Abstract
The cell surface of Candida albicans is enriched in highly glycosylated mannoproteins that are involved in the interaction with the host tissues. N glycosylation is a posttranslational modification that is initiated in the endoplasmic reticulum (ER), where the Glc3Man9GlcNAc2 N-glycan is processed by α-glucosidases I and II and α1,2-mannosidase to generate Man8GlcNAc2. This N-oligosaccharide is then elaborated in the Golgi to form N-glycans with highly branched outer chains rich in mannose. In Saccharomyces cerevisiae, CWH41, ROT2, and MNS1 encode for α-glucosidase I, α-glucosidase II catalytic subunit, and α1,2-mannosidase, respectively. We disrupted the C. albicans CWH41, ROT2, and MNS1 homologs to determine the importance of N-oligosaccharide processing on the N-glycan outer-chain elongation and the host-fungus interaction. Yeast cells of Cacwh41Δ, Carot2Δ, and Camns1Δ null mutants tended to aggregate, displayed reduced growth rates, had a lower content of cell wall phosphomannan and other changes in cell wall composition, underglycosylated β-N-acetylhexosaminidase, and had a constitutively activated PKC-Mkc1 cell wall integrity pathway. They were also attenuated in virulence in a murine model of systemic infection and stimulated an altered pro- and anti-inflammatory cytokine profile from human monocytes. Therefore, N-oligosaccharide processing by ER glycosidases is required for cell wall integrity and for host-fungus interactions.
Candida albicans is an opportunistic fungal pathogen of humans that can cause superficial infections of the mucosa and, in the immunocompromised host, life-threatening systemic infections (10, 52, 53, 61). The cell wall of C. albicans is the immediate point of contact between the fungus and host and therefore plays a key role in the host-fungus interaction. The cell wall is composed of an inner layer of chitin and β1,3- and β1,6-glucans and an outer layer that is rich in mannoproteins that accounts for 40% of the yeast form cell wall mass (39). The mannoproteins have important roles in adhesion, antigenicity, modulation of the host immune response, and recognition of this fungus by innate immune cells (4, 9, 11, 49, 56, 67, 78). Therefore, studies of cell wall glycosylation can provide insights into the molecular basis of the pathogenic and commensal interactions between C. albicans and the human host.
The N-glycosylation pathway has been studied extensively in Saccharomyces cerevisiae. N glycosylation occurs by a stepwise process involving the transfer of Glc3Man9GlcNAc2 from Glc3Man9GlcNAc2-PP-dolichol to specific asparagine residues of nascent proteins in the endoplasmic reticulum (ER). Subsequently, the Glc3Man9GlcNAc2 core oligosaccharide is processed by α-glucosidases I and II, which sequentially remove the terminal α1,2-linked and two remaining α1,3-linked glucose residues, respectively. Further processing by ER α1,2-mannosidase I generates Man8GlcNAc2 isomer B (27, 28). In the Golgi compartment, Och1 initiates the outer-chain branching of N-glycans by the addition of a single α1,6-linked mannose residue to Man8GlcNAc2 core. In the och1Δ null mutant of C. albicans, the Man8GlcNAc2 N-glycan core is subjected to further modification with three to eight mannose residues added to one or more of the antenna residues of the core (4). The α1,6-mannose backbone of the N-mannan outer chain is extended by the enzyme complexes mannan polymerase I and II, and branched side chains are attached by a range of Golgi mannosyltransferases to yield a high-mannose N-glycan that may represent 95% of the glycoprotein mass (15, 39, 47). Studies of glycosylation pathways in C. albicans are important because key differences exist between the O- and N-glycan structures of this fungus and S. cerevisiae. For example, in C. albicans, the terminal mannose residues of O-glycans are attached by α1,2-linkages rather than α1,3-mannose residues as in S. cerevisiae (46), and β1,2-linked mannose residues are present in both the acid-labile and acid-stable N-glycans (64, 73). Such changes are likely reflected in surface-to-surface interactions of these organisms in their natural environments and during human infection.
In S. cerevisiae, CWH41 (GLS1), ROT2 (GLS2), and MNS1 encode three ER enzymes: α-glucosidase I, α-glucosidase II catalytic subunit, and α1,2-mannosidase, respectively, which are involved in N-glycan core processing (27). Cwh41 removes the outermost α1,2-glucose residue of Glc3Man9GlcNAc2 before it is trimmed further by α-glucosidase II, which removes the α1,3-glucose residues from Glc2Man9GlcNAc2 (23, 62, 76). In S. cerevisiae, the α-glucosidase II is a heterodimer with the catalytic α-subunit encoded by ROT2, a member of the glycosyl hydrolase family 31. In higher eukaryotes the β-subunit normally contains a KDEL-type ER retention motif (74). In lower eukaryotes such as fungi, no gene with significant homology to the β-subunit has been found. Mns1 is an α1,2-mannosidase that trims the Man9GlcNAc2 oligosaccharide to Man8GlcNAc2 isomer B, the last product of the N-glycan processing carried out in the ER (8, 34). It has been suggested that removal of this unique mannose residue induces a conformational reorganization in the N-glycan core that is required for the outer-chain synthesis (8). The core-processing α-glycosidases are also important for glycoprotein folding and for the ER quality control during glycoprotein biosynthesis (24, 29).
Recent studies in C. albicans have shown that protein glycosylation is essential for fungal pathogenesis and immune recognition. Mannosyltransferases involved in N- and O-linked glycosylation have been shown to be required for virulence (4, 7, 46, 54, 66, 69, 70). Golgi proteins involved in the provision of GDP-mannose for glycosylation, which are encoded by CaVRG4 and CaSRB1, are essential for this fungus, indicating the overall importance of glycosylation to cell viability (26, 51, 79, 80). Also, CaPmr1, a Golgi P-type Ca2+/Mn2+-ATPase involved in the transport of Ca2+ and Mn2+ ions into the Golgi compartment, is necessary for normal O- and N-linked glycosylation and virulence (3). However, cell wall phosphomannan synthesis is apparently not required for full virulence (30).
To assess the importance of N-oligosaccharide processing and N-mannan structure on the host-fungus interaction, we disrupted the C. albicans CWH41, ROT2, and MNS1 homologs. The null mutants displayed a number of cell wall defects, were attenuated in virulence in a murine model of systemic infection, and stimulated an altered cytokine profile by human peripheral blood mononuclear cells (PBMC). Therefore, N-oligosaccharide processing by ER α-glycosidases to generate high-mannose N-glycans is vital for the host-fungus interaction and for virulence.
MATERIALS AND METHODS
Strains, media, and culture conditions.
All of the strains used and constructed in the present study are listed in Table 1. Strains were grown at 30°C in YPD medium (1% [wt/vol] yeast extract, 2% [wt/vol] mycological peptone, 2% [wt/vol] glucose) or in SD medium (0.67% [wt/vol] yeast nitrogen base with ammonium sulfate without amino acids, 2% [wt/vol] glucose, 0.077% [wt/vol] complete supplement mixture minus uracil [Qbiogene, Cambridge, United Kingdom]) with uridine (50 μg/ml) as required. Hyphal cells were grown in YPD medium plus 20% (vol/vol) newborn calf serum, Lee's medium (pH 6.5) (41), or salt base (0.45% [wt/vol] NaCl, 0.335% [wt/vol] yeast nitrogen base with ammonium sulfate without amino acids, 2.5 mM GlcNAc) (16) at 37°C or on solid Spider medium (43) at 30°C. To induce β-N-acetylhexosaminidase (HexNAcase) synthesis, cells were grown in SC+GlcNAc (0.67% [wt/vol] yeast nitrogen base with ammonium sulfate without amino acids, 0.077% [wt/vol] complete supplement mixture minus uracil, 25 mM GlcNAc) at 30°C. For virulence assays, the inoculum of yeast cells was grown in NGY medium (0.1% [wt/vol] neopeptone, 0.4% [wt/vol] glucose, and 0.1% [wt/vol] yeast extract) at 30°C.
TABLE 1.
