Abstract
Francisella tularensis subsp. holarctica is widely disseminated in North America and the boreal and temperate regions of the Eurasian continent. Comparative genomic analyses identified a 1.59-kb genomic deletion specific to F. tularensis subsp. holarctica isolates from Spain and France. Phylogenetic analysis of strains carrying this deletion by multiple-locus variable-number tandem repeat analysis showed that the strains comprise a highly related set of genotypes, implying that these strains were recently introduced or recently emerged by clonal expansion in France and the Iberian Peninsula.
The species Francisella tularensis is a gram-negative intracellular pathogen comprising four different subspecies: subsp. tularensis, subsp. holarctica, subsp. mediasiatica, and subsp. novicida. Of these four, the tularensis and holarctica subspecies are clinically significant for humans, with the tularensis subspecies causing a more serious disease in humans and with animal models (7, 25). In addition to virulence characteristics, F. tularensis subsp. tularensis and F. tularensis subsp. holarctica also display unique geographic distributions. F. tularensis subsp. holarctica is found in the Eastern and Western hemispheres, whereas F. tularensis subsp. tularensis appears to be limited to North America (7).
The basis for these apparent differences in ecological and virulence characteristics is not well understood. Comparative genomics studies show that genomic organization among the subspecies is distinct, with multiple rearrangements, translocations, inversions, and recombination events that have occurred during divergence of the subspecies (1, 6, 15, 20, 23). Nonetheless, the composition of the genomes is remarkably similar among the subspecies.
Though F. tularensis subsp. tularensis populations display a more limited geographic range, at least two different sublineages (AI and AII) have been detected in the Eastern and Western portions of the United States by at least two different genotyping methods (8, 13, 18, 24). In contrast, F. tularensis subsp. holarctica populations found throughout North America and the Palearctic ecozone of Eurasia show surprisingly little genetic diversity, leading many to speculate that these populations are derived from a relatively recent bottleneck or clonal expansion event (6, 18, 20-22, 25).
The dearth of genetic diversity in F. tularensis poses substantial epidemiological problems, particularly with F. tularensis subsp. holarctica, constraining molecular epidemiological analyses to the most rapidly evolving genetic markers, such as tandem repeats. Because these markers can be susceptible to homoplasy, additional genomic markers or events can greatly enhance both epidemiological and phylogenetic inference. With this problem in mind, we used our previously described shotgun DNA microarray (23) to search F. tularensis subsp. holarctica strains for genetic diversity by comparative genome hybridization (CGH). Our strain collection included a set of 42 F. tularensis subsp. holarctica strains derived from an epidemic of tularemia in animals and humans in Spain and Southern France in 1997 to 1999 (5, 10), as well as three strains from Czech Republic (Tu-28, Tu-29, and Tu-35) and one from the Russian Federation (Tu-42) (see Table S1 in the supplemental material). A set of 16 F. tularensis subsp. tularensis strains, an F. tularensis subsp. novicida strain, and a Francisella philomiragia strain that had previously been tested with this same array were also included as controls (23). DNA isolation, labeling, hybridization, washing, and data analyses were performed as we have previously described, with binary conversion of the ratios based on >2 standard deviations from the mean of each array (23, 26).
