Abstract
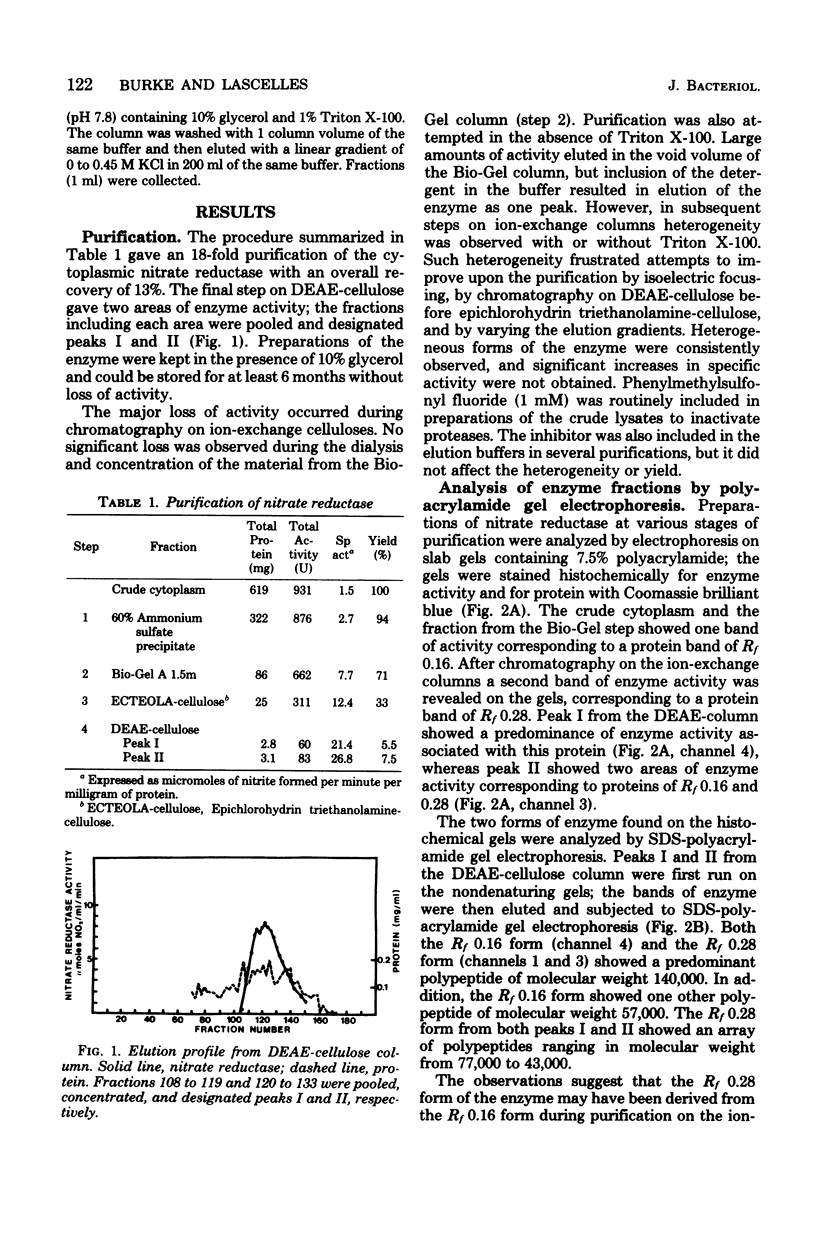
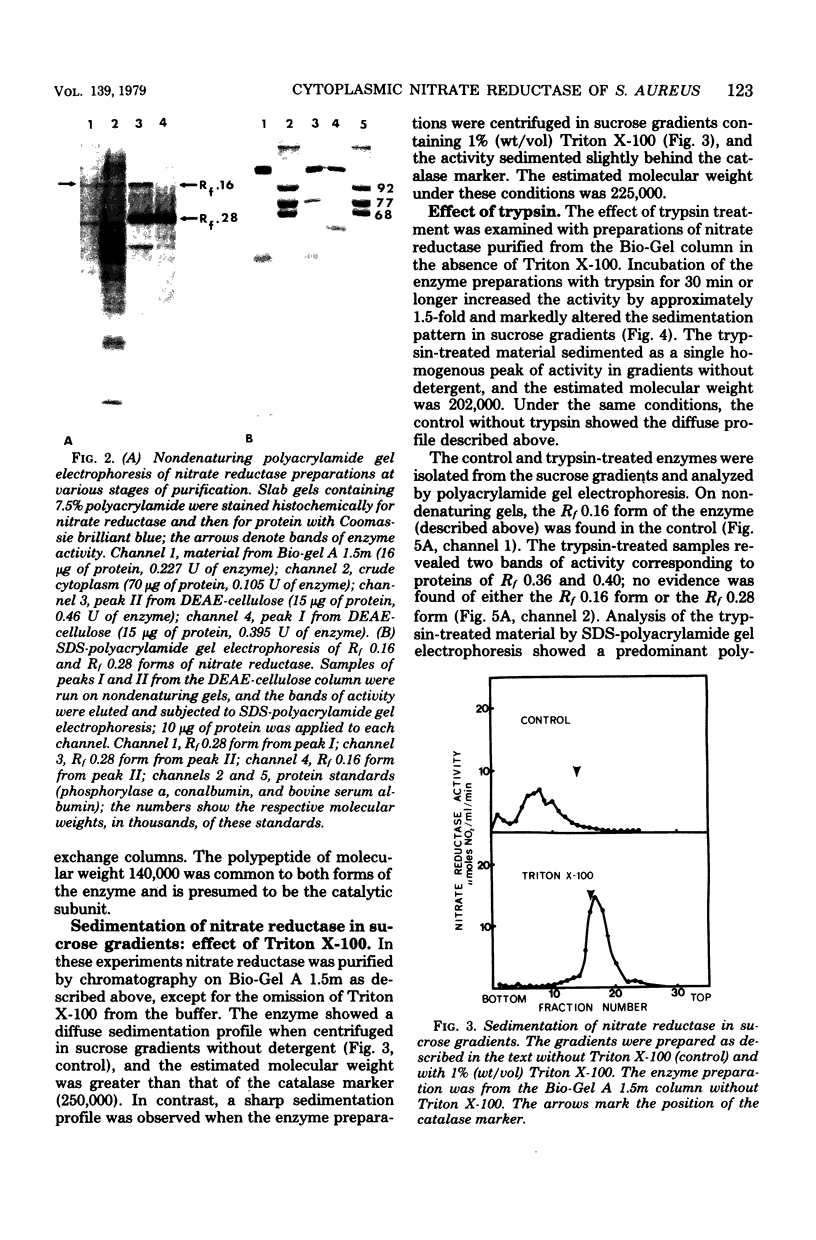
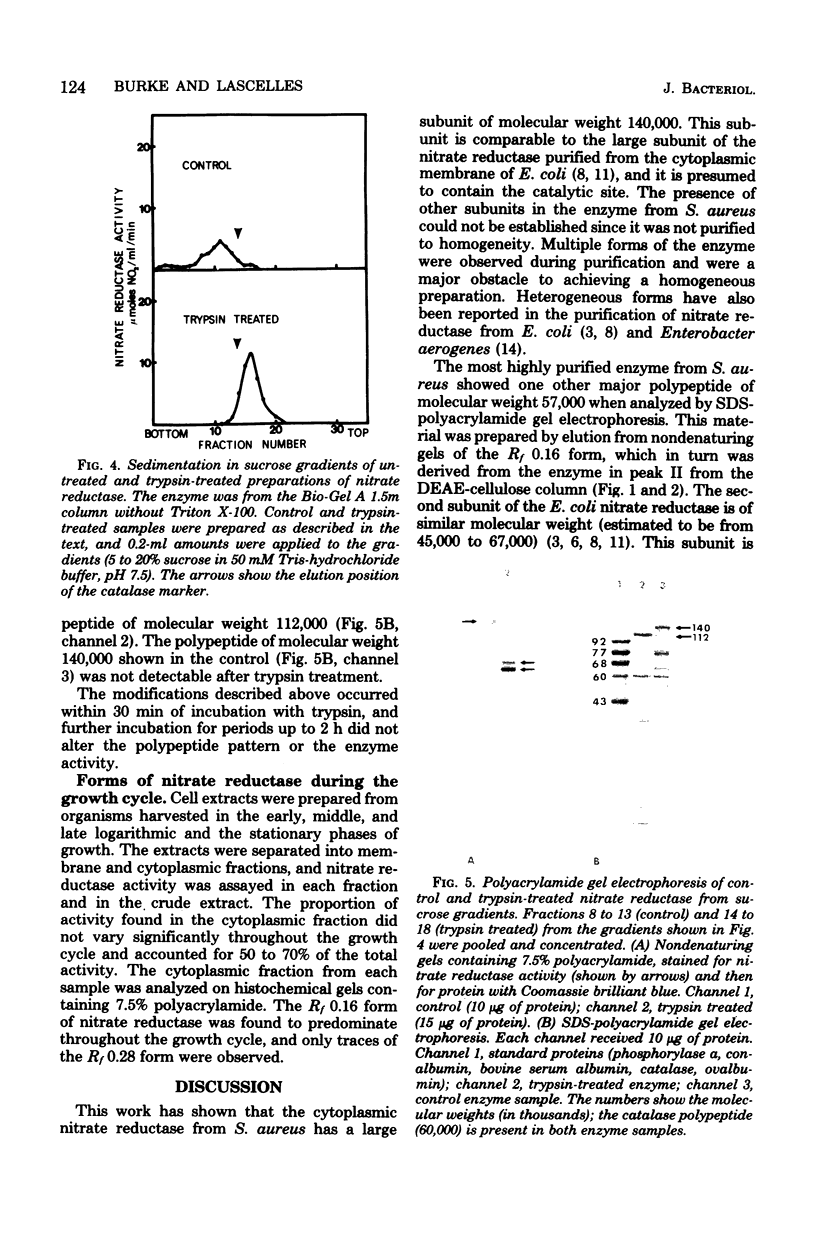
The cytoplasmic nitrate reductase in heme mutant H-14 of Staphylococcus aureus was partially purified by steps which included ammonium sulfate fractionation and chromatography on Bio-Gel A 1.5m and ion-exchange columns. The active fractions from the ion-exchange columns showed two forms of the enzyme upon electrophoresis in nondenaturing gels of polyacrylamide; these corresponded to proteins of Rf 0.16 and 0.28. Each form contained a predominant polypeptide of molecular weight 140,000, as shown by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The Rf 0.16 form contained another major polypeptide of molecular weight 57,000, but the Rf 0.28 form contained several other polypeptides. The sedimentation properties of the enzyme were examined after partial purification on Bio-Gel A 1.5m. In sucrose gradients containing Triton X-100 the enzyme sedimented as a homogeneous peak with an estimated molecular weight of 225,000; without detergent a heterogeneous profile was observed of molecular weight greater than 250,000. Treatment of the enzyme with trypsin increased the specific activity, and the enzyme sedimented as a homogeneous peak in sucrose gradients without Triton X-100, with an estimated molecular weight of 202,000. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis indicated that trypsin treatment converted the polypeptide of molecular weight 140,000 to a polypeptide of molecular weight 112,000. We conclude that the cytoplasmic nitrate reductase of S. aureus has a large subunit of molecular weight 140,000, which can be modified by trypsin to a polypeptide of molecular weight 112,000 without loss of catalytic activity.