Strains used in this study
| Strain | Parent strain | Genotype | Source or reference |
|---|---|---|---|
| CAI4 | CAF2-1 | ura3Δ::imm434/ura3Δ::imm434 | 22 |
| NGY152 | CAI4 | Same as CAI4 but RPS1/rps1Δ::CIp10 | 6 |
| HMY1 | CAI4 | Same as CAI4 but MNS1/mns1Δ::dp1200-URA3-dp1200 | This study |
| HMY2 | HMY1 | Same as CAI4 but MNS1/mns1Δ::dp1200 | This study |
| HMY3 | HMY2 | Same as CAI4 but mns1Δ::dp1200/mns1Δ::dp1200-URA3-dp1200 | This study |
| HMY4 | HMY3 | Same as CAI4 but mns1Δ::dp1200/mns1Δ::dp1200 | This study |
| HMY5 | HMY4 | Same as CAI4 but mns1Δ::dp1200/mns1Δ::dp1200, RPS1/rps1Δ::CIp10 | This study |
| HMY6 | HMY4 | Same as CAI4 but mns1Δ::dp1200/mns1Δ::dp1200, RPS1/rps1Δ::CIp10-MNS1 | This study |
| HMY7 | HMY2 | Same as CAI4 but MNS1/mns1Δ::dp1200, RPS1/rps1Δ::CIp10 | This study |
| HMY8 | CAI4 | Same as CAI4 but ROT2/rot2Δ::dp1200-URA3-dp1200 | This study |
| HMY9 | HMY8 | Same as CAI4 but ROT2/rot2Δ::dp1200 | This study |
| HMY10 | HMY9 | Same as CAI4 but rot2Δ::dp1200/rot2Δ::dp1200-URA3-dp1200 | This study |
| HMY11 | HMY10 | Same as CAI4 but rot2Δ::dp1200/rot2Δ::dp1200 | This study |
| HMY12 | HMY11 | Same as CAI4 but rot2Δ::dp1200/rot2Δ::dp1200, RPS1/rps1Δ::CIp10 | This study |
| HMY13 | HMY11 | Same as CAI4 but rot2Δ::dp1200/rot2Δ::dp1200, RPS1/rps1Δ::CIp10-ROT2 | This study |
| HMY14 | HMY9 | Same as CAI4 but ROT2/rot2Δ::dp1200, RPS1/rps1Δ::CIp10 | This study |
| HMY15 | CAI4 | Same as CAI4 but CWH41/cwh41Δ::dp1200-URA3-dp1200 | This study |
| HMY16 | HMY15 | Same as CAI4 but CWH41/cwh41Δ::dp1200 | This study |
| HMY17 | HMY16 | Same as CAI4 but cwh41Δ::dp1200/cwh41Δ::dp1200-URA3-dp1200 | This study |
| HMY18 | HMY17 | Same as CAI4 but cwh41Δ::dp1200/cwh41Δ::dp1200 | This study |
| HMY19 | HMY18 | Same as CAI4 but cwh41Δ::dp1200/cwh41Δ::dp1200, RPS1/rps1Δ::CIp10 | This study |
| HMY20 | HMY18 | Same as CAI4 but cwh41Δ::dp1200/cwh41Δ::dp1200, RPS1/rps1Δ::CIp10-CWH41 | This study |
| HMY21 | HMY16 | Same as CAI4 but CWH41/cwh41Δ::dp1200 RPS1/rps1Δ::CIp10 | This study |
Construction of null mutants and control strains.
The MNS1, ROT2, and CWH41 genes were disrupted by the “mini-ura-blaster” protocol (81). To make the disruption cassette, long primers containing 70-pb 5′ and 3′ regions of the open reading frames were used to amplify by PCR the dp1200-URA3-dp1200 cassette contained in plasmid pDDB57 (81): for MNS1, primer pair 5′-ATGCTATTAAAAGGTTTTATGTTGTCTTTAGTATTATATGCTGTGTACCATT TAGCATCAAATGGTGGGCTGTGGAATTGTGAGCGGATA-3′ and 5′-TTACCCAGCAATTTCTTCAATAATTTCTTTAGCTTCTTGATCAGCTGATTTATCAACTGGTTGAGCTTCCGTTTTCCCAGTCACGACGTT-3′; for ROT2, primer pair 5′-ATGAAATTATTTCTAACAATAATTTTTATAATTGCGTCAGTGAATGCTGTAAAGGAGTACTTGTTCAAATTGTGGAATTGTGAGCGGATA-3′ and 5′-TTATAGTTCGTCATGCTCGATTTTTCTGTGGCTGTCAAAACTAAAGGGGAGACTCCAATCCAGGTTGATGGTTTTCCCAGTCACGACGTT-3′; and for CWH41, primer pair 5′-ATGAGATTGTTATCGTGGTTGCCATTTGTATTTCTTTTGGAAGTGATATTTGCCAGCAATCAAA TAAAATTGTGGAATTGTGAGCGGATA-3′ and 5′-TCATTGCAAATGTTCAGGCATTGTCATCATTATCAAGACCAAAGATGACCAGCCCAAAAA GTTTTTAGCTGTTTTCCCAGTCACGACGTT-3′; the regions complementary to plasmid pDDB57 are underlined. The genes were disrupted by sequential rounds of transformation of strain CAI4 and the recycling of the URA3 marker by selection on SD medium plus 5-fluoroorotic acid (1 mg/ml) and uridine. To avoid the problems associated with the ectopic expression of URA3 (6), the Ura− mns1Δ, rot2Δ, and cwh41Δ null strains were transformed with StuI-digested CIp10 plasmid (48); hence, URA3 was expressed at the RPS1 locus. To construct reintegrant control strains, the MNS1 open reading frame plus 999 bp of its promoter and 597 bp of its terminator sequences (total of 3.3 kb) and ROT2 (979 bp of promoter and 693 bp of terminator [total of 4.3 kb]) and CWH41 (946 bp of promoter and 593 bp of terminator [total of 4.0 kb]) genes were amplified by PCR (primer pair for MNS1, 5′-GCGGCCGCTAAGATCAACTTTTTTCTATTC-3′ and 5′-GCGGCCGCAATGATACCAATAAGGA-3′; primer pair for ROT2, 5′-GCGGCCGCAATCTTATTAGCTCCAGAC-3′ and 5′-GCGGCCGCATACCGTACCAAGAAA-3′; and primer pair for CWH41, 5′-GCGGCCGCTTATCTAGACAAATGTTTAAAAT-3′ and 5′-GCGGCCGCTTAGATACTGAACGTCATT-3′, with the bases to generate a NotI site underlined), and the products were cloned into pGEM-T Easy vector (Promega, Ltd., Southampton, United Kingdom). The inserts were released by NotI digestion and subcloned into the NotI site of CIp10. The resulting plasmids were digested with StuI and used to transform the Ura− null strains. As a further control, strain CAI4 was transformed with StuI-digested CIp10. Therefore, all strains analyzed had the URA3 marker expressed at the RPS1 locus.
Assay of α-glycosidase activity.
The α-mannosidase and α-glucosidase activities were measured by using fluorogenic substrates as described previously (45, 71). Cells were collected by low-speed centrifugation, washed twice with 50 mM sodium phosphate buffer (pH 6.0) (buffer A), and broken with glass beads using a FastPrep machine (Qbiogene), and the homogenate was centrifuged at 21,500 × g for 10 min. The supernatant obtained was subjected to ultracentrifugation at 105,000 × g for 1 h, and the high-speed supernatant and the mixed membrane fraction were collected, freeze-dried, and kept at −20°C until use. Fractions (10 to 100 μg of protein) were incubated at 37°C with 40 μM MUαMan or MUαGlc and buffer A for assay of α-mannosidase or α-glucosidase, respectively, in a final volume of 200 μl. After 30 min, the reaction was stopped by adding 3.3 ml of 50 mM glycine-NaOH buffer (pH 11.0), and the fluorescence of 4-methylumbelliferone (MU) was read in a spectrofluorometer with excitation and emission set at 350 and 440 nm, respectively. Specific activity was expressed as nmoles of MU liberated per minute per milligram of protein (1 U = 1 nmol of MU). Total activity is referred as units of specific activity liberated by the total protein of the preparations.
Sensitivity testing.
Strains that were tested for sensitivity to specific cell-wall-perturbing agents were initially grown for 24 h in YPD medium and then washed with water and resuspended at an optical density at 600 nm (OD600) of 1.0. These cells were inoculated into YPD medium at an OD600 of 0.01, and 95-μl aliquots were spotted into microdilution plate wells. Test agents in 5-μl volumes were added at a range of doubling dilutions. Duplicate plates were incubated for 16 h at 30°C, and the OD600 was determined. The agents tested were Calcofluor White (100 μg/ml), Congo red (100 μg/ml), 0.1% sodium dodecyl sulfate (SDS), hygromycin B (500 μg/ml), salts (NaCl and KCl at 1 M), caffeine (50 mM), and tunicamycin (100 μg/ml).
Alcian Blue binding assays.
Alcian Blue affinity assays for phosphomannan content were carried out as described previously (4, 30). Assays were also performed on β-eliminated cells that were stripped of O-glycans, by overnight treatment with 100 mM NaOH at room temperature and subsequent washing of the cells with water.
Analysis of the cell wall composition.
Yeast cells were grown in YPD medium at 30°C and broken as described above. The homogenate was centrifuged at 1,000 × g for 10 min, and the pellet, containing the cell debris and walls, was washed with 1 M NaCl, resuspended in buffer (500 mM Tris-HCl buffer [pH 7.5], 2% [wt/vol] SDS, 0.3 M β-mercaptoethanol, 1 mM EDTA), boiled for 10 min, and freeze-dried. The β-glucan, mannan, and chitin levels were determined by hydrolysis of those polymers and quantification of glucose, mannose, and glucosamine, respectively. For quantification of glucose and mannose, cell walls were resuspended in 2 M trifluoroacetic acid, boiled for 3 h, washed, and centrifuged at 21,500 × g for 10 min. The hydrolysates were analyzed by high-performance anion-exchange chromatography as described previously (45). For the determination of chitin content, the cell walls were hydrolyzed and analyzed as described previously (37). For total protein determination, cell walls were resuspended in 1 N NaOH, boiled for 30 min, neutralized with 1 N HCl, and assayed by using the method described by Bradford (5).
In situ β-N-acetylhexosaminidase activity staining.