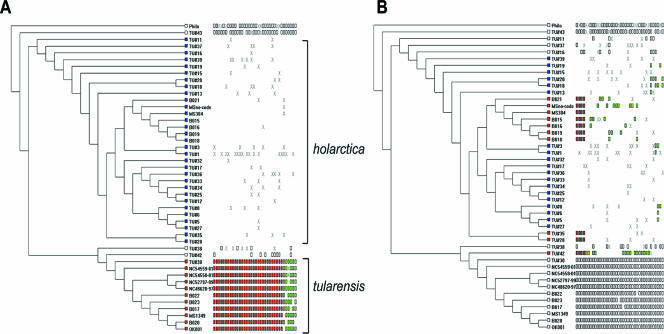
To validate performance of this microarray data set, the binary-converted data were sorted to identify probes hybridizing specifically to F. tularensis subsp. tularensis strains (Fig. 1A) but not the F. tularensis subsp. holarctica, F. tularensis subsp. novicida, or F. philomiragia strains. This yielded 36 probes, which DNA sequence and contig analysis subsequently showed to all be derived from the previously identified RDtularensis1 to RDtularensis13, validating that the array performance was similar to that in our previous study (23). To screen the data for diversity within F. tularensis subsp. holarctica, the data were then sorted among the F. tularensis subsp. holarctica strains according to geography. As shown in Fig. 1B, four of the microarray probes hybridized to F. tularensis subsp. holarctica strains isolated from the United States, the Czech Republic, and Russia but not to the large set of F. tularensis subsp. holarctica isolated from Spain. DNA sequence analysis revealed that all four probes formed a 1.8-kb contig (Fig. 2), corresponding to the FTL_1083 to FTL_1080 coding regions of the F. tularensis subsp. holartica LVS genome sequence and the corresponding region encoding FTT1006 to FTT1009 in the F. tularensis subsp. tularensis SCHU S4 genome sequence (15, 21). The CGH pattern, the fact that these probes formed a contig, and the fact that the Spanish strains are believed to comprise an epidemic (19) suggested that this genome alteration could represent a marker of a clonal population. This segment is referred to herein as the region of genomic difference 23 (RD23) in keeping with the previously used RD numbering system (1, 23, 25).
FIG. 1.
MARKFIND output identifying microarray addresses corresponding to RD23. The dendrograms were generated using the MARKFIND program (26). Dendrograms were generated using the unweighted-pair group method using average linkages algorithm of the binary converted comparative genome hybridization data. The data sets were converted to binary 1 if the ratio of test to reference was <2 standard deviations from the mean (segment present) and converted to 0 if the ratio of test to reference was >2 standard deviations from the mean (significant deviation in hybridization signal). Strain names are indicated to the right of the leaves of the tree. In panel A, the binary data were sorted in the MARKFIND program to identify segments unique to F. tularensis subsp. tularensis strains (red squares at leaves of branches) and absent in F. tularensis subsp. holarctica (blue squares at leaves of branches). The white or colored rectangles to the right of the branches illustrate individual array features that score binary 1 (segment present), whereas white space indicates binary 0 (significant deviation in hybridization signal). An X indicates missing data or instances where the standard deviation was <2 but >1. In panel B, the same UPGMA dendrogram is depicted, except the binary data were sorted to identify array features that score binary 0 for all F. tularensis subsp. holarctica strains derived from Spain and France (marked by blue squares at the leaves) but binary 1 for all other F. tularensis subsp. holarctica stains (red squares at leaves) and binary 1 for all F. tularensis subsp. tularensis strains and F. philomiragia strains (white rectangles at leaves).
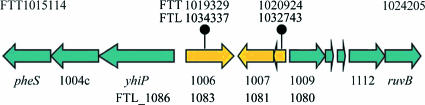
FIG. 2.
Map of the RD23 genomic region from the F. tularensis subsp. tularensis SCHU S4 genome. The arrows depict individual coding regions. Coordinates of the 3′ end of pheS and the 5′ end of ruvB (relative to the F. tularensis subsp. tularensis strain SCHU S4 genome sequence) are indicated above the respective arrows. Coordinates of the deletion endpoints are indicated above the ball and sticks for the corresponding F. tularensis subsp. tularensis SCHU S4 genome sequence (FTT) and the corresponding F. tularensis subsp. holarctica LVS genome sequence (FTL). Coding regions from the FTT and FTL genome sequences are indicated below the relevant portion of the map.
