Cholera, a severe disease caused by Vibrio cholerae bacteria, has had a central role in the history of infectious disease research. The cholera studies of John Snow and Robert Koch, among many others, largely gave birth to modern epidemiology and microbiology. Despite its long history as a research target, cholera continues to afflict approximately 5 million people each year and remains an important public health problem in many areas of the globe. Here we review the current knowledge of the complex regulatory network used by V. cholerae to control expression of its virulence determinants.
FEATURES OF VIBRIO CHOLERAE PATHOGENICITY
Cholera, which is characterized by voluminous watery diarrhea, is produced when the gram-negative curved bacillus V. cholerae colonizes the upper small intestine of its human host. V. cholerae is found throughout the world in coastal areas, most often associated with aquatic fauna such as copepods and shellfish, and is transmitted to humans by contaminated food or water. Since 1817, seven cholera pandemics have been recorded, with the most recent, ongoing pandemic having begun in 1961. Although more than 200 different serogroups have been isolated from the environment, the O1 serogroup of V. cholerae was responsible for all seven pandemics. However, beginning in 1992, O139 serogroup strains were found to cause outbreaks of cholera as well; O139 V. cholerae is sometimes referred to as the eighth cholera pandemic strain.
V. cholerae O1 exist in two biotypes: classical and El Tor. Classical V. cholerae was responsible for the first six cholera pandemics, whereas the seventh pandemic has been caused by El Tor V. cholerae. The two V. cholerae biotypes differ considerably. El Tor strains generally cause a milder form of cholera than that caused by classical strains and apparently evolved as better survivors in the aquatic environment; currently, El Tor strains are predominant everywhere in the world that V. cholerae O1 can be found. V. cholerae O139 likely arose by seroconversion of O1 El Tor strains (8, 72). In addition, there are subtle differences in the way that El Tor strains and classical strains regulate expression of key virulence factors, as will be discussed below.
The clinical aspects of cholera are primarily induced by the activity of cholera toxin (CT), a bipartite toxin that consists of a single active A subunit and five B subunits that bind the toxin to the GM1 ganglioside on the surface of the intestinal epithelium. Once inside epithelial cells, a proteolytically derived fragment of the CT-A subunit, CT-A1, ADP-ribosylates Gαs protein, resulting in constitutive cyclic AMP production. This leads to massive secretion of chloride and water into the lumen of the intestine. Cholera patients can lose up to 20 liters of fluid within a 24-h period, resulting in rapid dehydration, and >50% of cholera patients die without treatment. However, if patients are rehydrated orally and/or intravenously, mortality rates decrease to ∼1%.
Aside from CT, the other major V. cholerae virulence factor is the toxin-coregulated pilus (TCP), a type IV pilus that is required for intestinal colonization. The TCP causes aggregation of V. cholerae and induces microcolony formation within the intestine. However, the TCP is most likely not directly responsible for adhesion of V. cholerae to the intestinal epithelium. In addition to its role in colonization, the TCP acts as the receptor for the bacteriophage, CTXΦ, which harbors the ctxAB genes encoding the CT. The genes for TCP biogenesis are located in the Vibrio pathogenicity island in a large operon beginning with tcpA, which encodes the pilin subunit. The expression of other genes located within the Vibrio pathogenicity island is coordinately regulated with TCP expression by the same set of transcription regulators (see below). These other genes include the acfA-D genes, which encode accessory colonization factors. Insertional disruption of any of the acf genes results in a reduction in intestinal colonization by V. cholerae, as assessed in the infant mouse cholera model, but their function in colonization and/or pathogenesis is indeterminate.
Our purpose here is to review recent literature regarding the complex regulatory network that leads to expression of CT, TCP, and other virulence-associated genes. We explore V. cholerae virulence regulation by beginning with ToxT, the regulator most directly responsible for virulence factor expression, and then move on to describe the multiple regulatory inputs and pathways that contribute to whether the cell expresses ToxT or not.
TRANSCRIPTION ACTIVATION BY ToxT
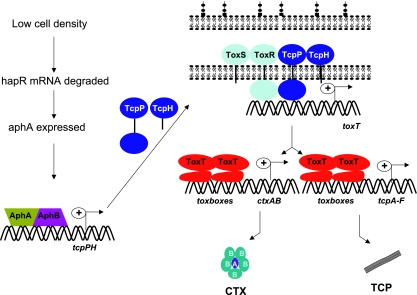
The primary direct transcriptional activator of the V. cholerae virulence genes is ToxT protein (Fig. 1). ToxT is a member of the large AraC/XylS protein family (40); this family shares an ∼100-amino-acid domain that conveys DNA binding, via two helix-turn-helix motifs, and transcription activation activities (31, 63). ToxT has the AraC family domain at its C terminus; the remaining N-terminal 176 amino acids presumably form a secondary domain within ToxT (NTD), but the function of this domain is unclear. BLAST searches with just the ToxT NTD sequence return no homology to any other protein, and significant variation is found within the NTD in environmental V. cholerae strains (24), whereas the AraC family domain is nearly invariant among all known ToxT sequences. AraC family proteins consisting of only the conserved family domain exist, as do other AraC family proteins with a secondary domain either N or C terminal to the family domain activities (31, 63). Commonly, these secondary domains are involved in effector binding and/or multimerization. The natural effector of ToxT is proposed to be bile, which causes a decrease in the expression of CT and TCP (34, 84). Mutational analysis indicates that bile likely interacts with the ToxT NTD (79). A synthetic compound, virstatin, identified as an inhibitor of ctx gene expression in a high-throughput screen also inhibits ToxT activity by interacting with the ToxT NTD in the same region where bile is hypothesized to bind, suggesting that there is an effector-binding patch in this portion of the NTD (43).
FIG. 1.
Pathway by which cell density regulates expression of the major V. cholerae virulence factors CT and TCP. See the text for details.
Results of in vitro experiments suggest that ToxT binds to DNA as individual monomers (93, 94). Nevertheless, bacterial two-hybrid experiments suggest that ToxT NTDs can interact with each other (18), although full-length ToxT did not behave as a dimer in one of these studies (85). If dimerization of the NTD does occur, perhaps it occurs subsequent to DNA binding. Further evidence of NTD dimerization is that a truncated toxT allele expressing residues 6 to 167 acts in a dominant-negative fashion (85). Virstatin inhibits dimerization of ToxT in a bacterial two-hybrid assay and causes monomer accumulation in gel filtration studies but does not strongly inhibit ToxT-dependent activation of genes other than ctx, suggesting that dimerization is not an absolute requirement for ToxT to activate gene expression (85). As for the mechanism of dimerization, alanine scanning mutagenesis identified only one NTD mutation, F151A, that decreased apparent dimerization by as much as 50% (18). This could reflect a complex interaction surface between the two monomers that is not easily disrupted by any single mutation.
