Abstract
Members of the class Epsilonproteobacteria are known to be of major importance in biogeochemical processes at oxic-anoxic interfaces. In pelagic redoxclines of the central Baltic Sea, an uncultured epsilonproteobacterium related to Sulfurimonas denitrificans was proposed to play a key role in chemolithotrophic denitrification (I. Brettar, M. Labrenz, S. Flavier, J. Bötel, H. Kuosa, R. Christen, and M. G. Höfle, Appl. Environ. Microbiol. 72:1364-1372, 2006). In order to determine the abundance, activity, and vertical distribution of this bacterium in high-resolution profiles, 16S rRNA cloning and catalyzed reporter deposition and fluorescence in situ hybridization (CARD-FISH) and quantitative PCR measurements were carried out. The results showed that 21% of the derived clone sequences, which in the present study were grouped together under the name GD17, had >99% similarity to the uncultured epsilonproteobacterium. A specific gene probe against GD17 (S-*-Sul-0090-a-A-18) was developed and used for enumeration by CARD-FISH. In different pelagic redoxclines sampled during August 2003, May 2005, and February 2006, GD17 cells were always detected from the lower oxic area to the sulfidic area. Maximal abundance was detected around the chemocline, where sulfide and nitrate concentrations were close to the detection limit. The highest GD17 numbers (2 × 105 cells ml−1), representing up to 15% of the total bacteria, were comparable to those reported for Epsilonproteobacteria in pelagic redoxclines of the Black Sea and the Cariaco Trench (X. Lin, S. G. Wakeham, I. F. Putnam, Y. M. Astor, M. I. Scranton, A. Y. Chistoserdov, and G. T. Taylor, Appl. Environ. Microbiol. 72:2679-2690, 2006). However, in the Baltic Sea redoxclines, Epsilonproteobacteria consisted nearly entirely of cells belonging to the distinct GD17 group. This suggested that GD17 was the best-adapted epsilonproteobacterium within this ecological niche.
Members of the class Epsilonproteobacteria are increasingly recognized as predominantly auto- to mixotrophic organisms globally ubiquitous in modern marine and terrestrial ecosystems (8). A number of studies have verified their significant role in biogeochemical cycles, particularly those that are sulfur dependent, as demonstrated for deep-sea hydrothermal fields (8, 33, 35), sulfidic cave springs (11, 12), and autotrophic episymbiotic associations (35, 40). Some studies have discovered the prevalence and diversity of Epsilonproteobacteria in pelagic marine redoxclines of the Black Sea (42) and the Cariaco Trench (31), two of the largest anoxic basins on this planet (8). Marine pelagic redoxclines, as the transition between oxic and anoxic milieus, are characterized by a gradient in the redox potential that is dependent on the activity of oxidizing versus reducing agents (16). Catalyzed reporter deposition and fluorescence in situ hybridization (CARD-FISH) analyses using gene probe EPS549 demonstrated maximal epsilonproteobacterial abundances of up to 30% of total cell counts in pelagic redoxclines of both basins (29), usually in zones where remarkable chemoautotrophic activity had been previously detected (20, 29, 41).
The Baltic Sea itself is among the largest brackish basins of the world. The Baltic proper comprises a number of deep areas with anoxic bottom water, of which the Gotland Deep is the largest and the Landsort Deep, at 495 m, is the deepest. Pelagic redoxclines in the central Baltic Sea are also generally characterized by high carbon dioxide (CO2) dark fixation rates, which may account for up to 30% of surface primary production (10). The simultaneous occurrence of high denitrification rates has already led to the conclusion that chemolithoautotrophic oxidation of sulfur compounds coupled to nitrate reduction likely plays an important role in these pelagic redoxclines (6). Furthermore, the epsilonproteobacterium “uncultured Helicobacteraceae G138eps,” which is related to Sulfurimonas denitrificans, was identified as a potentially important chemolithoautotrophic sulfide oxidizer and nitrate reducer responsible for denitrification processes at the redoxcline of the Gotland Deep (5). Quantitative PCR (qPCR) (24) and calculations based on stimulation experiments (26) showed that the abundance of this organism within the redoxcline is as high as 5 × 105 cells ml−1. However, compared to the Black Sea (42) or Cariaco Trench (31), the Gotland Deep contains a lower epsilonproteobacterial diversity (5, 26), which raises the question whether the previously identified, uncultured autotrophic denitrifier dominates this class in Baltic redoxclines. In order to resolve this question, we determined the vertical distribution and in situ abundance of this strain in high-resolution profiles of different redoxclines of the central Baltic Sea by using a newly developed gene probe. The results were compared to the overall abundance of Epsilonproteobacteria.
MATERIALS AND METHODS
Sampling.
The samples were obtained from the central Baltic Sea during research cruises onboard the RV Alexander von Humboldt in August 2003 (Baltic sea monitoring station 286, 58°0.00′N; 19°54.00′E, Farö Deep), the RV Alkor in May 2005 (station 271, 57°19.2′N; 20°03′E, Gotland Deep), and the RV Maria S. Merian in February 2006 (station 271). Water samples from different depths around the chemocline, which we define as the shallowest appearance of sulfide, were collected in free-flow bottles attached to a conductivity, temperature, and depth rosette. Electrodes for salinity and temperature measurements were also attached to the conductivity, temperature, and depth rosette. Concentrations of inorganic nutrients, oxygen, and hydrogen sulfide were analyzed immediately after sampling, as described elsewhere (15).
CO2 dark fixation.
CO2 dark fixation rates were determined using a modification of the method described by Steemann-Nielsen (39). Accordingly, 100 μCi [14C]bicarbonate in anoxic solution (specific activity, 53.0 mCi mmmol−1; Hartmann Analytic GmbH, Braunschweig, Germany) was added to 120-ml Winkler bottles containing the collected samples. After incubation at in situ temperatures for 24 h in the dark, the samples were filtered through 0.2-μm filters and then exposed to HCl fumes. Radioactivity was counted in a scintillation counter (Packard).