To precisely map the endpoints of the RD23 deletion, PCR primers were designed within the FTT1005c and FTT1009 coding regions abutting the 1.8-kb RD23 contig (RD23Forward, 5′-GTCTTGTTGAGCAAATGCCC-3′; RD23Reverse, 5′-CGGAGCAGGCTTAAATAGTGA-3′) and PCR was performed on F. tularensis subsp. tularensis and F. tularensis subsp. holarctica strains. As shown in Fig. 3, F. tularensis subsp. holarctica strains showing the RD23 CGH pattern (Tu-2, Tu-4, Tu-7, and Tu-10) gave rise to PCR products of the same size, 1.38 kb, whereas all other strains produced the expected 2.97-kb amplicon, in common with the SCHU S4 strain.
FIG. 3.
PCR analysis of RD23. Primers for the RD23 PCR assays were positioned an additional 1 to 2 kb from the left and right ends of the respective RD to ensure inclusion of the junctions in the amplicons. PCRs were performed with 25-μl volumes containing 5 mM MgCl2 and 160 μM of each deoxynucleoside triphosphate (Idaho Technology, Salt Lake, UT), 500 nM (each) of forward and reverse primer, and 2.5 U of Platinum Taq (Invitrogen). Thermocycling conditions were optimized and performed using both T-Gradient (Biometra, Göttingen, Germany) and Dyad (MJ Research, Reno, Nevada) thermocyclers according to the following cycling parameters: initial hold at 95°C for 2 min, 30 s; 30 cycles of 95°C for 30 s, 64°C for 1 min, and 72°C for 1 min; final extension at 72°C; and a final indefinite hold at 4°C. PCR products were electrophoresed on an agarose gel and stained with ethidium bromide. Strain designations are indicated above the respective lanes. The marker lane on the left contains the 1-kb ladder with sizes of the 1- to 3-kb fragments indicated to the left. (For more information on strain designations, see Table S1 in the supplemental material.)
DNA sequence analysis of the shorter, 1.38-kb fragment from strain Tu-19 showed that the RD23 deletion extends from the FTT1006 coding region to that of FTT1008, as predicted from the microarray data. The deletion begins in the central region of the FTT1006 (403 amino acids; hypothetical membrane protein) coding region at the Leu160 codon (SCHU S4 coordinate 1019329) and extends into the FTT1008c (54 amino acids; hypothetical protein) coding region, ending at codon Leu23 (SCHU S4 coordinate 1020924). The protein encoded by FTT1006 shares similarity with several different proteins, including transporter proteins and flgJ product-like amidases. FTT1007 and FTT1008 encode hypothetical proteins.
RD23 is limited to strains from Spain and Southern France.
To test whether the RD23 deletion is a clonal marker for an epidemic population, a real-time PCR assay was developed to specifically identify the RD23 deletion and subsequently used to test a large set of F. tularensis strains. The design of the assay was based on use of TaqMan fluorophore probes, which can be detected on the ABI 7900HT Fast real-time PCR system (Applied Biosystems, Foster City, CA). Primers were designed to generate amplicons approximately 200 bp in size using Primer Express v. 2.0 software (Applied Biosystems). The primers and probe for the leftward flank of the deletion were RD23Left Forward (5′-TTTGTTAGGATTTAGTTTTTGTTTACTTATAGGT- 3′), RD23Left Reverse (5′-ACTGACTCCCTTAGAACCAGAGTCA-3′), and RD23Left Probe (VIC-5′-AGTCGATACTAATCAAKAATTTGTTGCACC-3′-6-carboxytetramethylrhodamine) (where K = T or G). The oligonucleotide sequences for the rightward flank of the deletion were RD23Right Forward (5′-GAAATATTCCATCTCCATCAAAATGC-3′), RD23Right Reverse (5′-ATGGTTTAAAGATGACAATAGTAAGTCGA-3′), and RD23Right Probe (6-carboxyfluorescein-5′-ACTACTTTGATTARGCATAAAAGCAAG-3′-6-carboxytetramethylrhodamine) (where R = A or G). Because naturally occurring single nucleotide polymorphisms were discovered in the SCHU S4 and LVS genome sequences in the probe segments, the probes were constructed with degenerate bases at the “K” and “R” positions (shown in boldface).