ToxT activates transcription by binding to a degenerate 13-bp DNA sequence, the toxbox (94) (Fig. 1). Toxboxes are located upstream of all known ToxT-activated genes. However, the configurations of toxboxes differ at different promoters. For example, at tcpA, ToxT binds to two toxboxes organized as a direct repeat between positions −44 and −67 relative to the start of transcription. In contrast, between the divergent acfA and acfD genes, ToxT binds to two toxboxes organized as an inverted repeat (93). From this pair of toxboxes both acfA and acfD transcription is activated by ToxT. ToxT activates transcription of aldA, which encodes an aldehyde dehydrogenase of unknown function in pathogenesis, by binding to a single toxbox (95). Toxboxes are always located upstream of, and not overlapping, the core −35 promoter element, suggesting that ToxT-activated promoters are all of the class I variety, and thus ToxT directly interacts with the α subunit C-terminal domain (α-CTD) of RNA polymerase to activate transcription (11). Overexpression of α subunit deleted for the CTD causes a large decrease in activation of tcpA by ToxT in accordance with this model (41).
Activation of ctxAB and tcpA transcription by ToxT is counteracted by the histone-like protein H-NS, which binds to the same DNA region (33, 76, 101). At the ctxAB promoter, H-NS strongly represses transcription, and much of the function of ToxT at ctxAB is to antirepress by competing with H-NS for DNA binding. However, once bound to the DNA, ToxT also directly activates ctxAB expression, presumably by interacting with RNA polymerase (101). At the tcpA promoter, the effect of H-NS is much less dramatic, and ToxT is primarily a direct activator of transcription. H-NS further reduces virulence gene expression by binding to the toxT promoter region and repressing toxT transcription (76).
CONTROL OVER toxT EXPRESSION
TcpPH/ToxRS.
The complex pathway of V. cholerae regulating toxT expression and, consequently, downstream virulence genes including ctxAB and tcp is often referred to as the “ToxR regulon,” after the first identified positive regulator (77, 87) (Fig. 1). ToxR is a bitopic membrane protein containing a cytoplasmically localized DNA-binding/transcription activation domain, a transmembrane domain, and a periplasmic domain of unknown function. The amino-terminal DNA-binding domain is called a winged helix-turn-helix domain and is similar to that of the OmpR/PhoB family of transcriptional activators (64, 65). Wild-type ToxR activity requires the presence of another protein called ToxS. ToxS is also localized to the inner membrane but is thought to reside predominantly in the periplasm, where ToxR and ToxS are hypothesized to interact. The exact role of ToxS is unclear, but it appears to serve as an effector of ToxR function, perhaps by influencing stability and/or enhancing dimerization of ToxR (21, 23, 78).
To regulate expression of ToxT, ToxR acts in conjunction with a second transcription activator, TcpP, which, like ToxR, is membrane localized and has a cytoplasmic DNA-binding/activation and periplasmic domains. TcpP also requires the presence of a membrane-bound effector protein, TcpH, with which TcpP is thought to interact through its periplasmic domain (5, 15). Transcription of the operon encoding these two genes is responsive to environmental signals such as temperature and pH (6) and the production of specific autoinducers (discussed below).
Along with control over its gene expression, levels of TcpP are regulated by interaction with TcpH. In cells lacking TcpH TcpP is rapidly degraded (5). TcpP is also degraded in wild-type (TcpH+) cells under conditions unfavorable for virulence gene expression in vitro, such as growth at pH 8.5 and 37°C (66). TcpP degradation is a regulated proteolytic event that requires at least two proteases working in sequence and is thus similar to the process by which a transmembrane protein called RseA—and its homologues—are degraded in Escherichia coli and other bacteria (2, 66) In fact, two-site proteolytic liberation of transcription factors from the membrane—termed regulated intramembrane proteolysis—is broadly conserved, being found in both prokaryotes and eukaryotes (10, 62).
In E. coli, envelope damage (or indicators of it, such as the presence of unfolded outer membrane proteins) induces proteolysis of an anti-sigma factor called RseA. RseA works by sequestering an alternative sigma factor, sigma E, to the membrane, thereby inactivating it. Sigma E is required for the cell to respond to the consequences of a damaged envelope; thus, under envelope stress, RseA is degraded in two distinct and successive steps by DegS and YaeL (also called RseP) (2). Such proteases, working in sequence, are generally termed site-1 and site-2 proteases. After RseA is eliminated, sigma E is released from the membrane and activates genes whose products are needed for surviving the envelope stress (this pathway is reviewed in reference 1).
For TcpP degradation, the consequence of the regulated intramembrane proteolysis is to destroy a transcription factor rather than to release one from inhibition, but the degradation process is partially conserved nonetheless. In TcpP degradation, the V. cholerae YaeL homologue serves as the site-2 protease, as in the degradation of E. coli RseA. However, the site-1 protease for TcpP has yet to be identified; it is not DegS (66). That DegS is not the site-1 protease suggests that the initiating signal for TcpP degradation may be distinct from signals that trigger sigma-E-dependent gene expression. In addition to the identity of the site −1 protease, a key unanswered question is the mechanism by which TcpH is released, allowing access of the proteases to TcpP. Given that regulation of TcpPH occurs both transcriptionally and posttranslationally, it appears that the activity of these regulators is a critical checkpoint in the cell.
Binding sites for ToxR and TcpP on the toxT promoter are located close together, with TcpP binding from positions −54 to −32 and ToxR binding upstream, from positions −104 to −68 (53). ToxR and TcpP appear to bind with different affinities to the toxT promoter, with ToxR binding more avidly (53). Overexpression of TcpP obviates the requirement for ToxR in toxT activation, but the converse is not true (37, 39, 53, 74). This suggests that TcpP is more directly responsible for transcription activation (i.e., stimulation of RNA polymerase) and that ToxR plays an indirect role. A working hypothesis is that ToxR provides toxT promoter recognition, and interaction of ToxR with TcpP fixes the latter into position for stimulating RNA polymerase, thus activating transcription. ToxR may also directly activate the ctxAB promoter within the host; how that may occur is not clear, although bile acids can stimulate partial activation of ctxAB by ToxR in vitro, and this could perhaps also occur in vivo (42, 55, 71).