Bacterioplankton preparation.
For FISH and total cell counts, 100-ml portions of water samples were directly transferred to glass bottles and fixed with particle-free formaldehyde (2% final concentration) for 2 to 5 h at 4°C. Portions of 40 to 45 ml were filtered onto white polycarbonate membrane filters (type GTTP; pore size, 0.2 μm; diameter, 47 mm; Millipore). The filters were then rinsed with sterile seawater, air dried, and stored at −80°C until further processing. For DNA extraction, 1 to 2 liters of water samples was filtered onto white Durapore filters (type GVWP; pore size, 0.22 μm; diameter, 47 mm; Millipore) and frozen at −80°C.
16S rRNA gene clone library construction and RFLP.
A bacterial 16S rRNA gene clone library was prepared from a genomic DNA extract originating from the Farö Deep (depth of 110 m). Nucleic acids were extracted by the procedure of Schauer et al. (36). To amplify nearly full-length 16S rRNA genes from the total community DNA, the bacterial primer system 27f (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492r (5′-GGTTACCTTGTTACGACTT-3′) (27) was used for PCR. PCR mixtures (50 μl) contained 1× PCR buffer, 250 μM of each deoxynucleoside triphosphate, 0.15 μM of each forward and reverse primer, and 1.25 U Taq polymerase (Fermentas). The reaction mixtures were incubated in a MyCycler (Bio-Rad) under the following conditions: initial denaturation at 94°C for 5 min; 30 cycles, with 1 cycle consisting of 94°C for 1 min, 45°C for 45 s, and 72°C for 90 s; followed by one cycle of 72°C for 2 min. The PCR products were purified (MinElute PCR purification kit; Qiagen) and then cloned following the manufacturer's instructions using the pGEM-T Easy vector system (Promega) together with competent Escherichia coli JM109 cells. For restriction fragment length polymorphism (RFLP) and sequencing, the inserted fragment was PCR amplified with the vector-specific primers T7 and SP6. The unpurified PCR products were digested with the restriction enzymes Hin6I (HhaI) and MspI (HpaII) (Fermentas). Restricted fragments were analyzed by gel electrophoresis, and restriction patterns were compared visually. 16S rRNA gene clones with identical band patterns were merged into one RFLP group, and representative cloned fragments were sequenced by Seqlab (Göttingen, Germany) and JenaGen (Jena, Germany) using the primers 27f, 1492r, 533f [5′-GTGCCAGC(A/C)GCCGCGGTAA-3′] (27), and com2rpH (5′-CCGTCAATTCCTTTGAGTTT-5′) (37).
Phylogenetic analysis.
16S rRNA gene sequences were examined for accuracy using the software program SeqMan (DNAstar) and checked for chimeras using the Bellerophon program (19). Phylogenetic affiliations of the partial 16S rRNA sequences were initially estimated with the program BLAST (2). Sequences were aligned using the ARB software package (30). Sequences with similarity greater than 99% were grouped and named GD17. Phylogenetic trees were constructed based on sequences of approximately 1,400 nucleotides. These sequences were reduced to unambiguously alignable positions using group-specific filters. An evolutionary-distance dendrogram was constructed using the Jukes-Cantor correction and neighbor joining followed by maximum likelihood and maximum parsimony analyses.
Probe design.
A specific probe against GD17 cells was designed with the PROBE_FUNCTION tool of the ARB package. Probe specificity was validated by employing the PROBE_MATCH tool of the ARB package and BLAST. Horseradish peroxidase (HRP)-labeled probes used in this study were synthesized by Biomers (Ulm, Germany). The newly designed S-*-Sul-0090-a-A-18 probe, which was named according to the Oligonucleotide Probe Database nomenclature (1), is abbreviated hereafter as SUL90. The specificity of the newly designed SUL90 probe, was experimentally tested by whole-cell hybridization with positive- and negative-control bacterial strains. The epsilonproteobacteria Sulfuricurvum kujiense (ATCC BAA-921T) and Sulfurimonas denitrificans (ATCC 33889T) and the gammaproteobacterium Methylophaga marina (ATCC 35842T) served as negative controls (Table 1). All strains were cultivated following the suggestions of the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (DSMZ). The positive control, which consisted of a culture enriched in GD17 cells, was prepared as follows. Water samples originating from the Gotland Deep (May 2005) at a depth of 215 m were reinoculated in artificial brackish water medium (7). For anoxic cultivation, glass bottles were filled with the boiled medium, covered with butyl rubber stoppers, bubbled with N2, and then autoclaved. Subsequently, 1 ml of trace element solution SL10 (45), 2 ml of 10-vitamin solution (4), 0.2 ml of selenite-tungstate solution (44), and 10 ml of a solution containing 0.5 mmol KNO3 and Na2S2O3 liter−1 were added per liter of medium. Cloning, sequencing, and qPCR (24) demonstrated that the selective enrichment culture was dominated by cells phylogenetically identified as members of the GD17 group.
TABLE 1.