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Burke K. A., Lascelles J. Nitrate reductase activity in heme-deficient mutants of Staphylococcus aureus. J Bacteriol. 1976 Apr;126(1):225–231. doi: 10.1128/jb.126.1.225-231.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke K. A., Lascelles J. Nitrate reductase system in Staphylococcus aureus wild type and mutants. J Bacteriol. 1975 Jul;123(1):308–316. doi: 10.1128/jb.123.1.308-316.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clegg R. A. Purification and some properties of nitrate reductase (EC 1.7.99.4) from Escherichia coli K12. Biochem J. 1976 Mar 1;153(3):533–541. doi: 10.1042/bj1530533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMoss J. A. Limited proteolysis of nitrate reductase purified from membranes of Escherichia coli. J Biol Chem. 1977 Mar 10;252(5):1696–1701. [PubMed] [Google Scholar]
- Dulley J. R., Grieve P. A. A simple technique for eliminating interference by detergents in the Lowry method of protein determination. Anal Biochem. 1975 Mar;64(1):136–141. doi: 10.1016/0003-2697(75)90415-7. [DOI] [PubMed] [Google Scholar]
- Enoch H. G., Lester R. L. The purification and properties of formate dehydrogenase and nitrate reductase from Escherichia coli. J Biol Chem. 1975 Sep 10;250(17):6693–6705. [PubMed] [Google Scholar]
- Lascelles J., Burke K. A. Reduction of ferric iron by L-lactate and DL-glycerol-3-phosphate in membrane preparations from Staphylococcus aureus and interactions with the nitrate reductase system. J Bacteriol. 1978 May;134(2):585–589. doi: 10.1128/jb.134.2.585-589.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund K., DeMoss J. A. Association-dissociation behavior and subunit structure of heat-released nitrate reductase from Escherichia coli. J Biol Chem. 1976 Apr 25;251(8):2207–2216. [PubMed] [Google Scholar]