Zymograms of native polyacrylamide gel electrophoresis (PAGE) were assayed as reported previously (4). After the growth for 16 h in SC plus GlcNAc, the cells were washed, resuspended in 10 mM Tris-HCl buffer (pH 8.0) containing protease inhibitor cocktail (Roche, Lewes, United Kingdom), and broken with glass beads in a FastPrep machine, and the homogenate was centrifuged at 21,500 × g for 10 min. For deglycosylation treatment, the native sample was incubated with 25 mU of endoglycosidase H (Roche) for 16 h at 37°C in 50 mM sodium acetate buffer (pH 5.2). Samples were mixed with native loading dye (62.5 mM Tris-HCl buffer [pH 6.8], 0.01% [wt/vol] bromophenol blue, and 15% [vol/vol] glycerol) and run on a 3 to 8% Tris-acetate PAGE gel (Invitrogen, Paisley, United Kingdom) for 1 h at 19 V/cm under nondenaturing conditions. The gel was washed in 0.1 M citrate-KOH buffer (pH 4.0) for 10 min at room temperature and then incubated in the substrate solution (0.18 mM naphthyl-GlcNAc [Glycosynth, Ltd., Warrington, United Kingdom] in 0.1 M citrate-KOH buffer [pH 4.0]) for 30 min at 37°C. The reaction product was visualized by incubation in the substrate solution plus 0.7 mM Fast Blue at 60°C until color developed.
Protein extracts and Western blotting.
To test for activation of the cell integrity pathway, cells were grown in YPD medium at 30°C and collected in mid-exponential growth phase. As positive controls, strains were stressed by the addition of Calcofluor White (100 μg/ml) 2 h before collection. Cells were washed, resuspended in extraction buffer (100 mM Tris-HCl buffer [pH 7.5], 0.01% [wt/vol] SDS, 1 mM dithiothreitol, 10% [wt/vol] glycerol, protease inhibitor mixture [Roche]), and broken with glass beads in a FastPrep machine, and the lysate was centrifuged at 21,500 × g for 10 min. Then, 50 μg of protein was separated on a 4 to 12% NuPAGE Bis-Tris gel (Invitrogen) and electrotransferred to a polyvinylidene difluoride membrane. The membrane was blocked in phosphate-buffered saline plus 0.1% Tween 20 and 5 mg of bovine serum albumin/ml for 2 h at room temperature. Detection was then carried out with the PhosphoPlus p44/42 mitogen-activated protein kinase (Thr202/Tyr204) antibody kit (Cell Signaling Technology, Hertfordshire, United Kingdom), which cross-reacts with C. albicans Mkc1 (Slt2) in both its active (phosphorylated) and inactive (nonphosphorylated) forms.
Stimulation of cytokine production in human monocytes.
Isolation of human PBMC was performed as described elsewhere (20). Samples of 5 × 105 PBMC in a 100-μl volume were added to round-bottom 96-well plates (Greiner, Alphen a/d Rijn, The Netherlands) and incubated with 100 μl of various strains of heat-killed (30 min at 56°C) C. albicans yeast cells at a concentration 106 cells/ml. After 24 h of incubation at 37°C, the PBMC-C. albicans cell suspensions were centrifuged; supernatants were collected and stored at −70°C until assayed. Human tumor necrosis factor alpha (TNF-α) concentrations were determined by specific radioimmunoassays as described previously (19). Interleukin-6 (IL-6) and IL-10 concentrations were measured by commercial enzyme-linked immunosorbent assay kits (Pelikine Compact; Sanquin, Amsterdam, The Netherlands). The experiments were performed in duplicate with samples from four volunteers. The differences between strains were analyzed by using the Student t test, and the level of significance was set at P < 0.05.
Virulence tests.
Female, immunocompetent BALB/c mice (Harlan Sera-Lab, Ltd., Loughborough, United Kingdom) were challenged intravenously with yeasts grown for 18 to 24 h in NGY medium at 30°C. The cells were washed twice with water and resuspended in physiological saline to give a challenge inoculum of 1.8 × 104 CFU/g of mouse body weight in a 100-μl volume. For mutants with attenuated growth in vitro, virulence attenuation in vivo was confirmed in groups of two mice intravenously inoculated and monitored over 21 days. Mice showing signs of illness were humanely terminated, and their deaths recorded as occurring the following day.
Nucleotide sequence accession numbers.
The CaCWH41 and CaMNS1 sequences have been submitted to GenBank and were assigned accession numbers DQ295807 and AY167027, respectively.
RESULTS
Isolation, analysis, and deletion of CaCWH41, CaROT2, and CaMNS1.
The analysis of C. albicans genome databank identified two DNA fragments as the 5′ and 3′ ends of CaCWH41 (GenBank accession XM_718516 and XM_705221, respectively). To identify the complete coding sequence, primers aligning in the 5′ and 3′ of the putative open reading frame were used to amplify CaCWH41, and the fragment was cloned and sequenced. The CaCWH41 open reading frame of 2,493 bp (GenBank/EBI accession no. DQ295807) is predicted to encode a protein of 830 amino acids showing homology to other α-glucosidase I enzymes of the glycosyl hydrolase family 63 (60% homology to Cwh41 from S. cerevisiae). The predicted protein had a characteristic type II membrane protein domain structure with a single 20-amino-acid transmembrane region at the N terminus and a 2-amino-acid cytosolic tail. CaCwh41 includes a 606ELNVDLISW614 sequence similar to substrate binding motifs reported in vertebrate enzymes (58). Residues Ser421, Arg467, and Gly717 are homologous to those in human α-glucosidase I, which are necessary for catalytic activity (29, 31, 77).
CaROT2 was identified in the C. albicans genome (36) by homology to the S. cerevisiae homologue. The CaROT2 open reading frame of 2,616 bp (GenBank accession no. XM_711779) is predicted to encode a protein of 871 amino acids of the glycosyl hydrolase family 31 that is between 57 and 38% identical to other fungal and mammalian Rot21 proteins (61% homology to Rot2 from S. cerevisiae). Again, this sequence was typical of a type II membrane protein, having a predicted 2-amino-acid cytosolic tail and a 20-amino-acid transmembrane region. The glycosyl hydrolase family 31 members contain a short peptide segment of conserved amino acids (DGXWIDMNEXSXF), including a conserved Asp residue that is thought to be involved in catalysis (25, 32, 33, 38, 72). A similar peptide sequence (487IHLWNDMNEPSVF499) is present in CaRot2, indicating that this protein also belongs to the same protein family. Consistent with its putative role in N-glycan processing, CaRot2 contains the motif HDEL in the C-terminal region responsible for the receptor-mediated retrieval of a number of ER proteins from the Golgi compartment (68).
The CaMNS1 open reading frame of 1,698 bp (GenBank/EBI accession no. AY167027) is predicted to encode a protein of 565 amino acids with significant homology to other ER glycosyl hydrolase family 47 members (65% homology to Mns1 from S. cerevisiae). This protein is 71 and 42% identical to Mns1 from fungi and mammals, respectively, and also had a characteristic type II membrane protein structure with one cytosolic amino acid and 22 amino acids in the membrane region of the N-terminal domain. Conserved catalytic and metal ion coordinating amino acid residues typical of ER α1,2-mannosidases are present. These include Cys320 and Cys363 corresponding to ScMns1 Cys340 and Cys385, which are necessary for stabilization of the tertiary structure of the catalytic pocket (42), and Arg251, which would be predicted to be required for the specificity of the ER α1,2-mannosidase reaction (59).
The CaCWH41, CaROT2, and CaMNS1 genes were disrupted in strain CAI4 by sequential gene replacement using the mini-ura-blaster protocol (81). The resulting Cacwh41Δ (HMY-19), Carot2Δ (HMY-12), and Camns1Δ (HMY-5) null mutants (Table 1) had URA3 reintroduced at the neutral RSP1 locus to avoid problems due to ectopic expression of URA3 (6, 48). Reintegrant control strains were also constructed in which CaCWH41, CaROT2, or CaMNS1 were introduced into the null strains under the control of their own promoters at the RPS1 locus. Strain CAI4 transformed with CIp10 was used as a control in all experiments and is referred as the parent strain, equivalent to the wild type.
Growth and morphology of the null mutants.
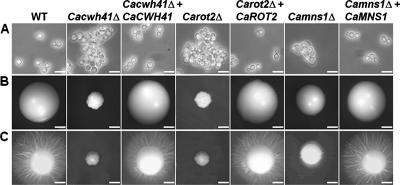
The Cacwh41Δ, Carot2Δ, and Camns1Δ null mutants had reduced specific growth rates in YPD medium at 30°C of 0.44, 0.47, and 0.56 h−1 respectively, compared to the parent strain (0.69 h−1). The reintegrant controls had specific growth rates identical to that of the parent strain. Yeast cells of the three null mutants tended to form small aggregates, and Cacwh41Δ and Carot2Δ yeast cells were swollen and enlarged (Fig. 1A) and small and crenulated (Fig. 1B) colonies. The Camns1Δ null mutant grew as normal hyphae in 20% (vol/vol) serum, GlcNAc-containing medium, and Lee's medium at pH 6.5, while the Carot2Δ and Cacwh41Δ null mutants had delayed filamentation and formed shorter and swollen germ tubes with decreased extension rates (data not shown). All three null mutants failed to induce filaments on solid Spider medium (Fig. 1C). In all cases the mutant phenotypes were fully complemented by reintegration of a wild-type copy of the respective gene.
FIG. 1.
Cell and colony morphology in the Cacwh41Δ, Carot2Δ, and Camns1Δ null mutants. (A) Cell morphology after growth at 30°C for 16 h in YPD medium, demonstrating clumping of cells in the Cacwh41Δ (HMY19), Carot2Δ (HMY12), and Camns1Δ (HMY5) null mutants. Scale bars, 10 μm. (B and C) Colony morphology after 5 days growth at 30°C on YPD agar plates (B) or solid Spider medium (C). Scale bars, 1 mm.