Probe sequence, appropriate target sites, and corresponding sequences in other bacteria, including reference organisms
| SUL90 probe,a target, and bacteria | No. of mismatched nucleotides | Sequenceb |
|---|---|---|
| Probe | AUACUAAUCACCGCGUGC | |
| Target | UAUGAUUAGUGGCGCACG | |
| Target/reference organisms | ||
| Uncultured Helicobacteraceae | 0 | ================== |
| Clone GD17b | 0 | ================== |
| Clone GD17a | 0 | ================== |
| Moraxella cuniculi | 1 | ==============A=== |
| Moraxella bovis | 1 | ==============A=== |
| Moraxella ovis | 1 | ==============A=== |
| Methylophaga sp. | 1 | ==============G=== |
| Sulfuricurvum kujiense YK-4 | 1 | C================= |
| Sulfuricurvum kujiense YK-2 | 1 | =U================ |
| Legionella sp. | 1 | ===A============== |
| Sulfurimonas denitrificans | 4 | GC===CG=========== |
| Methylophaga marina | 4 | ACG===========G=== |
The S-*-Sul-0090-a-A-18 probe was named according to the Oligonucleotide Probe Database nomenclature (1) and is abbreviated as SUL90.
The probe sequence is shown 3′ to 5′, and the target sequence is shown 5′ to 3′. For the target/reference organisms, nucleotides identical to those in the target sequence (=) are indicated.
Quantification with CARD-FISH.
Bacterial CARD-FISH enumeration was carried out according to protocols modified from the methods of Pernthaler et al. (34) and Sekar et al. (38). All HRP-labeled probes, appropriate sequences, target sites, and hybridization conditions used in this study are shown in Table 2 . Nonspecific binding was determined using the NonEUB probe. By changing the formamide concentration, the hybridization condition for the newly designed probe SUL90 was optimized (formamide concentrations tested, 55%, 60%, and 65%). A concentration of 55% formamide in the hybridization buffer was found to be optimal for probe specificity.
TABLE 2.
HRP-labeled oligonucleotide probes and hybridization conditions used in this studya
| Probe | Sequence (5′ to 3′) | Positionsb | Specificity | Reference |
|---|---|---|---|---|
| EUB338 I | GCTGCCTCCCGTAGGAGT | 338-355 | Most Bacteria | 3 |
| EUB338 II | GCAGCCACCCGTAGGTGT | 338-355 | Planctomycetales | 9 |
| EUB338 III | GCTGCCACCCGTAGGTGT | 338-355 | Verrucomicrobiales | 9 |
| NonEUB | ACTCCTACGGGAGGCAGC | None (negative control) | 43 | |
| EPS549 | CAGTGATTCCGAGTAACG | 548-565 | Epsilonproteobacteria | 29 |
| SUL90 | CGTGCGCCACTAATCATA | 90-107 | Subgroup of Epsilonproteobacteria | This study |
The formamide concentration in the hybridization buffer was 55% for all probes.
Escherichia coli 16S rRNA positions.
Discrimination of non-target organisms was successful for the close relative Sulfurimonas denitrificans and for the less-related Methylophaga marina, whereas nonspecific binding with the negative-control strain Sulfuricurvum kujiense was recorded at all formamide concentrations tested. However, the signal from this negative-control strain was always much weaker than that from the positive reference enrichment culture. We considered that the nonspecific binding with Sulfuricurvum kujiense did not raise a problem in this study, since Sulfuricurvum kujiense, isolated from an underground crude oil storage cavity (23), was assumed to not be present in the Baltic Sea and has never been detected in this habitat before.
In conformity with the protocol of Pernthaler et al. (34), filter sections embedded in low-gelling-point agarose were permeabilized with lysozyme and then placed in 1.5-ml reaction vials containing 400 μl of hybridization buffer and 2 μl of probe working solution (50 pmol μl−1). For probe mix EUB338 I to III, 600 μl of hybridization buffer and 3 μl of probe working solution were used. Hybridization was carried out at 35°C for 8 to 12 h on a rotary shaker in the dark. According to the procedure of Sekar et al. (38), filter sections were washed in prewarmed washing buffer and the tyramide signal was amplified with 5-(and 6-)carboxyfluorescein-labeled tyramides. The preparations were counterstained with a previously described mixture of 4′,6′-diamidino-2-phenylindole (DAPI), Citifluor, and VectaShield (34). Stained filter sections were examined with an epifluorescence microscope (Axioskop 2 mot plus; Zeiss) equipped with a 100× Plan Apochromat oil objective lens (Zeiss). Fluorescein isothiocyanate-stained cells were counted, followed by the determination of DAPI-stained cells as an indicator of total prokaryotic abundance. At least 1,000 DAPI-stained cells in randomly distributed microscopic fields were counted for each filter section. Negative-control counts with probe NonEUB averaged 0.11 to 0.28% for both the Farö Deep and Gotland Deep in May 2005 and February 2006. Bacterial counting of both DAPI-stained samples and hybridized samples was usually done with a standard deviation of less than 7.5%.
Quantification with real-time PCR.
An iCycler (Bio-Rad) and the iQ SYBR Green supermix were used for real-time PCR. A GD17-specific absolute DNA standard with a defined number of gene copies was prepared by amplifying inserted fragments of the GD17a clone (generated by the 27f/1492r primer pair [see above]) with the vector-specific primers T7 and SP6, resulting in a 1,661-bp PCR product. This was purified using the MinElute purification kit (Qiagen), and the PCR product concentration was determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). The copy number nanogram−1 was calculated based on the weight and length of the PCR product as described previously (24). Eventually, GD17 16S rRNA genes were quantified based on a nested PCR approach by using the specific primers OST 1F (5′-TCAGATGTGAAATCCAATGGCTCA-3′; Escherichia coli positions 663 to 686) and OST 1R (5′-CTTAGCGTCAGTTATGTTCCAGG-3′; E. coli positions 803 to 825) as previously described (24). The conversion of environmental GD17 16S rRNA gene copy numbers into cell numbers was calculated based on the assumption of 2.3 16S rRNA gene copies per cell, which was previously reported as the average epsilonproteobacterial gene copy number (22). GD17 quantification was done with standard deviations of less than 2%.
Statistical analyses.
Spearman rank correlations were used to investigate the relations among different parameters at significance levels of P ≤ 0.01.