The real-time (RT)-PCR assay was developed as a duplex assay, consisting of the leftward forward primer as the common forward primer, the leftward probe just inside the deletion and flanked by its reverse primer, and the rightward probe outside the right junction and flanked by its reverse primer. This scheme allows detection of the RD23Left Probe in instances of an intact region and detection of the RD23Right Probe in strains carrying the RD23 deletion. The RT-PCRs were performed in 25-μl reaction volumes containing 1× Universal Master Mix (Applied Biosystems), 400 nM of each primer, 600 nM of each probe, and 0.55 U of Platinum Taq (Invitrogen). The thermocycling conditions included 50°C for 2 min; a hold for 10 min at 95°C; 45 cycles of 95°C for 15 s, 56°C for 15 s, and 61°C for 1 min; and a final indefinite hold at 4°C. The RT-PCR was validated by first comparing a set of 90 strains, using the conventional PCR and the RT-PCR, where perfect correlation was observed (data not shown). We subsequently used the RT-PCR to screen a large strain collection comprising isolates of F. tularensis subsp. tularensis (n = 88), F. tularensis subsp. holarctica (n = 300), F. tularensis subsp. novicida (n = 7), F. tularensis subsp. mediasiatica (n = 4), and F. philomiragia (n = 6) strains.
Only 51 of 405 strains tested carried the RD23 deletion (see Table S1 in the supplemental material). All 51 of the RD23 deletion strains were F. tularensis subsp. holarctica strains, and 50 of the 51 RD23--positive strains were isolated from Spain or France. The six strains isolated from France were from Chateneaux (AFIP3 and F0295), St. Germaine (AFIP4), Lorraine (FR-LauR), Langres (FR-Syl-S), and Vosges (F0020), implying that RD23-positive strains may be widespread in France. The FR-LauR and FR-Syl-S strains were isolated in 1993 and 2000, respectively, with the FR-Syl-S strain being isolated from one of the first cases of human F. tularensis bacteremia reported in Southern Europe (12). However, the F0020 strain was isolated in 1952, implying that the RD23-positive strains may have been in France for some time.
The larger set of RD23-positive strains from Spain comprised isolates from the Valladolid, Palencia, Zamora, and Leon regions, representing a wide geographic distribution in this country. These strains were all isolated during a narrow time frame of 1997 to 1999, coinciding with an epidemic of tularemia found in humans and animals during that time (11). The only exception of a strain carrying RD23 that was isolated outside of Spain or France was F0228, which was isolated in Uppsala, Sweden. Thus, with the exception of the F0228 isolate, the RD23 allele appears to be concentrated among strains isolated from the Iberian Peninsula and France.
Strains carrying RD23 comprise a diverging clonal complex.
Many of the RD23-positive isolates in our collection from Spain have previously been shown to share a close genetic relationship by amplified fragment length polymorphism and other genotyping methods (5, 9). If the RD23 deletion is indeed a clonal marker of an epidemic population, then we would expect that the strains from France carrying RD23 should also be genetically related. The RD23-positive strains were therefore genotyped by multiple-locus variable-number tandem repeat analysis (MLVA) and normalized to MLVA data generated previously (8, 13). MLVA was performed as described previously (13) using primers amplifying regions Ft-M1 through Ft-M25. All reverse primers were labeled with IRD800 (Li-Cor, Lincoln, NE), and the reactions were resolved on 41-cm gels using a Li-Cor 4000L automated DNA sequencer. Control reactions were run using the SCHU S4 and LVS strains. Sizes of MLVA products were predicted from a combination of known SCHU S4 and LVS sizes, the 50- to 350-bp and 50- to 700-bp IRD800 size standards (Li-Cor), and a 1-bp sequencing ladder. The data from each strain were tabulated with the previously published MLVA data (13) run on an ABI 377 system (Applied Biosystems). Data from the two data sets were converted to numeric base pair values and normalized for allele size by the NAU Laboratory. An unweighted, midpoint-rooted, neighbor-joining tree was created using MEGA 3.1 software (14).