The null mutants have altered α-glycosidase activities.
The α-glucosidase or α-mannosidase activities in the null mutants were determined by using the fluorogenic substrates 4-methylumbellyferyl-α-d-glucopyranoside (MUαGlc) or 4-methylumbellyferyl-α-d-mannopyranoside (MUαMan), respectively. Homogenates of Cacwh41Δ and Carot2Δ null mutants had total α-glucosidase activities of 48 and 51%, respectively, compared to the total activity present in wild-type cells (Table 2). Complemented reintegrant controls recovered the wild-type activity. In wild-type cells, the α-glucosidase activity was distributed equally between the soluble and mixed membrane fractions. It was demonstrated previously that the soluble activity corresponds to α-glucosidase II (71). As predicted, the Cacwh41Δ null mutant had no measurable membrane-associated α-glucosidase activity, whereas Carot2Δ null mutant lacked soluble α-glucosidase activity (Table 2). No measurable α-mannosidase activity was found in soluble fraction or mixed membrane preparations of the Camns1Δ null mutant (data not shown).
TABLE 2.
α-Glucosidase activity in Cacwh41Δ and Carot2Δ null mutants and reintegrant strains
| Strain genotype | Mean α-glucosidase activity ± SDa
|
||
|---|---|---|---|
| Total | Soluble (%)b | Membrane bound (%)b | |
| WT | 35 ± 1 | 55 ± 3 | 45 ± 3 |
| Cacwh41Δ | 17 ± 0.3 | 99 ± 1 | 1 ± 1 |
| Cacwh41Δ + CaCWH41 | 28 ± 0.4 | 57 ± 3 | 43 ± 3 |
| Carot2Δ | 18 ± 0.3 | 2 ± 1 | 98 ± 1 |
| Carot2Δ + CaROT2 | 28 ± 1 | 42 ± 3 | 58 ± 3 |
Expressed as nmol of MU min−1 total protein−1 (n = 3).
Expressed as a percentage of the total activity.
Glycosylation defects.
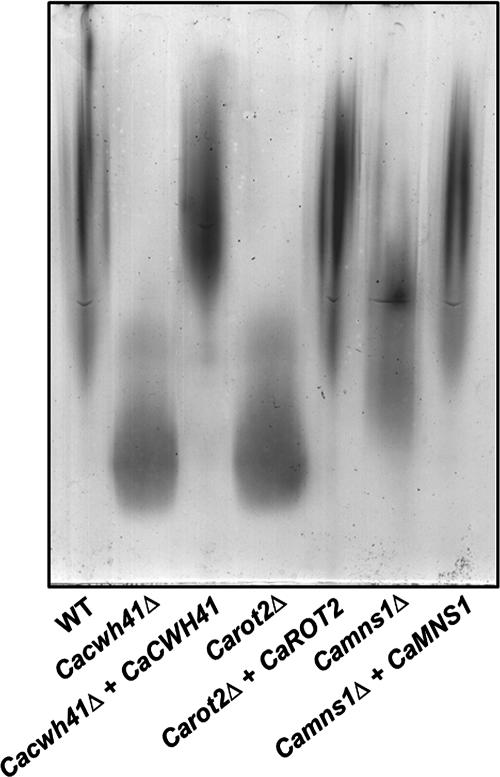
We determined the consequences of the alteration in N-glycan structure in Cacwh41Δ, Carot2Δ, and Camns1Δ null mutants by measuring changes in the electrophoretic mobility of secreted HexNAcase in native gels using an in situ activity assay (4). HexNAcase, encoded by CaHEX1, is induced in media containing GlcNAc as the sole carbon source and has been demonstrated to be highly N glycosylated (12, 44, 50). The HexNAcase from Cacwh41Δ, Carot2Δ, and Camns1Δ null mutants had an increased electrophoretic mobility. Mutants lacking α-glucosidase I and II had the greater mobility, indicating a more severe N-glycosylation defect than in Camns1Δ null mutant (Fig. 2). The electrophoretic mobility of HexNAcase of the reintegrant controls under inducing conditions was similar to wild type. After endoglycosidase H treatment to remove N-glycans, the HexNAcase of all mutant and parent strains migrated faster and with the same mobility (data not shown).
FIG. 2.
N-glycosylation defects in Cacwh41Δ, Carot2Δ, and Camns1Δ null mutants. The electrophoretic mobility of β-N-acetylhexosaminidase under nondenaturing conditions was examined. The strains tested include NGY152 (wild type), HMY19 (Cacwh41Δ), HMY20 (Cacwh41Δ + CaCWH41), HMY12 (Carot2Δ), HMY13 (Carot2Δ + CaROT2), HMY5 (Camns1Δ), and HMY6 (Camns1Δ + CaMNS1).
In C. albicans most of the acid-labile phosphomannan fraction is attached to N-linked mannan (30). Phosphomannan accounts for the negative charge of the cell wall, and this binds the cationic dye Alcian Blue. The Cacwh41Δ, Carot2Δ, and Camns1Δ null mutants bound 17.7, 18.5, and 41.1% of the Alcian Blue bound by the parent strain, respectively (Table 3). The reintegrant controls showed the wild-type levels of Alcian Blue bound. When the Alcian Blue binding assay was carried out after elimination of O-glycans by β-elimination, the phosphomannan levels in the Carot2Δ and Cacwh41Δ null mutants decreased from 17.7 and 18.5% to 3.2 and 3.9%, respectively (Table 3), indicating that the N-glycan phosphomannan residues were almost completely absent in these β-eliminated null mutants strains. These results indicate that CaCWH41 and CaROT2 have an important role in N-glycan outer-chain elaboration.
TABLE 3.
Alcian Blue binding of Cacwh41Δ, Carot2Δ, and Camns1Δ null mutants and reintegrant strains
| Strain genotype | Mean Alcian Blue binding ± SDa
|
|
|---|---|---|
| No treatment | After β-elimination | |
| WT | 111 ± 5 | 95 ± 6 |
| Cacwh41Δ | 20 ± 3 | 3 ± 2 |
| Cacwh41Δ + CaCWH41 | 109 ± 5 | 92 ± 5 |
| Carot2Δ | 21 ± 5 | 4 ± 3 |
| Carot2Δ + CaROT2 | 108 ± 8 | 91 ± 4 |
| Camns1Δ | 46 ± 5 | 29 ± 4 |
| Camns1Δ + CaMNS1 | 109 ± 7 | 92 ± 5 |
Expressed as μg bound/OD600 = 1 cells (n = 3).
Cell wall composition, sensitivity, and cell integrity pathway activation.
To determine the effect of the disruption of CaCWH41, CaROT2, and CaMNS1 on the overall cell wall composition, the content of total carbohydrates, and proteins was analyzed. Cacwh41Δ and Carot2Δ null mutants showed decreases of 30.4 and 29.7% and of 65.0 and 62.9% in the contents of glucan and mannan, respectively (Table 4). Also, an increase in the chitin and protein levels of 2.2- and 3.3-fold for the Cacwh41Δ null mutant and of 2.2- and 3.2-fold for the Carot2Δ null mutant was observed. Camns1Δ null mutant had an increase of 10.2, 16.9, and 53.0% in the content of glucan, chitin, and proteins, respectively (Table 4). These changes were reflected in an overall decrease of 51.4% in the mannan levels. The reintegrant controls had cell wall compositions similar to that of wild-type yeast cells.
TABLE 4.
Cell wall composition of Cacwh41Δ, Carot2Δ, and Camns1Δ null mutants and reintegrant strains
| Strain genotype | Mean amt (μg) of polymer ± SDa
|
|||
|---|---|---|---|---|
| Glucan | Mannan | Chitin | Protein | |
| WT | 546 ± 11 | 276 ± 15 | 18 ± 2 | 140 ± 7 |
| Cacwh41Δ | 380 ± 14 | 97 ± 11 | 41 ± 2 | 459 ± 6 |
| Cacwh41Δ + CaCWH41 | 591 ± 17 | 246 ± 11 | 18 ± 1 | 124 ± 8 |
| Carot2Δ | 384 ± 10 | 102 ± 13 | 40 ± 2 | 444 ± 8 |
| Carot2Δ + CaROT2 | 561 ± 15 | 277 ± 12 | 17 ± 1 | 120 ± 7 |
| Camns1Δ | 602 ± 15 | 134 ± 13 | 21 ± 2 | 215 ± 6 |
| Camns1Δ + CaMNS1 | 581 ± 14 | 258 ± 15 | 18 ± 4 | 128 ± 10 |
That is, per mg of cell wall dry weight (n = 3).