Nucleotide sequence accession numbers.
Sequences were deposited in the GenBank database under accession numbers EF405797 (GD17b) and EF405798 (GD17a).
RESULTS
Physicochemical features.
The chemocline in the Farö Deep in 2003 was located at a depth of 108 m (Fig. 1A). A small overlap of nitrate and sulfide was detected around the chemocline. In the Gotland Deep in May 2005, sulfide was first detected at a depth of 208 m (Fig. 2A). This water profile lacked a clear overlap of sulfide and nitrate. In 2006, the Gotland Deep chemocline was difficult to define because of the extended overlap of oxygen and sulfide at a depth of 121 to 127 m (Fig. 3A).
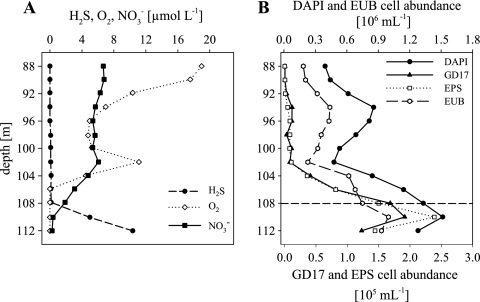
FIG. 1.
Chemical parameters and the distribution and abundance of bacteria detected by CARD-FISH along the Farö Deep redoxcline in August 2003. (A) Concentrations of H2S, O2, and NO3−. (B) Vertical distributions of total cell numbers (DAPI), eubacteria (EUB), GD17, and epsilonproteobacterial (EPS) cell numbers. The broken horizontal line in panel B indicates the chemocline.
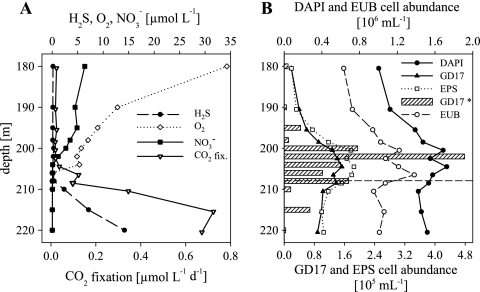
FIG. 2.
Chemical parameters and the distribution and abundance of bacteria detected by CARD-FISH along the Gotland Deep redoxcline in May 2005. (A) Concentrations of H2S, O2, NO3−, and CO2 and dark CO2 fixation rates. (B) Vertical distributions of total cell numbers (DAPI), eubacteria (EUB), GD17, and epsilonproteobacterial (EPS) cell numbers. GD17* cell numbers (bars) were determined by qPCR (as described in Materials and Methods). The broken horizontal line in panel B indicates the chemocline.
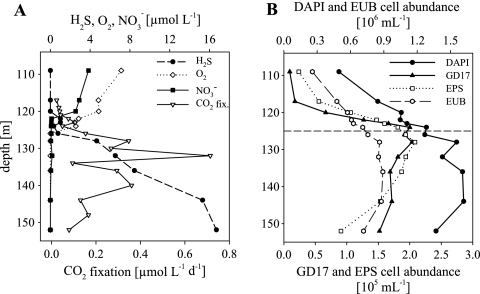
FIG. 3.
Chemical parameters and the distribution and abundance of bacteria detected by CARD-FISH along the Gotland Deep redoxcline in February 2006. (A) Concentrations of H2S, O2, NO3−, and CO2 dark fixation rates. (B) Vertical distributions of total cell numbers (DAPI), eubacteria (EUB), GD17, and epsilonproteobacterial (EPS) cell numbers. The broken horizontal line in panel B indicates the chemocline.
CO2 dark fixation rates.
In the oxic/sulfidic transition zone of the Gotland Deep, the CO2 fixation rates increased in May 2005 from 0.12 μmol liter−1 day−1 at 206.5 m to 0.72 μmol liter−1 day−1 at 215.5 m, with a maximum in the sulfidic area (Fig. 2A). In February 2006, the CO2 fixation rates increased from 0.1 μmol liter−1 day−1 at 123 m to the highest rate of 0.71 μmol liter−1 day−1 at a depth of 132 m in the sulfidic zone (Fig. 3A). No measurements were done in 2003.
16S rRNA gene clone library and probe design.
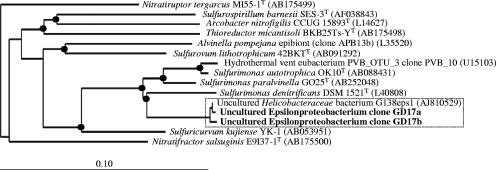
RFLP pattern evaluation of 159 16S rRNA gene clones showed 24 visually distinguishable RFLP groups. Representative clones of all RFLP groups which occurred more than once in the clone library were sequenced, accounting for a total of 21 RFLP patterns. Sequencing confirmed 12 phylogenetically different clones (see Table S1 in the supplemental material). The most abundant RFLP group (21%) consisted of clones phylogenetically closest related to the epsilonproteobacterium “uncultured Helicobacteraceae” (5) within the Sulfurimonas cluster and probably represented members of a new species (Fig. 4). The nearly identical full-length 16S rRNA gene sequences of two clones as well as the “uncultured Helicobacteraceae” sequence were pooled and named group GD17, and the group-specific rRNA-targeting oligonucleotide probe SUL90 was designed for the GD17 group. Probe details and number of mismatches with cultured organisms are presented in Table 1.
FIG. 4.
Unrooted tree showing relationships of subgroup GD17 and its closest phylogenetic relatives within the Epsilonproteobacteria. The tree was constructed using the neighbor-joining method based on a comparison of approximately 1,400 nucleotides. Escherichia coli K-12 was used as an outgroup. Branching points supported by neighbor-joining, maximum likelihood, and maximum parsimony algorithms are marked by a black circle. GenBank database accession numbers are given in parentheses. Bar, 10 substitutions per 100 nucleotides. The boxed area denotes subgroup GD17 as the target sequence of gene probe SUL90.