As shown in Fig. 4, all strains carrying the RD23 deletion comprised a single cluster based on MLVA genotypes. All strains in this cluster shared a unique 466-bp allele at M24 that was not observed in any other strain, supporting the conclusion that they are clonally related. Divergence within the cluster was due to multiple alleles at the M3, M6, M8, and M10 loci. Polymorphism at the M3 and M6 loci alone accounts for six of the nine different MLVA genotypes in the RD23 cluster. These loci are among the most highly polymorphic of the MLVA loci examined to date and would be expected to show some differentiation even within a clonal complex (13). Polymorphic alleles at the M8 and M10 loci within the RD23 cluster were detected only in strain F0228 (unique allele at M8) and strain Tu-31 (unique allele at M10). Thus, strains carrying the RD23 deletion appear to form a clone complex with divergence largely marked by events at the most rapidly evolving loci. The most divergent member of the RD23 clade was strain F0228, which carries unique alleles at M3, M6, and M8. The F0228 strain is an interesting case because it is the only isolate carrying RD23 which was not originally isolated from the Iberian Peninsula or France. It was isolated from a skin lesion of a human with tularemia in Uppsala, Sweden, in the year 2000 during a large epidemic in that country but was the only isolate among 28 tested from that epidemic that carries RD23. Further surveillance using a combination of the RD23 deletion and the MLVA genotypes identified in our study will now provide a basis on which to further track the distribution and spread of this apparent clone complex.
FIG. 4.
Cluster analysis of MLVA data from F. tularensis subsp. holarctica strains with or without the RD23 deletion. The dendrogram was generated by neighbor joining using a midpoint root with MEGA 3.1 software. Multiple strains in the same cluster are indicated by filled triangles at the tips of the branches. SCHU S4, an F. tularensis subsp. tularensis strain, was included as an outgroup.
Within the F. tularensis holarctica subsp., genetic variation is scarce, even among strains from unlinked geographies. This lack of diversity is perplexing given that the F. tularensis subsp. holarctica genome has undergone substantial rearrangements with respect to the F. tularensis subsp. tularensis genome (6, 20, 21). MLVA studies also bear this out, showing little allelic diversity in F. tularensis subsp. holarctica populations compared to F. tularensis subsp. tularensis populations (13). These findings have been taken to indicate that the F. tularensis subsp. holarctica population may have recently been through a global evolutionary bottleneck or, more likely, that a clone has emerged and spread globally within a relatively recent time frame.
Our study has now shown that a distinct but highly related clone complex of F. tularensis subsp. holarctica, carrying the distinct RD23 genomic signature, has emerged within France and the Iberian Peninsula. Because tularemia has not commonly been reported in Spain and Southern France, we believe the isolation of highly related strains carrying RD23 is a consequence of recent epidemic spread of a single founding population. Further differentiation of the clone during its spread across the peninsula likely resulted in the eight distinct genotypes that are observed within the RD23 clade of the MLVA tree (Fig. 4). The RD23 strain F0228 appears unusual among the other RD23 strains. It was isolated outside Spain or France, and it bears a relatively distantly related MLVA genotype. It is unclear whether F0228 share ancestry with isolates from France and Spain or if RD23 occurred independently in a separate F. tularensis subsp. holarctica subclone.