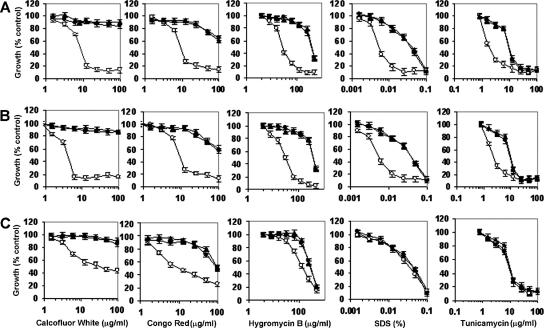
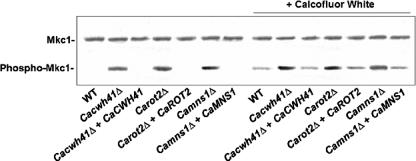
To investigate the effect of the ER α-glycosidase loss on the integrity of the cell wall, we tested the null mutants for their sensitivity to a range of cell-wall-perturbing agents and other compounds associated with glycosylation defects. The Camns1Δ null mutant was hypersensitive to Congo red, Calcofluor White, and hygromycin B, and Carot2Δ and Cacwh41Δ null mutants were hypersensitive to Calcofluor White, Congo red, hygromycin B, tunicamycin, and SDS (Fig. 3). Hypersensitivity to these agents is shared by other N-glycosylation mutants of S. cerevisiae and C. albicans (2-4, 14). There were no changes in the sensitivity to other stress-inducing agents such as caffeine, NaCl, or KCl (data not shown). The walls of the null mutants were therefore sensitive to cell wall stress but not to osmotic stress. We tested whether the PKC-Mkc1 cell integrity pathway was activated in the null mutants by Western analysis with an antibody that recognizes the phosphorylated form of the Mkc1 mitogen-activated protein kinase (17). Mkc1 was activated in the Cacwh41Δ, Carot2Δ, and Camns1Δ null mutants but not in the wild type and reintegrant controls (Fig. 4). As a positive control, the strains were stressed with 100 μg of Calcofluor White/ml, which is known to activate the pathway. These results reinforce the conclusion that N-mannan processing glycosidases are required for the assembly of a normal robust cell wall.
FIG. 3.
Sensitivity of Cacwh41Δ, Carot2Δ, and Camns1Δ null mutants to cell-wall-perturbing agents. Wild type (▴), null mutants (○), and reintegrant controls (•) strains were tested for sensitivity to cell-wall-perturbing agents by using the microdilution method. The strains tested were the Cacwh41Δ (HMY19) (A), Carot2Δ (HMY12) (B), and Camns1Δ (HMY5) (C) null mutants. Error bars indicate the means ± the standard deviation (n = 3). The results are pooled data from duplicate experiments.
FIG. 4.
Activation of the cell integrity pathway in glycosidase null mutants assessed by Western analysis. Protein extracts were prepared from cells in mid-exponential phase. As a positive control for activation of the cell integrity pathway, the strains were treated with Calcofluor White (100 μg/ml) as indicated. Extracts are from the following strains: NGY152 (wild type), HMY19 (Cacwh41Δ), HMY20 (Cacwh41Δ + CaCWH41), HMY12 (Carot2Δ), HMY13 (Carot2Δ + CaROT2), HMY5 (Camns1Δ), and HMY6 (Camns1Δ + CaMNS1). Equal loading was confirmed by Ponceau S staining and determining the intensity of nonspecific bands.
N-mannan processing is required for a pathogenic host-fungus interaction.
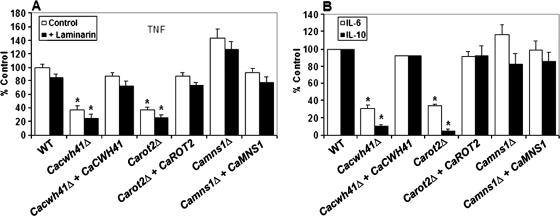
It has been demonstrated previously that the N-linked mannosyl residues of the C. albicans cell wall are involved in its recognition by monocyte and macrophages and in the induction of proinflammatory and anti-inflammatory cytokines by these mononuclear cells of the innate immune system (49). Therefore, cytokine production by human PBMC was investigated after stimulation by yeast cells of the Cacwh41Δ, Carot2Δ, and Camns1Δ mutants. In the Cacwh41Δ null mutant stimulation of TNF, IL-6, and IL-10 was reduced by 62, 70, and 90%, respectively (Fig. 5). Similar results were obtained with the Carot2Δ null mutant, with levels of TNF, IL-6, and IL-10 reduced by 63, 66, and 95%, respectively (Fig. 5). Normal cytokine release was recovered in the respective reintegrant controls. TNF, IL-6, and IL-10 levels stimulated by the Camns1Δ null mutant were not statistically different (P = 0.1001, P > 0.5, and P > 0.5, respectively) from those stimulated by wild-type cells (Fig. 5). In order to determine whether the changes in the cell wall of the null mutants led to exposure of elements present in the inner layers such as the β-glucans, human PBMC were treated with laminarin before the challenge with the yeast cells to block signaling via the β-glucan/dectin-1 receptor system (49). For the wild-type cells, there was a small but statistically insignificant decrease in the stimulation of TNF. A similar reduction was observed in the null mutants and reintegrants control tested (Fig. 5A). The results indicate that recognition of the Cacwh41Δ, Carot2Δ, and Camns1Δ null mutants by the dectin-1 receptor was not a significant factor in the recognition of these stains under these conditions.
FIG. 5.
Cytokine stimulation by Cacwh41Δ, Carot2Δ, and Camns1Δ null mutants. Human PBMC were stimulated 24 h with 106 yeast cells/ml and the TNF (A) and IL-6 and IL-10 (B) concentrations were determined. The experiments for panel A were carried out in the absence (□) or presence (▪) of laminarin, a blocking agent of the β-glucan/dectin-1 recognition pathway. The strains tested are NGY152 (wild type), HMY19 (Cacwh41Δ), HMY20 (Cacwh41Δ + CaCWH41), HMY12 (Carot2Δ), HMY13 (Carot2Δ + CaROT2), HMY5 (Camns1Δ), and HMY6 (Camns1Δ + CaMNS1). The results are pooled data from four volunteers. Error bars indicate the means ± the standard deviation. *, Significant differences in the mutant compared to the wild type (P < 0.05).
The effect of the N-mannan processing α-glycosidase loss encoded by CaCWH41, CaROT2, and CaMNS1 on virulence was assessed in a mouse model of systemic infection. Because mutants with reduced growth rates are usually attenuated in virulence, we simply confirmed the virulence loss in a systemic mouse model using only duplicate mice. The Cacwh41Δ, Carot2Δ, and Camns1Δ null mutants were highly attenuated in virulence with a mean survival time of 21 days, compared to 5.5 days for the wild-type control (data not shown). Virulence was restored in the reintegrant controls, which displayed mean survival times similar to the wild-type cells. Therefore, normal N-glycan processing is essential for pathogenesis and a normal host-fungus interaction and virulence.
DISCUSSION
In this study, we describe the importance of N-glycan processing for normal N glycosylation, structure, and integrity of the C. albicans cell wall and for interaction with the host. The Glc3Man9GlcNAc2 N-oligosaccharide is processed by ER α-glycosydases to generate the Man8GlcNAc2 core, which is a substrate for Golgi mannosyltransferases in charge of the highly branched outer-chain elaboration (27). Previous studies have demonstrated the importance of glycosylation for the cell wall structure, adherence, and virulence (3, 4, 7, 30, 46, 51, 54, 60, 69, 70, 80). For N-mannans, the N-glycan outer-chain elongation is necessary for the assembly of a normal cell wall structure and for virulence (4). We have extended these studies by analyzing the importance of N-glycan processing. Accordingly, we disrupted the C. albicans homologs of CWH41, ROT2, and MNS1 genes of S. cerevisiae, which encode the ER α-glucosidase I, the catalytic subunit of α-glucosidase II and α-mannosidase I, respectively. The Carot2Δ and Cacwh41Δ null mutants had defects in the N-glycan outer-chain elongation, evidenced by the loss of cell wall phosphomannan and increased mobility of HexNAcase, and had a weakened cell wall. Null mutants were strongly affected in virulence and in their ability to induce cytokine production by PBMC. These defects were similar but less dramatic than those displayed by Caoch1Δ null mutant, which lacks the entire N-glycan outer chain (4). The Camns1Δ null mutant had a milder phenotype than the Cacwh41Δ and Carot2Δ null mutants, in terms of specific growth rate, phosphomannan content, HexNAcase mobility, and cell wall integrity, but was still significantly affected in virulence and had an altered cytokine induction profile. Therefore, Ν-mannan processing is important for the host-fungus interaction of C. albicans, but alterations in core-mannan production at different steps resulted in different phenotypes.
We previously demonstrated, using the fluorogenic substrate MUαMan, that C. albicans α1,2-mannosidases belong to the glycosyl hydrolase family 47 and are present in both soluble and membrane-bound forms (45; H. M. Mora-Montes et al., unpublished data). The total absence of α-mannosidase activity in the Camns1Δ null mutant indicates that CaMNS1 is likely to encode both the soluble and the membrane-bound activities. The absence of soluble α-glucosidase activity in the Carot2Δ null mutant agrees with previous studies indicating that in C. albicans the α-glucosidase II activity is associated with a soluble 47-kDa polypeptide (71). Because the molecular mass of this soluble protein is lower than that predicted for CaRot2, it is possible that the α-glucosidase II activity is processed by a protease to generate a soluble catalytic domain.
The Cacwh41Δ, Carot2Δ, and Camns1Δ null mutants had 36, 32, and 19% reductions, respectively, in the growth rate of the yeast form. This contrasts with previous reports in S. cerevisiae, wherein no defects on the growth rates were observed in mutants lacking α-glucosidase I or II (21, 74). Yeast cells of the Cacwh41Δ, Carot2Δ, and Camns1Δ null mutants tended to clump as small aggregates. Aggregation may be the result of changes in the cell wall hydrophobicity due to decreased charged phosphomannan content. Alternatively, clumping may be the result of a cell separation defect due to alterations in the activity of glycosylated cell wall hydrolases that participate in cytokinesis. A similar clumping phenotype has been reported for other C. albicans glycosylation mutants such as the Camnt1Δ/Camnt2Δ (46), Capmr1Δ (3), Caoch1Δ (4), Camnn9Δ (66), and Cavrg4Δ (51) null mutants.