Independently of TcpP, ToxR reciprocally regulates the production of two outer membrane proteins, OmpU and OmpT (19, 58, 70). OmpU requires ToxR for expression and confers resistance to some antimicrobial peptides and may also function as an adhesin (88). OmpT is maximally expressed in cells lacking ToxR, and its role in virulence has not been determined. Appropriate expression of these porins by ToxR is important for V. cholerae bile resistance, for intestinal colonization in mice, and for resistance to organic acids (58, 80). ToxR activates expression of ompU by binding DNA within the ompU promoter without the need for additional cofactors other than ToxS and RNA polymerase (19). ompT repression requires ToxR binding to a region of the ompT promoter where CRP is predicted to bind, thereby interfering with CRP activation of ompT transcription (58, 59). Another feature of ToxR regulation at the ompU and ompT promoters is that the ToxR winged-helix domain does not have to be localized to the membrane to bind those promoters, unlike the case for activation at the toxT promoter (20).
AphAB.
The toxRS operon appears to be constitutively active, but tcpPH transcription is regulated by two activators, AphA and AphB, which are encoded by unlinked genes on the V. cholerae large chromosome (Fig. 1). AphA is in a family of regulatory proteins with homology to the PadR repressor, which controls expression of genes involved in the detoxification of phenolic acids (4). AphA binds the tcpPH promoter at a region of partial dyad symmetry between positions −101 and −71 relative to the transcriptional start site (51). AphA cannot activate transcription of tcpPH alone, requiring interaction with the LysR-type regulator AphB that binds downstream of AphA, between positions −78 and −43 relative to the transcriptional start site (48, 51). This interaction is thought to stabilize AphB binding to its recognition site and result in activation of the tcpPH promoter under appropriate environmental conditions. Thus, activation of tcpPH by AphA and AphB is conceptually similar in mechanism to activation of toxT by ToxR and TcpP. The observation that El Tor and classical biotype V. cholerae regulate toxT differently (22) was explained by the discovery of a single base pair difference between the classical and El Tor tcpPH promoters that alters the ability of AphB to bind (49, 50). As discussed in greater detail below, AphA serves as a link between quorum sensing and virulence gene expression in some strains of V. cholerae, since its expression is subject to control by a mechanism involving bacterial cell density.
AphA regulates the expression of several other genes not associated with the Vibrio pathogenicity island (47). A set of genes involved in the biosynthesis of acetoin under some conditions is strongly repressed by AphA. Acetoin is a metabolic end product synthesized by a variety of bacteria with a neutral pH, as opposed to the organic acids that can be produced when bacteria are grown in the presence of excess glucose or other carbohydrates. Because V. cholerae is very acid sensitive, accumulation of acidic fermentation products in the growth medium results in a loss of viability. This metabolic regulation pathway may contribute to an important difference between the classical and El Tor biotypes of V. cholerae. Classical strains grow poorly in media containing high levels of glucose and lose viability due to production of organic acids and the resulting acidification of the growth medium (100). However, strains of the El Tor biotype grow much better in the presence of excess carbohydrates due to production of the neutral end products acetoin and 2,3-butanediol. AphA may repress acetoin biosynthesis more actively in classical strains than in El Tor, leading to increased fitness of the El Tor biotype in the environment and in the host.
QUORUM SENSING, SMALL RNA MOLECULES, AND HapR
Multiple quorum-sensing systems in V. cholerae act in parallel to regulate virulence gene expression, biofilm formation, and protease production (69). The central regulator ultimately under the control of these multiple systems is HapR, which links quorum sensing and virulence regulation through its control over aphA expression (52) (Fig. 2).
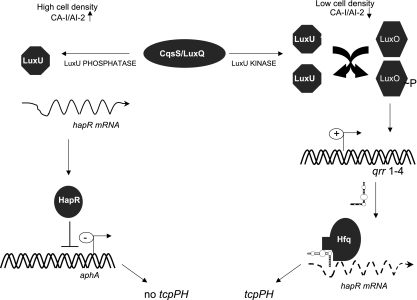
FIG. 2.
Mechanism of quorum sensing using small RNAs that controls HapR levels in V. cholerae. See the text for details.
Quorum sensing system 1 is composed of the CAI-1 autoinducer, the structure of which is unknown, and a two-component sensor-kinase CqsS. System 2 is composed of AI-2 (a furanosyl borate diester), the periplasmic binding protein LuxP, and the two-component sensor LuxQ (17) (Fig. 2). The sensory information from both of these systems is conveyed through a phosphorelay mechanism that leads to a transcription factor called LuxO. A third system involves the VarS/VarA sensor kinase/response regulator pair, which activates expression of three small RNAs (CsrB, CsrC, and CsrD) (56). These sRNAs serve to inhibit the activity of the global regulatory protein, CsrA, which activates LuxO function independently of LuxU (69). The action of CsrA on LuxO is not direct, and the molecular mechanism of this event is unknown.
At low cell density, corresponding to a low autoinducer concentration, LuxU is phosphorylated by the sensor kinases and transfers its phosphate to LuxO (26, 27). LuxO∼P activates the expression of four regulatory RNAs termed Qrr1 to -4 (for quorum-regulatory RNA) (57, 60). These sRNAs, together with the chaperone Hfq, bind the hapR mRNA and destabilize it. The result is that very little HapR protein is produced at low cell density. Alternatively, at high cell density the autoinducer concentrations increase, and the sensors CqsS and LuxQ act as phosphatases rather than kinases. This leads to dephosphorylation of LuxO, rendering it inactive and consequently the qrr genes are not expressed. The result is stable production of hapR mRNA and its translation into HapR protein (Fig. 2).
HapR, a LuxR homolog, directly activates or represses a variety of genes. Virulence activation is downregulated by HapR because it represses aphA transcription (52). Biofilm formation is downregulated by HapR because it represses the vps operon encoding the polysaccharide component of the V. cholerae biofilm (see below). HapR upregulates expression of hapA, encoding the secreted hemagglutinin/protease that is responsible for detachment of bacteria from the intestinal epithelium in a process, also controlled by the stationary-phase sigma factor RpoS, called the “mucosal escape response” (25, 45, 75). hapR expression itself is repressed at high cell densities by HapR protein; however, the significance of this autorepression is currently unknown (61).
While quorum sensing is important in a variety of Vibrio species, some V. cholerae strains (including the sequenced El Tor N16961 strain) are not able to respond to the sensory input due to a frameshift mutation in hapR (102). Therefore, a functional quorum-sensing system does not appear to be an absolute requirement for virulence and biofilm formation in V. cholerae.