Among isolates from France and Spain, phylogenetic inference from the clustering shown in Fig. 4 implies that two primary subpopulations carrying the RD23 signature are circulating. The F0020 strain, isolated from France in 1993, and the F0295 strain, isolated from France in 1952, are within one of these two large subpopulations. If a founder effect can explain the relative abundance of this subpopulation, then the F0295 strain implies that the RD23 clone or its immediate ancestor has been present in the region for >50 years. Although the exact origin of the clone is unknown, its emergence in Northern Spain is now well documented. The RD23 marker will be highly useful in tracking epidemiological and ecological characteristics of this clone complex as it spreads and in understanding how its genome is evolving over time.
It has generally been believed that F. tularensis subsp. holarctica strains are indeed virulent but less so than the F. tularensis subsp. tularensis strains (7). The outbreak of tularemia in northwest Spain, which occurred between 1997 and 1998, is the first recorded outbreak in that region, resulting in over 500 human cases (11). It is not yet clear whether the RD23 clone complex is a hypervirulent clone or whether other factors may explain its epidemic spread. Although a gain of virulence characteristics is usually associated with allelic diversity or gene acquisition, there are well-documented instances of specific deletions enhancing virulence of other species (2, 4, 16, 17) and of genome decay resulting in emergence of highly virulent species (3). The RD23 deletion itself does not include any obvious genes associated with virulence functions, but the adjacent genomic region contains several pseudogenes and genes that have been inactivated by multiple stop codons. One of these degraded coding regions encodes a member of the Mip family (macrophage infectivity potentiator), suggesting some type of association of the region with virulence. Clearly more study will be necessary to determine if the RD23 deletion itself influences any of the apparent virulence characteristics of the clone complex.
Supplementary Material
Acknowledgments
We thank the Centers for Disease Control and Prevention in Fort Collins, CO, for access to DNAs.
This work was supported in part by grant 1R21AI057755-01 to A.K.B. from the National Institutes of Health and a grant from the Homeland Security Advanced Research Projects Agency Bioinformatics and Assays Development Program to P.K. and D.M.W.
The views expressed in this article are those of the authors and do not reflect the official policy or position of the U.S. Air Force, the Department of Defense, or the U.S. Government.
Footnotes
Published ahead of print on 21 September 2007.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Broekhuijsen, M., P. Larsson, A. Johansson, M. Bystrom, U. Eriksson, E. Larsson, R. G. Prior, A. Sjostedt, R. W. Titball, and M. Forsman. 2003. Genome-wide DNA microarray analysis of Francisella tularensis strains demonstrates extensive genetic conservation within the species but identifies regions that are unique to the highly virulent F. tularensis subsp. tularensis. J. Clin. Microbiol. 41:2924-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casalino, M., M. C. Latella, G. Prosseda, P. Ceccarini, F. Grimont, and B. Colonna. 2005. Molecular evolution of the lysine decarboxylase-defective phenotype in Shigella sonnei. Int. J. Med. Microbiol. 294:503-512. [DOI] [PubMed] [Google Scholar]