Quantification of bacteria by CARD-FISH.
The total prokaryotic abundance in the Farö Deep redoxcline in August 2003 varied between 3.9 ×105 and 1.5 × 106 cells ml−1, with maximal cell numbers within the redoxcline (Fig. 1B). Bacterial cell numbers followed the prokaryotic pattern, with 57% EUB338-positive cells. GD17 cells were absent at depths less than 104 m but increased in cell number, with a peak of 1.9 × 105 cells ml−1 at 110 m, i.e., shortly below the chemocline in the small overlap of nitrate and sulfide (Fig. 1A). The highest percentage of GD17 cells accounted for 12.7% of all DAPI-stained cells. The distribution and abundance of Epsilonproteobacteria and GD17 cells were very similar (Fig. 2B). The highest epsilonproteobacterial abundance (2.4 × 105 cells ml−1), corresponding to 15.8% of all DAPI-stained cells, was located at 110 m.
In the Gotland Deep in May 2005, total prokaryotic abundance varied between 1.0 × 106 and 1.7 × 106 cells ml−1 (Fig. 2B). Comparable to the situation in the Farö Deep, cell numbers were highest around the chemocline. Bacterial cell numbers followed the prokaryotic pattern, with 66% EUB338-positive cells. GD17 cells were detected from the suboxic zone to the sulfidic zone; their numbers varied between 1.9 × 104 and 1.5 × 105 cells ml−1. The highest cell numbers were located directly at the chemocline (Fig. 2B), where they accounted for 9.1% of all DAPI-stained cells. The vertical distribution and abundance of Epsilonproteobacteria and GD17 cells were again very similar, with cell numbers ranging from 2.0 × 104 to 1.9 × 105 cells ml−1 (Fig. 2B).
In February 2006, total prokaryotic abundance in the Gotland Deep redoxcline varied between 5.0 × 105 and 1.6 × 106 cells ml−1. In contrast to the other two profiles, maximal cell numbers were reached below the chemocline (Fig. 3B). Bacterial cell numbers followed the prokaryotic pattern, with a proportion of 56% EUB338-positive cells. GD17 cells occurred from the oxic zone to the sulfidic zone; in this interval, cell numbers ranged between 9.4 × 103 and 2.0 × 105 cells ml−1. The highest cell numbers were detected within an extended suboxic-sulfidic transition zone at a depth of 124 to 128 m (Fig. 3A and B). The highest GD17 cell abundance which accounted for up to 15.6% of all DAPI-stained cells appeared in the prolonged overlap of oxygen and sulfide where nitrate could not be detected anymore (Fig. 3A). The distribution and abundance of Epsilonproteobacteria closely resembled those of GD17 (Fig. 3B). Cell abundances according to the different zones of the water column are summarized in Table S2 in the supplemental material. Based on Spearman rank correlation with a level of P ≤ 0.01, there was no significant correlation between GD17 cell numbers and CO2 fixation rates in 2005 and 2006.
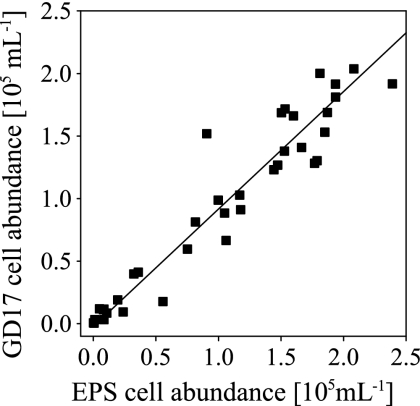
All GD17 cell numbers determined in the three profiles (n = 37) were plotted against the corresponding epsilonproteobacterial cell numbers (Fig. 5). The relationship between EPS and GD17 cell numbers was significant (Spearman's rho = 0.95, P ≤ 0.01). In order to estimate the proportion of GD17 cells, the slope of a regression line was calculated, assuming the same amount of error in GD17 and epsilonproteobacterial cell counts. Data points scattered around the regression line with most of the data points located slightly below it, resulting in an average GD17/Epsilonproteobacteria ratio of 0.94 ± 0.048.
FIG. 5.
Relationship between GD17 and Epsilonproteobacteria (EPS) cell numbers in central Baltic Sea redoxclines. Data points originated from the Farö Deep in 2003 and the Gotland Deep in 2005 and 2006. The straight line is the regression line.
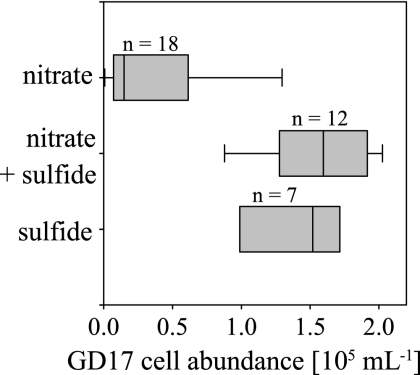
The GD17 cell abundances of all three profiles were compared to the respective occurrence of nitrate and sulfide in the water column (Fig. 6). Areas with a nitrate concentration of 6.7 to 1.2 μmol liter−1 were in general associated with low GD17 cell numbers. The highest GD17 cell numbers were recorded for depths with coexistence of nitrate and sulfide or concentrations near the detection limit. However, even in deeper layers where solely sulfide in concentrations of 5.0 to 16.7 μmol liter−1 could be measured, GD17 cell abundance remained high.
FIG. 6.
Box plot of grouped GD17 cell abundances from the Farö Deep in 2003, the Gotland Deep in 2005 and 2006 versus the occurrence of nitrate and sulfide in the water column. The number of data points included in each box is indicated.