BIOFILM FORMATION AND THE VIRULENCE CONNECTION
Because of the HapR-dependent link between virulence expression and biofilm formation, it is worth reviewing what is understood of the process of biofilm formation. V. cholerae can survive both in environmental reservoirs and in human hosts. The ability to switch between two different phase variants, termed smooth and rugose, is thought to play a role in this survival. Phenotypic traits associated with the rugose phenotype include increased ability to form biofilms. Rugose variants produce VPS (for Vibrio polysaccharide), an exopolysaccharide that enables them to form well-developed biofilms and resist a variety of environmental stresses (73, 82, 92). The vps genes are located on the large chromosome of V. cholerae in two clusters: vpsI (vpsA-K) and vpsII (vpsL-Q) (92, 99); mutation in any of these genes results in a smooth-colony phenotype and reduced ability to form biofilms.
Transcription of the vps genes is controlled by VpsR, a σ54-dependent two-component response regulator (98). The signals that lead to VpsR phosphorylation, and thus activation, are not known, and its cognate histidine kinase has not been identified. Transcription of the vps genes is also activated by another two-component response regulator, VpsT (16). VpsR and VpsT activate vpsA and vpsL expression and positively regulate their own and each other's transcription. Disruption of vpsR or vpsT in the rugose variant yields smooth colonies and prevents the formation of mature biofilms.
V. cholerae significantly enhances biofilm formation in response to bile acids (44). This is dependent on the vps genes and requires posttranslational activation of VpsR, and VpsT is not required. As noted above, HapR, the previously described quorum-sensing transcriptional regulator, represses the vps genes, as does CytR, a repressor of nucleoside uptake and catabolism (38).
VieSAB AND SECOND MESSENGER SIGNALING
Through HapR, quorum-sensing regulation controls both expression of virulence factors and formation of biofilms. The VieSAB signal transduction system also couples virulence gene expression and biofilm formation. The VieSAB proteins share similarities to sensor-kinase/response regulator two-component regulatory systems but deviate from the norm by having two putative response regulators, VieA and VieB. VieA appears to be a typical response regulator, with both a helix-turn-helix DNA-binding domain and a phosphoreceiver domain, while VieB contains only the phosphoreceiver domain and lacks a DNA-binding motif. VieS, the sensor kinase, was first identified in an in vivo promoter-trap experiment screening for positive regulators of ctxA expression (54). The screen identified a transposon insertion mutation in vieS that failed to induce expression of a ctxA:tnpR fusion during infection of infant mice. Although the vieSAB genes are located together on the large chromosome, it appears that the genes are differentially expressed. vieB expression only occurs in vivo during infection and requires TCP-mediated colonization (55). vieS is constitutively expressed during in vitro growth, whereas vieA expression is dependent on the presence of VieA during in vitro growth (54). A microarray study showed that VieA regulates 401 genes in the classical strain O395 but only 5 genes in the El Tor strain A1552. In the classical strain, toxT and ToxT-regulated genes were underexpressed in the vieA mutant, relative to the wild type. In addition, vieA mutant O395 expressed lower levels of genes for motility regulation, flagellum production, and the sigma-E regulon (7).
VieA works through a second messenger whose importance in bacterial cell signaling is becoming increasingly apparent. This molecule is 3′,5′-cyclic diguanylic acid (c-di-GMP), which controls a range of physiological and behavioral properties in both gram-negative and gram-positive bacteria (for a review, see reference 14). Biosynthesis and degradation of c-di-GMP is carried out through the activities of diguanylate cyclases and phosphodiesterases A, respectively. Two specific protein motifs have been associated with these activities: diguanylate cyclases typically have the conserved amino acid motif GGDEF, while phosphodiesterases A have the motif EAL (3). VieA contains an EAL domain, which acts as a c-di-GMP phosphodiesterase in vitro and in vivo (89). Through this activity, VieA maintains low intracellular c-di-GMP levels, negatively regulating biofilm formation and positively regulating virulence gene expression, including that of toxT (91).
CHITIN BINDING, REGULATION, AND COMPETENCE
V. cholerae is often found in the aquatic environment attached to the chitinous exoskeletons of zooplankton. This attachment likely enhances the survival of Vibrio species in the environment by providing a source of carbon and nitrogen, as well as a surface for the formation of biofilms. V. cholerae expresses a protein called GbpA, which binds to GlcNAc, a widespread compound that is both the constituent of chitin and a common modification of glycoproteins and lipids located on the intestinal epithelium (46). GbpA is required for efficient intestinal colonization by V. cholerae and therefore links an environmental survival strategy—binding to chitinous structures—and human disease. Also, TCP production contributes to the colonization of chitinaceous surfaces due to the fact that V. cholerae TCP mutants are not able to form differentiated biofilms on chitin and do not form microcolonies (81). This suggests that V. cholerae producing TCP may have a fitness advantage in the environment over those that do not.
Various V. cholerae strains can become competent for natural transformation when grown on chitin (67). HapR is required for transformation competence, and high cell density positively controls this phenotype by relieving LuxO-dependent repression of HapR synthesis. Chitin-induced serogroup conversion mediated by natural transformation has been demonstrated in an experimental setting designed to mimic aquatic reservoirs, suggesting the importance of this process in the natural history of V. cholerae (9).
MOTILITY, CHEMOTAXIS, VIRULENCE, AND HOST-HOST TRANSMISSION
V. cholerae have a single, polar flagellum and are highly motile. There is significant dispute in the literature over the importance of motility for V. cholerae colonization of the intestine. Conflicting results have been obtained using different animal models and with El Tor versus classical biotype V. cholerae (28, 30, 32, 83, 90, 96, 97). Evidence from experiments using defined nonmotile mutant V. cholerae strains suggests that motility is indeed required for pathogenesis (54, 86). Work from many labs using both the infant mouse and rabbit ligated ileal loop models has indicated than nonmotile V. cholerae are defective at intestinal colonization. Furthermore, motility and expression of virulence genes are inversely correlated (32, 34-36). The currently favored model for V. cholerae infection is that motile vibrios localize to the crypts of the small intestine, after which their motility is reduced and virulence genes are expressed.
The role of chemotaxis in V. cholerae colonization has also been disputed in the literature and remains a subject of debate. Nonchemotactic V. cholerae colonize the intestine well but do so aberrantly (12, 29, 54). In the infant mouse, chemotactic V. cholerae colonize the lower small intestine, whereas nonchemotactic V. cholerae colonize throughout the small intestine, which results in higher numbers of colonizing bacteria compared to the wild type.