- 3.Chain, P. S., E. Carniel, F. W. Larimer, J. Lamerdin, P. O. Stoutland, W. M. Regala, A. M. Georgescu, L. M. Vergez, M. L. Land, V. L. Motin, R. R. Brubaker, J. Fowler, J. Hinnebusch, M. Marceau, C. Medigue, M. Simonet, V. Chenal-Francisque, B. Souza, D. Dacheux, J. M. Elliott, A. Derbise, L. J. Hauser, and E. Garcia. 2004. Insights into the evolution of Yersinia pestis through whole-genome comparison with Yersinia pseudotuberculosis. Proc. Natl. Acad. Sci. USA 101:13826-13831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Day, W. A., Jr., R. E. Fernandez, and A. T. Maurelli. 2001. Pathoadaptive mutations that enhance virulence: genetic organization of the cadA regions of Shigella spp. Infect. Immun. 69:7471-7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de la Puente-Redondo, V. A., N. G. del Blanco, C. B. Gutierrez-Martin, F. J. Garcia-Pena, and E. F. Rodriguez Ferri. 2000. Comparison of different PCR approaches for typing of Francisella tularensis strains. J. Clin. Microbiol. 38:1016-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dempsey, M. P., J. Nietfeldt, J. Ravel, S. Hinrichs, R. Crawford, and A. K. Benson. 2006. Paired-end sequence mapping detects extensive genomic rearrangement and translocation during divergence of Francisella tularensis subsp. tularensis and Francisella tularensis subsp. holarctica populations. J. Bacteriol. 188:5904-5914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellis, J., P. C. Oyston, M. Green, and R. W. Titball. 2002. Tularemia. Clin. Microbiol. Rev. 15:631-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Farlow, J., D. M. Wagner, M. Dukerich, M. Stanley, M. Chu, K. Kubota, J. Petersen, and P. Keim. 2005. Francisella tularensis in the United States. Emerg. Infect. Dis. 11:1835-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia Del Blanco, N., M. E. Dobson, A. I. Vela, V. A. De La Puente, C. B. Gutierrez, T. L. Hadfield, P. Kuhnert, J. Frey, L. Dominguez, and E. F. Rodriguez Ferri. 2002. Genotyping of Francisella tularensis strains by pulsed-field gel electrophoresis, amplified fragment length polymorphism fingerprinting, and 16S rRNA gene sequencing. J. Clin. Microbiol. 40:2964-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.García Peña, F. J., P. Suárez Mayoral, C. Cogolludo Cogolludo, C. Arriola Garrote, and E. Anadón Navarro. 1998. An outbreak of tularemia in Castilla-León. First isolation of Francisella tularensis in Spain. Med. Vet. 15:418-423. [Google Scholar]
- 11.Gutierrez, M. P., M. A. Bratos, J. I. Garrote, A. Duenas, A. Almaraz, R. Alamo, H. Rodriguez Marcos, M. J. Rodriguez Recio, M. F. Munoz, A. Orduna, and A. Rodriguez-Torres. 2003. Serologic evidence of human infection by Francisella tularensis in the population of Castilla y Leon (Spain) prior to 1997. FEMS Immunol. Med. Microbiol. 35:165-169. [DOI] [PubMed] [Google Scholar]
- 12.Haristoy, X., A. Lozniewski, C. Tram, D. Simeon, L. Bevanger, and C. Lion. 2003. Francisella tularensis bacteremia. J. Clin. Microbiol. 41:2774-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johansson, A., J. Farlow, P. Larsson, M. Dukerich, E. Chambers, M. Bystrom, J. Fox, M. Chu, M. Forsman, A. Sjostedt, and P. Keim. 2004. Worldwide genetic relationships among Francisella tularensis isolates determined by multiple-locus variable-number tandem repeat analysis. J. Bacteriol. 186:5808-5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 15.Larsson, P., P. C. Oyston, P. Chain, M. C. Chu, M. Duffield, H. H. Fuxelius, E. Garcia, G. Halltorp, D. Johansson, K. E. Isherwood, P. D. Karp, E. Larsson, Y. Liu, S. Michell, J. Prior, R. Prior, S. Malfatti, A. Sjostedt, K. Svensson, N. Thompson, L. Vergez, J. K. Wagg, B. W. Wren, L. E. Lindler, S. G. Andersson, M. Forsman, and R. W. Titball. 2005. The complete genome sequence of Francisella tularensis, the causative agent of tularemia. Nat. Genet. 37:153-159. [DOI] [PubMed] [Google Scholar]