Quantification of 16S rRNA genes by real-time PCR.
For the Gotland Deep redoxcline in May 2005, GD17 16S rRNA gene numbers in the range of 9.3 × 103 to 1.1 × 106 copies ml−1 were determined. The highest numbers (3.5 to 105 to 1.1 × 106 copies ml−1) were detected directly at the chemocline. Conversion into GD17 cell numbers showed that these were particularly comparable to the cell numbers determined by CARD-FISH in the area of the highest cell numbers (Fig. 2B). An exception was at a depth of 202 m, where the cell counts determined by qPCR were approximately three times higher. However, based on Spearman rank correlation with a level of P ≤ 0.01, cell counts determined by qPCR and FISH correlated significantly.
DISCUSSION
Specificity and applicability of probe SUL90.
High-resolution in situ quantification of an already identified epsilonproteobacterium from Baltic Sea redoxclines (5, 18, 24) was obtained by designing a specific 16S rRNA-targeting oligonucleotide probe based on a culture-independent PCR clone library approach. Probe SUL90 was selected, since it was fully complementary to the three target sequences. Furthermore, the target region in the 16S rRNA of the probe was shown to be accessible for probe binding in Escherichia coli (13). There have already been two attempts to estimate absolute GD17-related cell numbers within pelagic redoxclines of the central Baltic Sea using methods independent of gene probes (24, 26). These attempts consisted of highly specific qPCR and calculations based on stimulation experiments. With maximal cell numbers of 1 × 105 to 5 × 105 cells ml−1, these indirect methods revealed cell numbers for the redoxcline that were comparable to those determined by the CARD-FISH approach (Fig. 1B to 3B), a finding that further confirms the applicability of gene probe SUL90.
Potential ecological role of epsilonproteobacterium GD17 in Baltic Sea redoxclines.
The vertical distribution of Epsilonproteobacteria in central Baltic Sea redoxclines was similar to the profiles described by Lin et al. (29) for redoxclines of the Black Sea and the Cariaco Basin. Analogous to these habitats, Epsilonproteobacteria in the Baltic Sea redoxclines were nearly absent in the upper oxygenated layer, reached relatively high abundances of 11 to 16% of total cell numbers around the chemocline, and were still present in significant numbers in the sulfidic zone. Even higher epsilonproteobacterial abundance of up to 30% were recorded for the Cariaco Basin and the Black Sea redoxclines (29). Nevertheless, absolute epsilonproteobacterial cell numbers were comparable because of generally higher total cell counts in Baltic Sea redoxclines (Fig. 1B to 3B).
The presence of specific epsilonproteobacterial taxa related to the Sulfurimonas group was also demonstrated for the Cariaco Basin by 16S rRNA gene clone libraries (31) and for the Black Sea by terminal RFLP fingerprinting (42). The results of both studies suggested that the Sulfurimonas group is globally distributed in this type of habitat. For the central Baltic Sea redoxclines, we were able to demonstrate that nearly all of the Epsilonproteobacteria (on average 94%) detectable by CARD-FISH belong to a single phylogenetic subgroup (Fig. 5). Special physicochemical conditions affect the availability of electron donors and acceptors in these pelagic redoxclines, and their availability may not be stable over time. Bacteria that are active along a suboxic-sulfidic transition zone should possess adaptations to react to changes in energy sources, and it seems that the best-adapted Epsilonproteobacteria in Baltic Sea redoxclines were members of group GD17.
Epsilonproteobacteria have been suggested to be strongly involved in the cycling of carbon, nitrogen and sulfur compounds in sulfidic habitats, especially at oxic-anoxic interfaces (8). Additionally, the ecological significance of chemoautotrophic Epsilonproteobacteria concerning chemoautotrophic production fueled by reduced sulfur species at oxic-anoxic interfaces has been demonstrated (31, 41). Likewise, previous studies (5) pointed to a key role for group GD17 in catalyzing autotrophic denitrification, which is the main process for nitrogen removal in oxic-anoxic interfaces of the central Baltic Sea (17).
The group GD17 distribution, as determined by CARD-FISH (Fig. 1B to 3B) but especially by the qPCR data for the 2005 sampling (Fig. 2B), supported a restricted high-activity zone of this organism around the chemocline. If all CO2 fixation activity attributed to this area of the Baltic Sea was due to GD17 cells, maximal CO2 fixation rates would account for 2 to 11 fg of C day−1 cell−1. Based on an individual cell biomass of 20 fg (14), this would result in cell doubling times of 2 to 12 days, which is much longer than previously determined by stimulation experiments (5, 26). Due to substantially higher CO2 fixation rates in 2006 (Fig. 3A), analogous calculations for the chemocline yield maximal CO2 fixation of 3 to 20 fg C day−1 cell−1, corresponding to cell doubling times of 1 to 7 days. These differences in the potential CO2 fixation activity of GD17 cells could be due to different nutrient availabilities in situ. Advective water transport into depths of 100 to 150 m in the central Gotland Basin is a frequent phenomenon and also occurs in addition to the major inflow events into the Baltic Sea (32). Lateral intrusions as well as internal waves result in small-scale turbulence, which can potentially produce local mixing events (28) and, in turn, temporal mixing of H2S- and NO3−-containing water. The frequency and magnitude of such mixing events, however, are presently unknown.