V. cholerae appears to become more transmissible by growing within the host during natural infection, a phenotype associated with changes in chemotaxis gene expression. This hypothesis derives from experiments showing that V. cholerae in human cholera stool outcompeted laboratory-grown, stationary-phase V. cholerae by 10- to 100-fold in an infant mouse model (68). This phenotype was maintained when the stool V. cholerae was incubated in pond water for up to 5 h but not after growth in LB medium. Competitive advantage of the stool V. cholerae was associated with repression of the cheW and cheR loci, whose products are required for chemotaxis (12, 13). Based on these studies, it was been proposed that V. cholerae repress chemotaxis when leaving the host. In contrast, work with rabbit ileal loops studying the mucosal escape response mentioned above showed that bacteria detach from the epithelial surface in a process that requires the stationary-phase sigma factor RpoS (75). One function of RpoS is to activate motility and chemotaxis functions. Further work is required to determine the precise role of chemotaxis during different phases of V. cholerae infection and escape from the host.
CONCLUSIONS
As befitting a pathogen that also is very successful living outside of human hosts, virulence gene regulation in V. cholerae is complex and linked very closely to other regulatory pathways in the cell. While expression of ToxT is the final committed step in virulence gene expression, there is not a direct, linear pathway from any one particular signal leading to transcription of toxT. Unlike other pathogens in which a principal difference in lifestyle is simply whether they are in the host or outside of the host, modulation of virulence gene expression in V. cholerae is intimately tied to whether or not the microbe is growing at high cell density or low cell density, with HapR levels serving as a measure of the two states. Each state might conceivably be achieved within a single infection, since a study of temporal and spatial patterns of virulence gene expression in vivo demonstrated that only a small fraction of the infecting inoculum may ultimately reach a site required to stimulate virulence gene expression to high levels (55). However, V. cholerae is also capable of growing to high levels during the course of an infection, after which it exits very efficiently and in large numbers. That virulence expression and the proposed mucosal escape response is so closely tied to HapR, which is controlled by cell density, makes sense in this “colonization/growth/escape” view of the infection. Even though we are coming to some broad understanding of this regulatory network, numerous questions remain. Are there yet-uncharacterized physiological states controlled by any of the major regulators in this system? The answer to that is almost certainly yes, as transcription profiling experiments to study the global effects of many of these regulators make clear. Further, the connection between motility and virulence is conceptually very appealing, but a precise mechanism linking these two complex traits has yet to be definitively established. Finally, the genetic program that controls the transmission state of V. cholerae is just now becoming appreciated, and future work will surely uncover interesting new knowledge in this important stage of V. cholerae regulation.
Acknowledgments
Work on Vibrio cholerae in the authors' laboratories is supported by the NIAID. J.S.M. is a recipient of an individual Kirschstein National Research Service Award.
Editor: J. B. Kaper
Footnotes
Published ahead of print on 17 September 2007.
REFERENCES
- 1.Alba, B. M., and C. A. Gross. 2004. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol. Microbiol. 52:613-619. [DOI] [PubMed] [Google Scholar]
- 2.Alba, B. M., J. A. Leeds, C. Onufryk, C. Z. Lu, and C. A. Gross. 2002. DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)-dependent extracytoplasmic stress response. Genes Dev. 16:2156-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausmees, N., R. Mayer, H. Weinhouse, G. Volman, D. Amikam, M. Benziman, and M. Lindberg. 2001. Genetic data indicate that proteins containing the GGDEF domain possess diguanylate cyclase activity. FEMS Microbiol. Lett. 204:163-167. [DOI] [PubMed] [Google Scholar]
- 4.Barthelmebs, L., B. Lecomte, C. Divies, and J. F. Cavin. 2000. Inducible metabolism of phenolic acids in Pediococcus pentosaceus is encoded by an autoregulated operon which involves a new class of negative transcriptional regulator. J. Bacteriol. 182:6724-6731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck, N. A., E. S. Krukonis, and V. J. DiRita. 2004. TcpH influences virulence gene expression in Vibrio cholerae by inhibiting degradation of the transcription activator TcpP. J. Bacteriol. 186:8309-8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behari, J., L. Stagon, and S. B. Calderwood. 2001. pepA, a gene mediating pH regulation of virulence genes in Vibrio cholerae. J. Bacteriol. 183:178-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beyhan, S., A. D. Tischler, A. Camilli, and F. H. Yildiz. 2006. Differences in gene expression between the classical and El Tor biotypes of Vibrio cholerae O1. Infect. Immun. 74:3633-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bik, E. M., A. E. Bunschoten, R. D. Gouw, and F. R. Mooi. 1995. Genesis of the novel epidemic Vibrio cholerae O139 strain: evidence for horizontal transfer of genes involved in polysaccharide synthesis. EMBO J. 14:209-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blokesch, M., and G. K. Schoolnik. 2007. Serogroup conversion of Vibrio cholerae in aquatic reservoirs. PLoS Pathog. 3:e81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown, M. S., and J. L. Goldstein. 1999. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Natl. Acad. Sci. USA 96:11041-11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Busby, S., and R. H. Ebright. 1994. Promoter structure, promoter recognition, and transcription activation in prokaryotes. Cell 79:743-746. [DOI] [PubMed] [Google Scholar]