- MARTIN R. G., AMES B. N. A method for determining the sedimentation behavior of enzymes: application to protein mixtures. J Biol Chem. 1961 May;236:1372–1379. [PubMed] [Google Scholar]
- MacGregor C. H. Anaerobic cytochrome b1 in Escherichia coli: association with and regulation of nitrate reductase. J Bacteriol. 1975 Mar;121(3):1111–1116. doi: 10.1128/jb.121.3.1111-1116.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacGregor C. H., Schnaitman C. A., Normansell D. E. Purification and properties of nitrate reductase from Escherichia coli K12. J Biol Chem. 1974 Aug 25;249(16):5321–5327. [PubMed] [Google Scholar]
- MacGregor C. H. Synthesis of nitrate reductase components in chlorate-resistant mutants of Escherichia coli. J Bacteriol. 1975 Mar;121(3):1117–1121. doi: 10.1128/jb.121.3.1117-1121.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne W. J. Reduction of nitrogenous oxides by microorganisms. Bacteriol Rev. 1973 Dec;37(4):409–452. doi: 10.1128/br.37.4.409-452.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouthamer A. H. Biochemistry and genetics of nitrate reductase in bacteria. Adv Microb Physiol. 1976;14(11):315–375. doi: 10.1016/s0065-2911(08)60230-1. [DOI] [PubMed] [Google Scholar]
- Tanaka Y., Anraku Y., Futai M. Escherichia coli membrane D-lactate dehydrogenase. Isolation of the enzyme in aggregated from and its activation by Triton X-100 and phospholipids. J Biochem. 1976 Oct;80(4):821–830. doi: 10.1093/oxfordjournals.jbchem.a131343. [DOI] [PubMed] [Google Scholar]