- 16.Maurelli, A. T., R. E. Fernandez, C. A. Bloch, C. K. Rode, and A. Fasano. 1998. “Black holes” and bacterial pathogenicity: a large genomic deletion that enhances the virulence of Shigella spp. and enteroinvasive Escherichia coli. Proc. Natl. Acad. Sci. USA 95:3943-3948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McCormick, B. A., M. I. Fernandez, A. M. Siber, and A. T. Maurelli. 1999. Inhibition of Shigella flexneri-induced transepithelial migration of polymorphonuclear leucocytes by cadaverine. Cell Microbiol. 1:143-155. [DOI] [PubMed] [Google Scholar]
- 18.Nubel, U., R. Reissbrodt, A. Weller, R. Grunow, M. Porsch-Ozcurumez, H. Tomaso, E. Hofer, W. Splettstoesser, E.-J. Finke, H. Tschape, and W. Witte. 2006. Population structure of Francisella tularensis. J. Bacteriol. 188:5319-5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez-Castrillon, J. L., P. Bachiller-Luque, M. Martin-Luquero, F. J. Mena-Martin, and V. Herreros. 2001. Tularemia epidemic in northwestern Spain: clinical description and therapeutic response. Clin. Infect. Dis. 33:573-576. [DOI] [PubMed] [Google Scholar]
- 20.Petrosino, J. F., Q. Xiang, S. E. Karpathy, H. Jiang, S. Yerrapragada, Y. Liu, J. Gioia, L. Hemphill, A. Gonzalez, T. M. Raghavan, A. Uzman, G. E. Fox, S. Highlander, M. Reichard, R. J. Morton, K. D. Clinkenbeard, and G. M. Weinstock. 2006. Chromosome rearrangement and diversification of Francisella tularensis revealed by the type B (OSU18) genome sequence. J. Bacteriol. 188:6977-6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rohmer, L., M. Brittnacher, K. Svensson, D. Buckley, E. Haugen, Y. Zhou, J. Chang, R. Levy, H. Hayden, M. Forsman, M. Olson, A. Johansson, R. Kaul, and S. I. Miller. 2006. Potential source of Francisella tularensis live vaccine strain attenuation determined by genome comparison. Infect. Immun. 74:6895-6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rohmer, L., C. Fong, S. Abmayr, M. Wasnick, T. Larson Freeman, M. Radey, T. Guina, K. Svensson, H. Hayden, M. Jacobs, L. Gallagher, C. Manoil, R. Ernst, B. Drees, D. Buckley, E. Haugen, D. Bovee, Y. Zhou, J. Chang, R. Levy, R. Lim, W. Gillett, D. Guenthener, A. Kang, S. Shaffer, G. Taylor, J. Chen, B. Gallis, D. D'Argenio, M. Forsman, M. Olson, D. Goodlett, R. Kaul, S. Miller, and M. Brittnacher. 2007. Comparison of Francisella tularensis genomes reveals evolutionary events associated with the emergence of human pathogenic strains. Genome Biol. 8:R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samrakandi, M. M., C. Zhang, M. Zhang, J. Nietfeldt, J. Kim, P. C. Iwen, M. E. Olson, P. D. Fey, G. E. Duhamel, S. H. Hinrichs, J. D. Cirillo, and A. K. Benson. 2004. Genome diversity among regional populations of Francisella tularensis subspecies tularensis and Francisella tularensis subspecies holarctica isolated from the US. FEMS Microbiol. Lett. 237:9-17. [DOI] [PubMed] [Google Scholar]
- 24.Staples, J. E., K. A. Kubota, L. G. Chalcraft, P. S. Mead, and J. M. Petersen. 2006. Epidemiologic and molecular analysis of human tularemia, United States, 1964-2004. Emerg. Infect. Dis. 12:1113-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Svensson, K., P. Larsson, D. Johansson, M. Bystrom, M. Forsman, and A. Johansson. 2005. Evolution of subspecies of Francisella tularensis. J. Bacteriol. 187:3903-3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang, C., M. Zhang, J. Ju, J. Nietfeldt, J. Wise, P. M. Terry, M. Olson, S. D. Kachman, M. Wiedmann, M. Samadpour, and A. K. Benson. 2003. Genome diversification in phylogenetic lineages I and II of Listeria monocytogenes: identification of segments unique to lineage II populations. J. Bacteriol. 185:5573-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.