- 12.Butler, S. M., and A. Camilli. 2004. Both chemotaxis and net motility greatly influence the infectivity of Vibrio cholerae. Proc. Natl. Acad. Sci. USA 101:5018-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butler, S. M., E. J. Nelson, N. Chowdhury, S. M. Faruque, S. B. Calderwood, and A. Camilli. 2006. Cholera stool bacteria repress chemotaxis to increase infectivity. Mol. Microbiol. 60:417-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camilli, A., and B. L. Bassler. 2006. Bacterial small-molecule signaling pathways. Science 311:1113-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carroll, P. A., K. T. Tashima, M. B. Rogers, V. J. DiRita, and S. B. Calderwood. 1997. Phase variation in tcpH modulates expression of the ToxR regulon in Vibrio cholerae. Mol. Microbiol. 25:1099-1111. [DOI] [PubMed] [Google Scholar]
- 16.Casper-Lindley, C., and F. H. Yildiz. 2004. VpsT is a transcriptional regulator required for expression of vps biosynthesis genes and the development of rugose colonial morphology in Vibrio cholerae O1 El Tor. J. Bacteriol. 186:1574-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen, X., S. Schauder, N. Potier, A. Van Dorsselaer, I. Pelczer, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 18.Childers, B. M., G. G. Weber, M. G. Prouty, M. M. Castaneda, F. Peng, and K. E. Klose. 2007. Identification of residues critical for the function of the Vibrio cholerae virulence regulator ToxT by scanning alanine mutagenesis. J. Mol. Biol. 367:1413-1430. [DOI] [PubMed] [Google Scholar]
- 19.Crawford, J. A., J. B. Kaper, and V. J. DiRita. 1998. Analysis of ToxR-dependent transcription activation of ompU, the gene encoding a major envelope protein in Vibrio cholerae. Mol. Microbiol. 29:235-246. [DOI] [PubMed] [Google Scholar]
- 20.Crawford, J. A., E. S. Krukonis, and V. J. DiRita. 2003. Membrane localization of the ToxR winged-helix domain is required for TcpP-mediated virulence gene activation in Vibrio cholerae. Mol. Microbiol. 47:1459-1473. [DOI] [PubMed] [Google Scholar]
- 21.DiRita, V. J., and J. J. Mekalanos. 1991. Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cell 64:29-37. [DOI] [PubMed] [Google Scholar]
- 22.DiRita, V. J., M. Neely, R. K. Taylor, and P. M. Bruss. 1996. Differential expression of the ToxR regulon in classical and E1 Tor biotypes of Vibrio cholerae is due to biotype-specific control over toxT expression. Proc. Natl. Acad. Sci. USA 93:7991-7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dziejman, M., and J. J. Mekalanos. 1994. Analysis of membrane protein interaction: ToxR can dimerize the amino terminus of phage lambda repressor. Mol. Microbiol. 13:485-494. [DOI] [PubMed] [Google Scholar]
- 24.Faruque, S. M., M. Kamruzzaman, I. M. Meraj, N. Chowdhury, G. B. Nair, R. B. Sack, R. R. Colwell, and D. A. Sack. 2003. Pathogenic potential of environmental Vibrio cholerae strains carrying genetic variants of the toxin-coregulated pilus pathogenicity island. Infect. Immun. 71:1020-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finkelstein, R. A., M. Boesman-Finkelstein, Y. Chang, and C. C. Hase. 1992. Vibrio cholerae hemagglutinin/protease, colonial variation, virulence, and detachment. Infect. Immun. 60:472-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freeman, J. A., and B. L. Bassler. 1999. A genetic analysis of the function of LuxO, a two-component response regulator involved in quorum sensing in Vibrio harveyi. Mol. Microbiol. 31:665-677. [DOI] [PubMed] [Google Scholar]
- 27.Freeman, J. A., and B. L. Bassler. 1999. Sequence and function of LuxU: a two-component phosphorelay protein that regulates quorum sensing in Vibrio harveyi. J. Bacteriol. 181:899-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freter, R. 1981. Mechanisms of association of bacteria with mucosal surfaces. CIBA Found. Symp. 80:36-55. [DOI] [PubMed] [Google Scholar]
- 29.Freter, R., and P. C. O'Brien. 1981. Role of chemotaxis in the association of motile bacteria with intestinal mucosa: chemotactic responses of Vibrio cholerae and description of motile nonchemotactic mutants. Infect. Immun. 34:215-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freter, R., P. C. O'Brien, and M. S. Macsai. 1981. Role of chemotaxis in the association of motile bacteria with intestinal mucosa: in vivo studies. Infect. Immun. 34:234-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gardel, C. L., and J. J. Mekalanos. 1996. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect. Immun. 64:2246-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ghosh, A., K. Paul, and R. Chowdhury. 2006. Role of the histone-like nucleoid structuring protein in colonization, motility, and bile-dependent repression of virulence gene expression in Vibrio cholerae. Infect. Immun. 74:3060-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gupta, S., and R. Chowdhury. 1997. Bile affects production of virulence factors and motility of Vibrio cholerae. Infect. Immun. 65:1131-1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hase, C. C. 2001. Analysis of the role of flagellar activity in virulence gene expression in Vibrio cholerae. Microbiology 147:831-837. [DOI] [PubMed] [Google Scholar]
- 36.Hase, C. C., and J. J. Mekalanos. 1999. Effects of changes in membrane sodium flux on virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 96:3183-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hase, C. C., and J. J. Mekalanos. 1998. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 95:730-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haugo, A. J., and P. I. Watnick. 2002. Vibrio cholerae CytR is a repressor of biofilm development. Mol. Microbiol. 45:471-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Higgins, D. E., and V. J. DiRita. 1994. Transcriptional control of toxT, a regulatory gene in the ToxR regulon of Vibrio cholerae. Mol. Microbiol. 14:17-29. [DOI] [PubMed] [Google Scholar]
- 40.Higgins, D. E., E. Nazareno, and V. J. DiRita. 1992. The virulence gene activator ToxT from Vibrio cholerae is a member of the AraC family of transcriptional activators. J. Bacteriol. 174:6974-6980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hulbert, R. R., and R. K. Taylor. 2002. Mechanism of ToxT-dependent transcriptional activation at the Vibrio cholerae tcpA promoter. J. Bacteriol. 184:5533-5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hung, D. T., and J. J. Mekalanos. 2005. Bile acids induce cholera toxin expression in Vibrio cholerae in a ToxT-independent manner. Proc. Natl. Acad. Sci. USA 102:3028-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hung, D. T., E. A. Shakhnovich, E. Pierson, and J. J. Mekalanos. 2005. Small-molecule inhibitor of Vibrio cholerae virulence and intestinal colonization. Science 310:670-674. [DOI] [PubMed] [Google Scholar]