Evidence was found for defects in N glycosylation in the three null mutants generated. This was more severe in the Cacwh41Δ and Carot2Δ null mutants, as demonstrated by the underglycosylation of HexNAcase. HexNAcase is exclusively N glycosylated, is readily detected in nondenaturing PAGE gels, and has been used as a sensitive marker of N-glycosylation defects (4). Loss of CaCWH41 or CaROT2 resulted in defects in N glycosylation demonstrated by the increased mobility of HexNAcase and a reduction in N-glycan-linked phosphomannan content of the cell wall. The remaining phosphomannan present in the null mutants was attached to O-mannan and could be removed by β-elimination. A similar result was observed in the Caoch1Δ null mutant, where the N-glycan outer-chain elongation is blocked (4). Therefore, the presence of glucose residues on the N-glycan core may inhibit the ability of the CaOch1 α1,6-mannosyltransferase to initiate the outer-N-chain elongation. The absence of α-glucosidase I activity in S. cerevisiae did not prevent outer-chain formation or the addition of α1,3-mannose residues to the core oligosaccharides (75), suggesting that the importance of the N-glycan core glucose residues for subsequent outer-chain elongation may be different in C. albicans and S. cerevisiae. The N-glycosylation defect in Camns1Δ null mutant was not as severe as in Carot2Δ and Cacwh41Δ null mutants, indicating that partial elongation of the N-glycan core occurred. This indicates that the removal of mannose from the N-glycan core is not required for outer-chain synthesis, as found in S. cerevisiae (55).
The Cacwh41Δ, Carot2Δ, and Camns1Δ null mutants had an altered and weakened cell wall, as demonstrated by changes in cell wall composition and hypersensitivity to a range of cell-wall-perturbing agents and other agents whose action is indicative of glycosylation defects. The consequences of CaCwh41, CaRot2, or CaMns1 loss resulted in the constitutive activation of the PKC-Mkc1 cell integrity pathway. A similar result was observed in other C. albicans null mutants with defects in glycosylation (3, 4, 14). Similar observations have been made for S. cerevisiae, where the lack α-glucosidase I or II leads to alterations in the cell wall composition (35, 63, 65). The Camns1Δ null mutant was least affected by cell-wall-perturbing agents, correlating with a milder N-glycosylation mutant phenotype. Disruption of CaCWH41 and CaROT2 resulted in a 30% reduction in the cell wall β-glucan content, but loss of CaMNS1 did not affect the levels of this polymer. This suggests that proteins involved in the biosynthesis of β-glucans are sensitive to changes in the N-glycosylation pathway. However, C. albicans och1Δ mutants that lack N-glycan outer chain do not have reduced β-glucan levels (4). This suggests that the β-glucan reduction in the walls of Cacwh41Δ and Carot2Δ null mutants may be due to a failure in the glycoprotein quality control/protein refolding pathways that are dependent on glucosylation of the N-mannan core rather than the shortening of the N-glycan outer chain. Indeed, it has been demonstrated in S. cerevisiae that the presence of glucose residues on the N-mannan core led to an instability of Kre6, a protein required for β1,6-glucan biosynthesis, resulting in decreased levels of β1,6-glucan in the cell wall (1). There was also no change in sensitivity to high salt conditions in the three null mutants, indicating that the mutants were not osmotically fragile and that the SDS sensitivity of the Cacwh41Δ and Carot2Δ null mutants is likely to be the result of the cell wall perturbation rather than effects on the plasma membrane.
The Cacwh41Δ, Carot2Δ, and Camns1Δ mutants were significantly attenuated in virulence in a mouse model of systemic infection, as has been demonstrated for other C. albicans N-glycosylation null mutants (4, 66).
The balance of pro- and anti-inflammatory response is known to be important for the outcome of a number of fungal infections (13, 18, 40, 57). Decreased levels of TNF, IL-6, and IL-10 were observed with human PBMC challenged with Cacwh41Δ and Carot2Δ null mutants. This finding is in agreement with the observation that approximately 70% less cytokine production was stimulated in PBMC by the Caoch1Δ null mutant (49). The disruption of CaMNS1 did not affect the release of IL-6 and IL-10 but did result in increased release of TNF. This suggests that IL-6 and IL-10 may be stimulated by a different cell wall epitope than that which activates PBMC to release TNF. These effects on cytokine production are not likely to be mediated via the dectin-1 receptor since laminarin-treated PBMC gave similar responses to untreated PBMC in all cases. Therefore, N-glycan core processing is vital to the cell wall architecture of C. albicans, to its pathogenesis, and for host-fungus interactions.
Acknowledgments
This study was supported by grant CONACyT-2002-CO1-39528/A-1 from the Consejo Nacional de Ciencia y Tecnología, México, and the Dirección de Investigación y Posgrado, Universidad de Guanajuato, and by Wellcome Trust Programme grants 063204 and 080088 to N.A.R.G., A.J.P.B., and F.C.O.
Footnotes
Published ahead of print on 12 October 2007.
REFERENCES
- 1.Abeijon, C., and L. Y. Chen. 1998. The role of glucosidase I (Cwh41p) in the biosynthesis of cell wall β-1,6-glucan is indirect. Mol. Biol. Cell 9:2729-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballou, L., R. A. Hitzeman, M. S. Lewis, and C. E. Ballou. 1991. Vanadate-resistant yeast mutants are defective in protein glycosylation. Proc. Natl. Acad. Sci. USA 88:3209-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bates, S., D. M. MacCallum, G. Bertram, C. A. Munro, H. B. Hughes, E. T. Buurman, A. J. P. Brown, F. C. Odds, and N. A. R. Gow. 2005. Candida albicans Pmr1p, a secretory pathway P-type Ca2+/Mn2+-ATPase, is required for glycosylation and virulence. J. Biol. Chem. 280:23408-23415. [DOI] [PubMed] [Google Scholar]
- 4.Bates, S., H. B. Hughes, C. A. Munro, W. P. H. Thomas, D. M. MacCallum, G. Bertram, A. Atrih, M. A. J. Ferguson, A. J. P. Brown, F. C. Odds, and N. A. R. Gow. 2006. Outer chain N-glycans are required for cell wall integrity and virulence of Candida albicans. J. Biol. Chem. 281:90-98. [DOI] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Brand, A., D. M. MacCallum, A. J. P. Brown, N. A. R. Gow, and F. C. Odds. 2004. Ectopic expression of URA3 can influence the virulence phenotypes and proteome of Candida albicans but can be overcome by targeted reintegration of URA3 at the RPS10 locus. Eukaryot. Cell 3:900-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buurman, E. T., C. Westwater, B. Hube, A. J. P. Brown, F. C. Odds, and N. A. R. Gow. 1998. Molecular analysis of CaMnt1p, a mannosyl transferase important for adhesion and virulence of Candida albicans. Proc. Natl. Acad. Sci. USA 95:7670-7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byrd, J. C., A. L. Tarentino, F. Maley, P. H. Atkinson, and R. B. Trimble. 1982. Glycoprotein synthesis in yeast. Identification of Man8GlcNAc2 as an essential intermediate in oligosaccharide processing. J. Biol. Chem. 257:14657-14666. [PubMed] [Google Scholar]
- 9.Calderone, R. A. 1993. Recognition between Candida albicans and host cells. Trends Microbiol. 1:55-58. [DOI] [PubMed] [Google Scholar]
- 10.Calderone, R. A. 2002. Candida and candidiasis. ASM Press, Washington, DC.
- 11.Calderone, R. A., and N. A. R. Gow. 2002. Host recognition by Candida species, p. 67-86. In R. A. Calderone (ed.), Candida and candidiasis. ASM Press, Washington, DC.