Interestingly, the CO2 fixation maxima were usually located several meters below the chemocline (Fig. 2 and 3), which is similar to findings from the Black Sea (20) and the Cariaco Trench (41). Moreover, for the Baltic Sea, it has been demonstrated that 20 to 40% of the total cell numbers within this area can consist of chemoautotrophic cells (21). GD17 cell numbers were still relatively high within these maxima (Fig. 2, 3, and 6), but it is not very probable that group GD17 was responsible for this deep peak in CO2 dark fixation based on autotrophic denitrification because nitrate does not occur in this layer. This could indicate other physiological capacities of GD17 cells as, e.g., microbial sulfur oxidation via reduction of particulate metal oxides, such as manganese oxide or iron oxide. However, it is unknown whether GD17 cells could be capable of using other inorganic electron donors and acceptors or even organic substances for gaining energy in these layers. So far, 16S rRNA gene and 16S rRNA (24) distribution data still indicate a more restricted activity zone of group GD17 closely around the chemocline. Still, our results support the previous proposed broader physiological capacity of this organism (25, 26).
Supplementary Material
Acknowledgments
We are very grateful to the captains and crews of the RV Alexander von Humboldt, RV Alkor, and RV Maria S. Merian for their excellent support during sampling cruises. The excellent technical assistance of Heike Brockmöller, Bärbel Buuk, and Annett Grüttmüller is greatly appreciated. Evgeniy Yakushev helped with chemical fieldwork.
This work was funded by the Leibniz-Institut für Ostseeforschung Warnemünde and a DFG grant (LA 1466/4-1) to M.L.
Footnotes
Published ahead of print on 5 October 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The Oligonucleotide Probe Database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brettar, I., M. Labrenz, S. Flavier, J. Bötel, H. Kuosa, R. Christen, and M. G. Höfle. 2006. Identification of a Thiomicrospira denitrificans-like epsilonproteobacterium as a catalyst for autotrophic denitrification in the central Baltic Sea. Appl. Environ. Microbiol. 72:1364-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brettar, I., and G. Rheinheimer. 1991. Denitrification in the Central Baltic: evidence for H2S-oxidation as motor of denitrification at the oxic-anoxic interface. Mar. Ecol. Prog. Ser. 77:157-169. [Google Scholar]
- 7.Bruns, A., H. Cypionka, and J. Overmann. 2002. Cyclic AMP and acyl homoserine lactones increase the cultivation efficiency of heterotrophic bacteria from the central Baltic Sea. Appl. Environ. Microbiol. 68:3978-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Campbell, B. J., A. S. Engel, M. L. Porter, and K. Takai. 2006. The versatile ɛ-proteobacteria: key players in sulphidic habitats. Nat. Rev. Microbiol. 4:458-468. [DOI] [PubMed] [Google Scholar]
- 9.Daims, H., A. Bruhl, R. Amann, K.-H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 10.Detmer, A. E., H. C. Giesenhagen, V. M. Trenkel, H. Auf dem Venne, and F. J. Jochem. 1993. Phototrophic and heterotrophic pico- and nanoplankton in anoxic depths of the central Baltic Sea. Mar. Ecol. Prog. Ser. 99:197-203. [Google Scholar]
- 11.Engel, A. S., N. Lee, M. L. Porter, L. A. Stern, P. C. Bennett, and M. Wagner. 2003. Filamentous “Epsilonproteobacteria” dominate microbial mats from sulfidic cave springs. Appl. Environ. Microbiol. 69:5503-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Engel, A. S., M. L. Porter, L. A. Stern, S. Quinlan, and P. C. Bennett. 2004. Bacterial diversity and ecosystem function of filamentous microbial mats from aphotic (cave) sulfidic springs dominated by chemolithoautotrophic “Epsilonproteobacteria.” FEMS Microbiol. Ecol. 51:31-53. [DOI] [PubMed] [Google Scholar]
- 13.Fuchs, B. M., G. Wallner, W. Beisker, I. Schwippl, W. Ludwig, and R. Amann. 1998. Flow cytometric analysis of the in situ accessibility of Escherichia coli 16S rRNA for fluorescently labeled oligonucleotide probes. Appl. Environ. Microbiol. 64:4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuhrman, J. A., T. D. Sleeter, C. A. Carlson, and L. M. Proctor. 1989. Dominance of bacterial biomass in the Sargasso Sea and its ecological implications. Mar. Ecol. Prog. Ser. 57:207-217. [Google Scholar]
- 15.Grasshoff, K., M. Erhardt, and K. Kremling (ed.). 1983. Methods of seawater analysis. Verlag Chemie, Weinheim, Germany.
- 16.Hallberg, R. O. 1972. Sedimentary sulfide mineral formation—an energy circuit system approach. Mineral Deposita 7:189-201. [Google Scholar]
- 17.Hannig, M., G. Lavik, M. M. M. Kuypers, D. Woebken, W. Martens-Habbena, and K. Jürgens. 2007. Shift from denitrification to anammox after inflow events in the central Baltic Sea. Limnol. Oceanogr. 52:1336-1345. [Google Scholar]
- 18.Höfle, M. G., S. Flavier, R. Christen, J. Bötel, M. Labrenz, and I. Brettar. 2005. Retrieval of nearly complete 16S rRNA gene sequences from environmental DNA following 16S rRNA-based community fingerprinting. Environ. Microbiol. 7:670-675. [DOI] [PubMed] [Google Scholar]
- 19.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 20.Jørgensen, B. B., H. Fossing, C. O. Wirsen, and H. W. Jannasch. 1991. Sulfide oxidation in the anoxic Black Sea chemocline. Deep-Sea Res. 38:S1083-S1103. [Google Scholar]
- 21.Jost, G., M. V. Zubkov, E. Yakushev, M. Labrenz, and K. Jürgens. High abundance and dark CO2 fixation of chemolithoautotrophic prokaryotes in anoxic waters of the Baltic Sea. Limnol. Oceanogr., in press.