- 44.Hung, D. T., J. Zhu, D. Sturtevant, and J. J. Mekalanos. 2006. Bile acids stimulate biofilm formation in Vibrio cholerae. Mol. Microbiol. 59:193-201. [DOI] [PubMed] [Google Scholar]
- 45.Jobling, M. G., and R. K. Holmes. 1997. Characterization of hapR, a positive regulator of the Vibrio cholerae HA/protease gene hap, and its identification as a functional homologue of the Vibrio harveyi luxR gene. Mol. Microbiol. 26:1023-1034. [DOI] [PubMed] [Google Scholar]
- 46.Kirn, T. J., B. A. Jude, and R. K. Taylor. 2005. A colonization factor links Vibrio cholerae environmental survival and human infection. Nature 438:863-866. [DOI] [PubMed] [Google Scholar]
- 47.Kovacikova, G., W. Lin, and K. Skorupski. 2005. Dual regulation of genes involved in acetoin biosynthesis and motility/biofilm formation by the virulence activator AphA and the acetate-responsive LysR-type regulator AlsR in Vibrio cholerae. Mol. Microbiol. 57:420-433. [DOI] [PubMed] [Google Scholar]
- 48.Kovacikova, G., W. Lin, and K. Skorupski. 2004. Vibrio cholerae AphA uses a novel mechanism for virulence gene activation that involves interaction with the LysR-type regulator AphB at the tcpPH promoter. Mol. Microbiol. 53:129-142. [DOI] [PubMed] [Google Scholar]
- 49.Kovacikova, G., and K. Skorupski. 2002. Binding site requirements of the virulence gene regulator AphB: differential affinities for the Vibrio cholerae classical and El Tor tcpPH promoters. Mol. Microbiol. 44:533-547. [DOI] [PubMed] [Google Scholar]
- 50.Kovacikova, G., and K. Skorupski. 2000. Differential activation of the tcpPH promoter by AphB determines biotype specificity of virulence gene expression in Vibrio cholerae. J. Bacteriol. 182:3228-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kovacikova, G., and K. Skorupski. 2001. Overlapping binding sites for the virulence gene regulators AphA, AphB, and cAMP-CRP at the Vibrio cholerae tcpPH promoter. Mol. Microbiol. 41:393-407. [DOI] [PubMed] [Google Scholar]
- 52.Kovacikova, G., and K. Skorupski. 2002. Regulation of virulence gene expression in Vibrio cholerae by quorum sensing: HapR functions at the aphA promoter. Mol. Microbiol. 46:1135-1147. [DOI] [PubMed] [Google Scholar]
- 53.Krukonis, E. S., R. R. Yu, and V. J. Dirita. 2000. The Vibrio cholerae ToxR/TcpP/ToxT virulence cascade: distinct roles for two membrane-localized transcriptional activators on a single promoter. Mol. Microbiol. 38:67-84. [DOI] [PubMed] [Google Scholar]
- 54.Lee, S. H., S. M. Butler, and A. Camilli. 2001. Selection for in vivo regulators of bacterial virulence. Proc. Natl. Acad. Sci. USA 98:6889-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee, S. H., D. L. Hava, M. K. Waldor, and A. Camilli. 1999. Regulation and temporal expression patterns of Vibrio cholerae virulence genes during infection. Cell 99:625-634. [DOI] [PubMed] [Google Scholar]
- 56.Lenz, D. H., M. B. Miller, J. Zhu, R. V. Kulkarni, and B. L. Bassler. 2005. CsrA and three redundant small RNAs regulate quorum sensing in Vibrio cholerae. Mol. Microbiol. 58:1186-1202. [DOI] [PubMed] [Google Scholar]
- 57.Lenz, D. H., K. C. Mok, B. N. Lilley, R. V. Kulkarni, N. S. Wingreen, and B. L. Bassler. 2004. The small RNA chaperone Hfq and multiple small RNAs control quorum sensing in Vibrio harveyi and Vibrio cholerae. Cell 118:69-82. [DOI] [PubMed] [Google Scholar]
- 58.Li, C. C., J. A. Crawford, V. J. DiRita, and J. B. Kaper. 2000. Molecular cloning and transcriptional regulation of ompT, a ToxR-repressed gene in Vibrio cholerae. Mol. Microbiol. 35:189-203. [DOI] [PubMed] [Google Scholar]
- 59.Li, C. C., D. S. Merrell, A. Camilli, and J. B. Kaper. 2002. ToxR interferes with CRP-dependent transcriptional activation of ompT in Vibrio cholerae. Mol. Microbiol. 43:1577-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lilley, B. N., and B. L. Bassler. 2000. Regulation of quorum sensing in Vibrio harveyi by LuxO and sigma-54. Mol. Microbiol. 36:940-954. [DOI] [PubMed] [Google Scholar]
- 61.Lin, W., G. Kovacikova, and K. Skorupski. 2005. Requirements for Vibrio cholerae HapR binding and transcriptional repression at the hapR promoter are distinct from those at the aphA promoter. J. Bacteriol. 187:3013-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Makinoshima, H., and M. S. Glickman. 2006. Site-2 proteases in prokaryotes: regulated intramembrane proteolysis expands to microbial pathogenesis. Microbes Infect. 8:1882-1888. [DOI] [PubMed] [Google Scholar]
- 63.Martin, R. G., and J. L. Rosner. 2001. The AraC transcriptional activators. Curr. Opin. Microbiol. 4:132-137. [DOI] [PubMed] [Google Scholar]
- 64.Martinez-Hackert, E., and A. M. Stock. 1997. Structural relationships in the OmpR family of winged-helix transcription factors. J. Mol. Biol. 269:301-312. [DOI] [PubMed] [Google Scholar]
- 65.Martinez-Hackert, E., and A. M. Stock. 1997. The DNA-binding domain of OmpR: crystal structures of a winged helix transcription factor. Structure 5:109-124. [DOI] [PubMed] [Google Scholar]
- 66.Matson, J. S., and V. J. DiRita. 2005. Degradation of the membrane-localized virulence activator TcpP by the YaeL protease in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 102:16403-16408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meibom, K. L., M. Blokesch, N. A. Dolganov, C. Y. Wu, and G. K. Schoolnik. 2005. Chitin induces natural competence in Vibrio cholerae. Science 310:1824-1827. [DOI] [PubMed] [Google Scholar]
- 68.Merrell, D. S., S. M. Butler, F. Qadri, N. A. Dolganov, A. Alam, M. B. Cohen, S. B. Calderwood, G. K. Schoolnik, and A. Camilli. 2002. Host-induced epidemic spread of the cholera bacterium. Nature 417:642-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Miller, M. B., K. Skorupski, D. H. Lenz, R. K. Taylor, and B. L. Bassler. 2002. Parallel quorum sensing systems converge to regulate virulence in Vibrio cholerae. Cell 110:303-314. [DOI] [PubMed] [Google Scholar]
- 70.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miller, V. L., R. K. Taylor, and J. J. Mekalanos. 1987. Cholera toxin transcriptional activator toxR is a transmembrane DNA binding protein. Cell 48:271-279. [DOI] [PubMed] [Google Scholar]
- 72.Mooi, F. R., and E. M. Bik. 1997. The evolution of epidemic Vibrio cholerae strains. Trends Microbiol. 5:161-165. [DOI] [PubMed] [Google Scholar]