- 12.Cannon, R. D., K. Niimi, H. F. Jenkinson, and M. G. Shepherd. 1994. Molecular cloning and expression of the Candida albicans β-N-acetylglucosaminidase (HEX1) gene. J. Bacteriol. 176:2640-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casadevall, A., and L. A. Pirofski. 2003. The damage-response framework of microbial pathogenesis. Nat. Rev. Microbiol. 1:17-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean, N. 1995. Yeast glycosylation mutants are sensitive to aminoglycosides. Proc. Natl. Acad. Sci. USA 92:1287-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dean, N. 1999. Asparagine-linked glycosylation in the yeast Golgi. Biochim. Biophys. Acta 1426:309-322. [DOI] [PubMed] [Google Scholar]
- 16.Delbruck, S., and J. F. Ernst. 1993. Morphogenesis-independent regulation of actin transcript levels in the pathogenic yeast Candida albicans. Mol. Microbiol. 10:859-866. [DOI] [PubMed] [Google Scholar]
- 17.de Nobel, H., C. Ruiz, H. Martin, W. Morris, S. Brul, M. Molina, and F. M. Klis. 2000. Cell wall perturbation in yeast results in dual phosphorylation of the Slt2/Mpk1 MAP kinase and in an Slt2-mediated increase in FKS2-lacZ expression, glucanase resistance, and thermotolerance. Microbiology 146:2121-2132. [DOI] [PubMed] [Google Scholar]
- 18.Djeu, J. Y. 1990. Role of tumor necrosis factor and colony-stimulating factors in phagocyte function against Candida albicans. Diagn. Microbiol. Infect. Dis. 13:383-386. [DOI] [PubMed] [Google Scholar]
- 19.Drenth, J. P., S. H. Van Uum, M. Van Deuren, G. J. Pesman, J. Van der Ven-Jongekrijg, and J. W. Van der Meer. 1995. Endurance run increases circulating IL-6 and IL-1ra but downregulates ex vivo TNF-α and IL-1β production. J. Appl. Physiol. 79:1497-1503. [DOI] [PubMed] [Google Scholar]
- 20.Endres, S., R. Ghorbani, G. Lonnemann, J. W. M. Van der Meer, and C. A. Dinarello. 1988. Measurement of immunoreactive interleukin-1β from human mononuclear cells: optimization of recovery, intrasubject consistency, and comparison with interleukin-1α and tumor necrosis factor. Clin. Immunol. Immunopathol. 49:424-438. [DOI] [PubMed] [Google Scholar]
- 21.Esmon, B., P. C. Esmon, and R. Schekman. 1984. Early steps in processing of yeast glycoproteins. J. Biol. Chem. 259:10322-10327. [PubMed] [Google Scholar]
- 22.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grinna, L. S., and P. W. Robbins. 1980. Substrate specificities of rat liver microsomal glucosidases which process glycoproteins. J. Biol. Chem. 255:2255-2258. [PubMed] [Google Scholar]
- 24.Helenius, A., and M. Aebi. 2004. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73:1019-1049. [DOI] [PubMed] [Google Scholar]
- 25.Hermans, M. M., M. A. Kroos, J. Van Beeumen, B. A. Oostra, and A. J. Reuser. 1991. Human lysosomal alpha-glucosidase: characterization of the catalytic site. J. Biol. Chem. 266:13507-13512. [PubMed] [Google Scholar]
- 26.Herrero, A. B., D. Uccelletti, C. B. Hirschberg, A. Dominguez, and C. Abeijon. 2002. The Golgi GDPase of the fungal pathogen Candida albicans affects morphogenesis, glycosylation, and cell wall properties. Eukaryot. Cell 1:420-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herscovics, A. 1999. Processing glycosidases of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1426:275-285. [DOI] [PubMed] [Google Scholar]
- 28.Herscovics, A., and P. Orlean. 1993. Glycoprotein biosynthesis in yeast. FASEB J. 7:540-550. [DOI] [PubMed] [Google Scholar]
- 29.Hitt, R., and D. H. Wolf. 2004. DER7, encoding alpha-glucosidase I is essential for degradation of malfolded glycoproteins of the endoplasmic reticulum. FEMS Yeast Res. 4:815-820. [DOI] [PubMed] [Google Scholar]
- 30.Hobson, R. P., C. A. Munro, S. Bates, D. M. MacCallum, J. E. Cutler, S. E. Heinsbroek, G. D. Brown, F. C. Odds, and N. A. R. Gow. 2004. Loss of cell wall mannosylphosphate in Candida albicans does not influence macrophage recognition. J. Biol. Chem. 279:39628-39635. [DOI] [PubMed] [Google Scholar]
- 31.Hong, Y., S. Sundaram, D. J. Shin, and P. Stanley. 2004. The Lec23 Chinese hamster ovary mutant is a sensitive host for detecting mutations in alpha-glucosidase I that give rise to congenital disorder of glycosylation IIb (CDG IIb). J. Biol. Chem. 279:49894-49901. [DOI] [PubMed] [Google Scholar]
- 32.Hülseweh, B., U. M. Dahlems, J. Dohmen, A. W. Strasser, and C. P. Hollenberg. 1997. Characterization of the active site of Schwanniomyces occidentalis glucoamylase by in vitro mutagenesis. Eur. J. Biochem. 244:128-133. [DOI] [PubMed] [Google Scholar]
- 33.Iwanami, S., H. Matsui, A. Kimura, H. Ito, H. Mori, M. Honma, and S. Chiba. 1995. Chemical modification and amino acid sequence of active site in sugar beet alpha-glucosidase. Biosci. Biotechnol. Biochem. 59:459-463. [DOI] [PubMed] [Google Scholar]
- 34.Jelinek-Kelly, S., and A. Herscovics. 1988. Glycoprotein biosynthesis in Saccharomyces cerevisiae: purification of the α-mannosidase which removes one specific mannose residue from Man9GlcNAc. J. Biol. Chem. 263:14757-14763. [PubMed] [Google Scholar]
- 35.Jiang, B., J. Sheraton, A. F. J. Ram, G. J. P. Dijkgraaf, F. M. Klis, and H. Bussey. 1996. CWH41 encodes a novel endoplasmic reticulum membrane N-glycoprotein involved in β-1,6-glucan assembly. J. Bacteriol. 178:1162-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jones, T., N. A. Federspiel, H. Chibana, J. Dungan, S. Kalman, B. B. Magee, G. Newport, Y. R. Thorstenson, N. Agabian, P. T. Magee, R. W. Davis, and S. Scherer. 2004. The diploid genome sequence of Candida albicans. Proc. Natl. Acad. Sci. USA 101:7329-7334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kapteyn, J. C., L. L. Hoyer, J. E. Hecht, W. H. Muller, A. Andel, A. J. Verkleij, M. Makarow, H. Van Den Ende, and F. M. Klis. 2000. The cell wall architecture of Candida albicans wild-type cells and cell wall-defective mutants. Mol. Microbiol. 35:601-611. [DOI] [PubMed] [Google Scholar]
- 38.Kimura, A., M. Takata, O. Sakai, H. Matsui, N. Takai, T. Taksyanagi, I. Nishimura, T. Uozumi, and S. Chiba. 1992. Complete amino acid sequence of crystalline alpha-glucosidase from Aspergillus niger. Biosci. Biotechnol. Biochem. 56:1368-1370. [DOI] [PubMed] [Google Scholar]
- 39.Klis, F. M., P. de Groot, and K. Hellingwerf. 2001. Molecular organization of the cell wall of Candida albicans. Med. Mycol. 39:1-8. [PubMed] [Google Scholar]
- 40.Kullberg, B. J., J. W. van't Wout, C. Hoogstraten, and R. van Furth. 1993. Recombinant interferon-gamma enhances resistance to acute disseminated Candida albicans infection in mice. J. Infect. Dis. 168:436-443. [DOI] [PubMed] [Google Scholar]
- 41.Lee, K. L., H. R. Buckley, and C. C. Campbell. 1975. An amino acid liquid synthetic medium for the development of mycelial and yeast forms of Candida albicans. Sabouraudia 13:148-153. [DOI] [PubMed] [Google Scholar]
- 42.Lipari, F., and A. Herscovics. 1996. Role of the cysteine residues in the alpha1,2-mannosidase involved in N-glycan biosynthesis in Saccharomyces cerevisiae: the conserved Cys340 and Cys385 residues form an essential disulfide bond. J. Biol. Chem. 271:27615-27622. [DOI] [PubMed] [Google Scholar]
- 43.Liu, H., J. Kohler, and G. R. Fink. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723-1726. [DOI] [PubMed] [Google Scholar]
- 44.Molloy, C., R. D. Cannon, P. A. Sullivan, and M. G. Shepherd. 1994. Purification and characterization of two forms of N-acetylglucosaminidase from Candida albicans showing widely different outer chain glycosylation. Microbiology 140:1543-1553. [DOI] [PubMed] [Google Scholar]
- 45.Mora-Montes, H. M., E. López-Romero, S. Zinker, P. Ponce-Noyola, and A. Flores-Carreón. 2004. Hydrolysis of Man9GlcNAc2 and Man8GlcNAc2 oligosaccharides by a purified alpha-mannosidase from Candida albicans. Glycobiology 14:593-598. [DOI] [PubMed] [Google Scholar]
- 46.Munro, C. A., S. Bates, E. T. Buurman, H. B. Hughes, D. M. MacCallum, G. Bertram, A. Atrih, M. A. Ferguson, J. M. Bain, A. Brand, S. Hamilton, C. Westwater, L. M. Thomson, A. J. P. Brown, F. C. Odds, and N. A. R. Gow. 2005. Mnt1p and Mnt2p of Candida albicans are partially redundant α-1,2-mannosyltransferases that participate in O-linked mannosylation and are required for adhesion and virulence. J. Biol. Chem. 280:1051-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Munro, S. 2001. What can yeast tell us about N-linked glycosylation in the Golgi apparatus? FEBS Lett. 498:223-227. [DOI] [PubMed] [Google Scholar]
- 48.Murad, A. M., P. R. Lee, I. D. Broadbent, C. J. Barelle, and A. J. P. Brown. 2000. CIp10, an efficient and convenient integrating vector for Candida albicans. Yeast 16:325-327. [DOI] [PubMed] [Google Scholar]
- 49.Netea, M. G., N. A. R. Gow, C. A. Munro, S. Bates, C. Collins, G. Ferwerda, R. P. Hobson, G. Bertram, H. B. Hughes, T. Jansen, L. Jacobs, E. T. Buurman, K. Gijzen, D. L. Williams, R. Torensma, A. McKinnon, D. M. MacCallum, F. C. Odds, J. W. Van der Meer, A. J. P. Brown, and B. J. Kullberg. 2006. Immune sensing of Candida albicans requires cooperative recognition of mannans and glucans by lectin and Toll-like receptors. J. Clin. Investig. 116:1642-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Niimi, K., M. Niimi, M. G. Shepherd, and R. D. Cannon. 1997. Regulation of N-acetylglucosaminidase production in Candida albicans. Arch. Microbiol. 168:464-472. [DOI] [PubMed] [Google Scholar]
- 51.Nishikawa, A., J. B. Poster, Y. Jigami, and N. Dean. 2002. Molecular and phenotypic analysis of CaVRG4, encoding an essential Golgi apparatus GDP-mannose transporter. J. Bacteriol. 184:29-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Odds, F. C. 1988. Candida and candidosis, 2nd ed. Bailliere Tindall, London, United Kingdom.