- 22.Klappenbach, J. A., P. R. Saxman, J. R. Cole, and T. M. Schmidt. 2001. rrndb: the Ribosomal RNA Operon Copy Number Database. Nucleic Acids Res. 29:181-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kodama, Y., and K. Watanabe. 2004. Sulfuricurvum kujiense gen. nov., sp. nov., a facultatively anaerobic, chemolithoautotrophic, sulfur-oxidizing bacterium isolated from an underground crude-oil storage cavity. Int. J. Syst. Evol. Microbiol. 54:2297-2300. [DOI] [PubMed] [Google Scholar]
- 24.Labrenz, M., I. Brettar, R. Christen, S. Flavier, J. Bötel, and M. G. Höfle. 2004. Development and application of a real-time PCR approach for quantification of uncultured bacteria in the central Baltic Sea. Appl. Environ. Microbiol. 70:4971-4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Labrenz, M., G. Jost, and K. Jürgens. 2007. Distribution of abundant prokaryotic organisms in the water column of the central Baltic Sea with an oxic-anoxic interface. Aquat. Microb. Ecol. 46:177-190. [Google Scholar]
- 26.Labrenz, M., G. Jost, C. Pohl, S. Beckmann, W. Martens-Habbena, and K. Jürgens. 2005. Impact of different in vitro electron donor/acceptor conditions on potential chemolithoautotrophic communities from marine pelagic redoxclines. Appl. Environ. Microbiol. 71:6664-6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lane, D. J. 1991. 16S/23S rRNA sequencing. p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, Chichester, United Kingdom.
- 28.Lass, H. U., H. Prandke, and B. Liljebladh. 2003. Dissipation in the Baltic proper during winter stratification. J. Geophys. Res. Oceans 108:3187. doi: 10.1029/2002JC001401. [DOI] [Google Scholar]
- 29.Lin, X., S. G. Wakeham, I. F. Putnam, Y. M. Astor, M. I. Scranton, A. Y. Chistoserdov, and G. T. Taylor. 2006. Comparison of vertical distributions of prokaryotic assemblages in the anoxic Cariaco Basin and Black Sea by use of fluorescence in situ hybridization. Appl. Environ. Microbiol. 72:2679-2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüβmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K.-H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madrid, V. M., G. T. Taylor, M. I. Scranton, and A. Y. Chistoserdov. 2001. Phylogenetic diversity of bacterial and archaeal communities in the anoxic zone of the Cariaco Basin. Appl. Environ. Microbiol. 67:1663-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matthäus, W., and H. Schinke. 1999. The influence of river runoff on deep water conditions of the Baltic Sea. Hydrobiologia 393:1-10. [Google Scholar]
- 33.Nakagawa, S., K. Takai, F. Inagaki, H. Hirayama, T. Nunoura, K. Horikoshi, and Y. Sako. 2005. Distribution, phylogenetic diversity and physiological characteristics of epsilon-Proteobacteria in a deep-sea hydrothermal field. Environ. Microbiol. 7:1619-1632. [DOI] [PubMed] [Google Scholar]
- 34.Pernthaler, A., J. Pernthaler, and R. Amann. 2002. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl. Environ. Microbiol. 68:3094-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Polz, M. F., and C. M. Cavanaugh. 1995. Dominance of one bacterial phylotype at a mid-Atlantic ridge hydrothermal vent site. Proc. Natl. Acad. Sci. USA 92:7232-7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schauer, M., R. Massana, and C. Pedrós-Alió. 2000. Spatial differences in bacterioplankton composition along the Catalan coast (NW Mediterranean) assessed by molecular fingerprinting. FEMS Microbiol. Ecol. 33:51-59. [DOI] [PubMed] [Google Scholar]
- 37.Schwieger, F., and C. C. Tebbe. 1998. A new approach to utilize PCR-single-strand-conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl. Environ. Microbiol. 64:4870-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sekar, R., A. Pernthaler, J. Pernthaler, F. Warnecke, T. Posch, and R. Amann. 2003. An improved protocol for quantification of freshwater Actinobacteria by fluorescence in situ hybridization. Appl. Environ. Microbiol. 69:2928-2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steemann-Nielsen, E. 1952. The use of radioactive carbon 14C for measuring organic production in the sea. J. Cons. Cons. Int. Explor. Mer. 18:117-140. [Google Scholar]
- 40.Suzuki, Y., S. Kojima, T. Sasaki, M. Suzuki, T. Utsumi, H. Watanabe, H. Urakawa, S. Tsuchida, T. Nunoura, H. Hirayama, K. Takai, K. H. Nealson, and K. Horikoshi. 2006. Host-symbiont relationships in hydrothermal vent gastropods of the genus Alviniconcha from the Southwest Pacific. Appl. Environ. Microbiol. 72:1388-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor, G. T., M. Iabichella, T.-Y. Ho, M. I. Scranton, R. C. Thunell, F. Muller-Karger, and R. Varela. 2001. Chemoautotrophy in the redox transition zone of the Cariaco Basin: a significant midwater source of organic carbon production. Limnol. Oceanogr. 46:148-163. [Google Scholar]
- 42.Vetriani, C., H. V. Tran, and L. J. Kerkhof. 2003. Fingerprinting microbial assemblages from the oxic/anoxic chemocline of the Black Sea. Appl. Environ. Microbiol. 69:6481-6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wallner, G., R. Amann, and W. Beisker. 1993. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry 14:136-143. [DOI] [PubMed] [Google Scholar]
- 44.Widdel, F., and F. Bak. 1992. Gram-negative mesophilic sulphate-reducing bacteria, p. 3353-3378. In T. H. G. Balows, A. M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes. Springer, New York, NY.
- 45.Widdel, F., G.-W. Kohring, and F. Mayer. 1983. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. III. Characterization of the filamentous gliding Desulfonema limicola gen. nov. sp. nov., and Desulfonema magnum sp. nov. Arch. Microbiol. 134:286-294. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.