- 73.Morris, J. G., Jr., M. B. Sztein, E. W. Rice, J. P. Nataro, G. A. Losonsky, P. Panigrahi, C. O. Tacket, and J. A. Johnson. 1996. Vibrio cholerae O1 can assume a chlorine-resistant rugose survival form that is virulent for humans. J. Infect. Dis. 174:1364-1368. [DOI] [PubMed] [Google Scholar]
- 74.Murley, Y. M., P. A. Carroll, K. Skorupski, R. K. Taylor, and S. B. Calderwood. 1999. Differential transcription of the tcpPH operon confers biotype-specific control of the Vibrio cholerae ToxR virulence regulon. Infect. Immun. 67:5117-5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nielsen, A. T., N. A. Dolganov, G. Otto, M. C. Miller, C. Y. Wu, and G. K. Schoolnik. 2006. RpoS controls the Vibrio cholerae mucosal escape response. PLoS Pathog. 2:e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nye, M. B., J. D. Pfau, K. Skorupski, and R. K. Taylor. 2000. Vibrio cholerae H-NS silences virulence gene expression at multiple steps in the ToxR regulatory cascade. J. Bacteriol. 182:4295-4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Peterson, K. M., and J. J. Mekalanos. 1988. Characterization of the Vibrio cholerae ToxR regulon: identification of novel genes involved in intestinal colonization. Infect. Immun. 56:2822-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pfau, J. D., and R. K. Taylor. 1998. Mutations in toxR and toxS that separate transcriptional activation from DNA binding at the cholera toxin gene promoter. J. Bacteriol. 180:4724-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Prouty, M. G., C. R. Osorio, and K. E. Klose. 2005. Characterization of functional domains of the Vibrio cholerae virulence regulator ToxT. Mol. Microbiol. 58:1143-1156. [DOI] [PubMed] [Google Scholar]
- 80.Provenzano, D., and K. E. Klose. 2000. Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc. Natl. Acad. Sci. USA 97:10220-10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Reguera, G., and R. Kolter. 2005. Virulence and the environment: a novel role for Vibrio cholerae toxin-coregulated pili in biofilm formation on chitin. J. Bacteriol. 187:3551-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rice, E. W., C. J. Johnson, R. M. Clark, K. R. Fox, D. J. Reasoner, M. E. Dunnigan, P. Panigrahi, J. A. Johnson, and J. G. Morris, Jr. 1992. Chlorine and survival of “rugose” Vibrio cholerae. Lancet 340:740. [DOI] [PubMed] [Google Scholar]
- 83.Richardson, K. 1991. Roles of motility and flagellar structure in pathogenicity of Vibrio cholerae: analysis of motility mutants in three animal models. Infect. Immun. 59:2727-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schuhmacher, D. A., and K. E. Klose. 1999. Environmental signals modulate ToxT-dependent virulence factor expression in Vibrio cholerae. J. Bacteriol. 181:1508-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shakhnovich, E. A., D. T. Hung, E. Pierson, K. Lee, and J. J. Mekalanos. 2007. Virstatin inhibits dimerization of the transcriptional activator ToxT. Proc. Natl. Acad. Sci. USA 104:2372-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Silva, A. J., G. J. Leitch, A. Camilli, and J. A. Benitez. 2006. Contribution of hemagglutinin/protease and motility to the pathogenesis of El Tor biotype cholera. Infect. Immun. 74:2072-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Skorupski, K., and R. K. Taylor. 1997. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol. Microbiol. 25:1003-1009. [DOI] [PubMed] [Google Scholar]
- 88.Sperandio, V., J. A. Giron, W. D. Silveira, and J. B. Kaper. 1995. The OmpU outer membrane protein, a potential adherence factor of Vibrio cholerae. Infect. Immun. 63:4433-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tamayo, R., A. D. Tischler, and A. Camilli. 2005. The EAL domain protein VieA is a cyclic diguanylate phosphodiesterase. J. Biol. Chem. 280:33324-33330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Teppema, J. S., P. A. Guinee, A. A. Ibrahim, M. Paques, and E. J. Ruitenberg. 1987. In vivo adherence and colonization of Vibrio cholerae strains that differ in hemagglutinating activity and motility. Infect. Immun. 55:2093-2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tischler, A. D., and A. Camilli. 2005. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect. Immun. 73:5873-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wai, S. N., Y. Mizunoe, A. Takade, S. I. Kawabata, and S. I. Yoshida. 1998. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl. Environ. Microbiol. 64:3648-3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Withey, J. H., and V. J. DiRita. 2005. Activation of both acfA and acfD transcription by Vibrio cholerae ToxT requires binding to two centrally located DNA sites in an inverted repeat conformation. Mol. Microbiol. 56:1062-1077. [DOI] [PubMed] [Google Scholar]
- 94.Withey, J. H., and V. J. DiRita. 2006. The toxbox: specific DNA sequence requirements for activation of Vibrio cholerae virulence genes by ToxT. Mol. Microbiol. 59:1779-1789. [DOI] [PubMed] [Google Scholar]
- 95.Withey, J. H., and V. J. Dirita. 2005. Vibrio cholerae ToxT independently activates the divergently transcribed aldA and tagA genes. J. Bacteriol. 187:7890-7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yancey, R. J., and L. J. Berry. 1978. Motility of the pathogen and intestinal immunity of the host in experimental cholera. Adv. Exp. Med. Biol. 107:447-455. [DOI] [PubMed] [Google Scholar]
- 97.Yancey, R. J., D. L. Willis, and L. J. Berry. 1978. Role of motility in experimental cholera in adult rabbits. Infect. Immun. 22:387-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yildiz, F. H., N. A. Dolganov, and G. K. Schoolnik. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPS(ETr)-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 183:1716-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 96:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yoon, S. S., and J. J. Mekalanos. 2006. 2,3-Butanediol synthesis and the emergence of the Vibrio cholerae El Tor biotype. Infect. Immun. 74:6547-6556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yu, R. R., and V. J. DiRita. 2002. Regulation of gene expression in Vibrio cholerae by ToxT involves both antirepression and RNA polymerase stimulation. Mol. Microbiol. 43:119-134. [DOI] [PubMed] [Google Scholar]
- 102.Zhu, J., M. B. Miller, R. E. Vance, M. Dziejman, B. L. Bassler, and J. J. Mekalanos. 2002. Quorum-sensing regulators control virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 99:3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]