- 53.Pappas, P. G., J. H. Rex, J. Lee, R. J. Hamill, R. A. Larsen, W. Powderly, C. A. Kauffman, N. Hyslop, J. E. Mangino, S. Chapman, H. W. Horowitz, J. E. Edwards, and W. E. Dismukes. 2003. A prospective observational study of candidemia: epidemiology, therapy, and influences on mortality in hospitalized adult and pediatric patients. Clin. Infect. Dis. 37:634-643. [DOI] [PubMed] [Google Scholar]
- 54.Prill, S. K., B. Klinkert, C. Timpel, C. A. Gale, K. Schroppel, and J. F. Ernst. 2005. PMT family of Candida albicans: five protein mannosyltransferase isoforms affect growth, morphogenesis and antifungal resistance. Mol. Microbiol. 55:546-560. [DOI] [PubMed] [Google Scholar]
- 55.Puccia, R., B. Grondin, and A. Herscovics. 1993. Disruption of the processing alpha-mannosidase gene does not prevent outer chain synthesis in Saccharomyces cerevisiae. Biochem. J. 290:21-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Romani, L. 2002. Immunology of invasive candidiasis, p. 223-241. In R. A. Calderone (ed.), Candida and candidiasis. ASM Press, Washington, DC.
- 57.Romani, L. 2004. Immunity to fungal infections. Nat. Rev. Immunol. 4:1-23. [DOI] [PubMed] [Google Scholar]
- 58.Romaniouk, A., and I. K. Vijay. 1997. Structure-function relationships in glucosidase I: amino acids involved in binding the substrate to the enzyme. Glycobiology 7:399-404. [DOI] [PubMed] [Google Scholar]
- 59.Romero, P. A., F. Vallée, P. L. Howell, and A. Herscovics. 2000. Mutation of Arg273 to Leu alters the specificity of the yeast N-glycan processing class I α1,2-mannosidase. J. Biol. Chem. 275:11071-11074. [DOI] [PubMed] [Google Scholar]
- 60.Rouabhia, M., M. Schaller, C. Corbucci, A. Vecchiarelli, S. K. Prill, L. Giasson, and J. F. Ernst. 2005. Virulence of the fungal pathogen Candida albicans requires the five isoforms of protein mannosyltransferases. Infect. Immun. 73:4571-4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sandven, P. 2000. Epidemiology of candidemia. Rev. Iberoam. Micol. 17:73-81. [PubMed] [Google Scholar]
- 62.Saunier, B., R. D. Kilker, Jr., J. S. Tkacz, A. Quaroni, and A. Herscovics. 1982. Inhibition of N-linked complex oligosaccharide formation by 1-deoxynojirimycin, an inhibitor of processing glucosidases. J. Biol. Chem. 257:14155-14161. [PubMed] [Google Scholar]
- 63.Shahinian, S., G. J. P. Dijkgraaf, A. M. Sdicu, D. Y. Thomas, C. A. Jakob, M. Aebi, and H. Bussey, H. 1998. Involvement of protein N-glycosyl chain glucosylation and processing in the biosynthesis of cell wall beta-1,6-glucan of Saccharomyces cerevisiae. Genetics 149:843-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shibata, N., M. Arai, E. Haga, T. Kikuchi, M. Najima, T. Satoh, H. Kobayashi, and S. Suzuki. 1992. Structural identification of an epitope of antigenic factor 5 in mannans of Candida albicans NIH B-792 (serotype B) and J-1012 (serotype A) as β-1,2-linked oligomannosyl residues. Infect. Immun. 60:4100-4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Simons, J. F., M. Ebersold, and A. Helenius. 1998. Cell wall 1,6-β-glucan synthesis in Saccharomyces cerevisiae depends on ER glucosidases I and II, and the molecular chaperone BiP/Kar2p. EMBO J. 17:396-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Southard, S. B., C. A. Specht, C. Mishra, J. Chen-Weiner, and P. W. Robbins. 1999. Molecular analysis of the Candida albicans homolog of Saccharomyces cerevisiae MNN9, required for glycosylation of cell wall mannoproteins. J. Bacteriol. 181:7439-7448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sundstrom, P. 2002. Adhesion in Candida spp. Cell. Microbiol. 4:461-469. [DOI] [PubMed] [Google Scholar]
- 68.Teasdale, R. D., and M. R. Jackson. 1996. Signal-mediated sorting of membrane proteins between the endoplasmic reticulum and the Golgi apparatus. Annu. Rev. Cell Dev. Biol. 12:27-54. [DOI] [PubMed] [Google Scholar]
- 69.Timpel, C., S. Strahl-Bolsinger, K. Ziegelbauer, and J. F. Ernst. 1998. Multiple functions of Pmt1p-mediated protein O mannosylation in the fungal pathogen Candida albicans. J. Biol. Chem. 273:20837-20846. [DOI] [PubMed] [Google Scholar]
- 70.Timpel, C., S. Zink, S. Strahl-Bolsinger, K. Schroppel, and J. Ernst. 2000. Morphogenesis, adhesive properties, and antifungal resistance depend on the Pmt6 protein mannosyltransferase in the fungal pathogen Candida albicans. J. Bacteriol. 182:3063-3071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Torre-Bouscoulet, M. E., E. López-Romero, R. Balcázar-Orozco, C. Calvo-Méndez, and A. Flores-Carreón. 2004. Partial purification and biochemical characterization of a soluble alpha-glucosidase II-like activity from Candida albicans. FEMS Microbiol. Lett. 236:123-128. [DOI] [PubMed] [Google Scholar]
- 72.Treml, K., D. Meimaroglou, A. Hentges, and E. Bause. 2000. The alpha- and beta-subunits are required for expression of catalytic activity in the hetero-dimeric glucosidase II complex from human liver. Glycobiology 10:493-502. [DOI] [PubMed] [Google Scholar]
- 73.Trinel, P. A., G. Lepage, T. Jouault, G. Strecker, and D. Poulain. 1997. Definitive chemical evidence for the constitutive ability of Candida albicans serotype A strains to synthesize β-1,2 linked oligomannosides containing up to 14 mannose residues. FEBS Lett. 416:203-206. [DOI] [PubMed] [Google Scholar]
- 74.Trombetta, E. S., J. F. Simons, and A. Helenius. 1996. Endoplasmic reticulum glucosidase II is composed of a catalytic subunit, conserved from yeast to mammals, and a tightly bound noncatalytic HDEL-containing subunit. J. Biol. Chem. 271:27509-27516. [DOI] [PubMed] [Google Scholar]
- 75.Tsai, P. K., L. Ballou, B. Esmon, R. Schekman, and C. E. Ballou. 1984. Isolation of glucose-containing high-mannose glycoprotein core oligosaccharides. Proc. Natl. Acad. Sci. USA 81:6340-6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ugalde, R. A., R. J. Staneloni, and L. F. Leloir. 1979. Microsomal glucosidases acting on the saccharide moiety of the glucose-containing dolichyl diphosphate oligosaccharide. Biochem. Biophys. Res. Commun. 91:1174-1181. [DOI] [PubMed] [Google Scholar]
- 77.Völker, C., C. M. De Praeter, B. Hardt, W. Breuer, B. Kalz-Füller, R. N. Van Coster, and E. Bause. 2002. Processing of N-linked carbohydrate chains in a patient with glucosidase I deficiency (CDG type IIb). Glycobiology 12:473-483. [DOI] [PubMed] [Google Scholar]
- 78.Wang, Y., S. P. Li, S. A. Moser, K. L. Bost, and J. E. Domer. 1998. Cytokine involvement in immunomodulatory activity affected by Candida albicans mannan. Infect. Immun. 66:1384-1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Warit, S., R. M. Walmsley, and L. I. Stateva. 1998. Cloning and sequencing of the Candida albicans homologue of SRB1/PSA1/VIG9, the essential gene encoding GDP-mannose pyrophosphorylase in Saccharomyces cerevisiae. Microbiology 144:2417-2426. [DOI] [PubMed] [Google Scholar]
- 80.Warit, S., N. Zhang, A. Short, R. M. Walmsley, S. G. Oliver, and L. I. Stateva. 2000. Glycosylation deficiency phenotypes resulting from depletion of GDP-mannose pyrophosphorylase in two yeast species. Mol. Microbiol. 36:1156-1166. [DOI] [PubMed] [Google Scholar]
- 81.Wilson, R. B., D. Davis, B. M. Enloe, and A. P. Mitchell. 2000. A recyclable Candida albicans URA3 cassette for PCR product-directed gene disruptions. Yeast 16:65-70. [DOI] [PubMed] [Google Scholar]