effect of temperature and salinity on Vibrio (Beneckea) vulnificus occurrence in a Gulf Coast environment. Appl. Environ. Microbiol. 44:820-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Linkous, D. A., and J. D. Oliver. 1999. Pathogenesis of Vibrio vulnificus. FEMS Microbiol. Lett. 174:207-214. [DOI] [PubMed] [Google Scholar]
- 15.Macián, M. C., C. R. Arias, R. Aznar, E. Garay, and J. J. Pujalte. 2000. Identification of Vibrio spp. (other than V. vulnificus) recovered from CPC agar from marine natural samples. Int. Microbiol. 3:51-53. [PubMed] [Google Scholar]
- 16.Motes, M. L., A. DePaola, D. W. Cook, J. E. Veazey, J. C. Hunsucker, W. E. Garthright, R. J. Blodgett, and S. J. Chirtel. 1998. Influence of water temperature and salinity on Vibrio vulnificus in Northern Gulf and Atlantic Coast oysters (Crassostrea virginica). Appl. Environ. Microbiol. 64:1459-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliver, J. D., R. A. Warner, and D. R. Cleland. 1983. Distribution of Vibrio vulnificus and other lactose-fermenting vibrios in the marine environment. Appl. Environ. Microbiol. 45:985-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panicker, G., and A. K. Bej. 2005. Real-time PCR detection of Vibrio vulnificus in oysters: comparison of oligonucleotide primers and probes targeting vvhA. Appl. Environ. Microbiol. 71:5702-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panicker, G., M. L. Myers, and A. K. Bej. 2004. Rapid detection of Vibrio vulnificus in shellfish and Gulf of Mexico water by real-time PCR. Appl. Environ. Microbiol. 70:498-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panicker, G., M. L. Myers, and A. K. Bej. 2004. Multiplex PCR detection of clinical and environmental strains of Vibrio vulnificus in shellfish. Can. J. Microbiol. 50:911-922. [DOI] [PubMed] [Google Scholar]
- 21.Randa, M. A., M. F. Polz, and E. Lim. 2004. Effects of temperature and salinity on Vibrio vulnificus population dynamics as assessed by quantitative PCR. Appl. Environ. Microbiol. 70:5469-5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ririe, K. M., R. P. Rasmussen, and C. T. Wittwer. 1997. Product differentiation by analysis of DNA melting curves during the polymerase chain reaction. Anal. Biochem. 245:154-160. [DOI] [PubMed] [Google Scholar]
- 23.Strom, M. S., and R. N. Paranjpye. 2000. Epidemiology and pathogenesis of Vibrio vulnificus. Microbes Infect. 2:177-188. [DOI] [PubMed] [Google Scholar]
- 24.Tamplin, M. L., G. E. Rodrick, N. J. Blake, and T. Cuba. 1982. Isolation and identification of Vibrio vulnificus from two Florida estuaries. Appl. Environ. Microbiol. 44:1466-1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wright, A. C., R. T. Hill, J. A. Johnson, M. C. Roghman, R. R. Colwell, and J. G. Morris, Jr. 1996. Distribution of Vibrio vulnificus in the Chesapeake Bay estuaries. Appl. Environ. Microbiol. 62:717-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wright, A. C., G. A. Miceli, W. L. Landry, J. B. Christy, W. D. Watkins, and J. G. Morris, Jr. 1993. Rapid identification of Vibrio vulnificus on nonselective media with an alkaline phosphatase-labeled oligonucleotide probe. Appl. Environ. Microbiol. 59:541-546. [DOI] [PMC free article] [PubMed] [Google Scholar]