Abstract
Rotavirus genotype G12 strains were detected for the first time among newborns with asymptomatic rotavirus infection (74% of 39 rotavirus strains isolated from the infected infants were genotype G12) in the nursery of the All India Institute of Medical Sciences during a period from 2005 to 2006. Sequence analysis of the VP7 genes from these neonatal strains indicated a high level of homology to other G12 strains reported worldwide, suggesting the recent emergence of these strains in humans. Such nosocomial infections of newborns represent a potential source of introduction of novel rotavirus serotypes into the community.
Each year, rotavirus (RV) accounts for an estimated 100,000 deaths and 400,000 hospitalizations in children <5 years of age in India and 454,000 to 705,000 deaths worldwide (13). The accelerated development and introduction of an RV vaccine have become a global health priority, and two vaccines have been recently licensed and may soon address this problem (1). At the same time, two neonatal RV strains are being developed as vaccine candidates because preliminary studies indicate that they are well adapted to the neonatal gut, induce systemic and mucosal immune responses, and protect children against severe RV-induced diarrhea upon reinfection (2-4, 10).
Our previous report on an outbreak of asymptomatic RV infection at the All India Institute of Medical Sciences (AIIMS), New Delhi, in 1985, as well as the multicenter surveillance study conducted from 1986 to 1993, demonstrated that two novel strains, G9P[11], a human-bovine reassortant strain, and G9P[6], were common causes of infections in the hospital nurseries in Delhi (2, 6). Since then, no further surveillance of RV infection in newborns has been conducted. Yet knowledge of circulating RV strains in local populations is important for the development and evaluation of new candidate vaccines, and early warning of the emergence of novel strains may help in the establishment of a follow-up strategy that may be useful in future vaccine development. RV strains infecting neonates may be a source of emergence of new strains that subsequently spread into the community. For example, novel strains of genotype P[6] were found worldwide in newborns years before the P[6] strains were identified as a cause of diarrhea in older children (7, 11). In this study, a decade after our previous survey examining newborns, we again conducted surveillance in the nursery at AIIMS to see if neonatal RV strains related to 116E or any other RV strains were present in the nursery.
Fecal specimens were collected twice during days 2 to 9 after birth from all the neonates born during the period from September 2005 to August 2006 at the AIIMS nursery in New Delhi, India. RV testing of the stool was performed using a monoclonal antibody-based enzyme immunoassay kit (Premier Rotaclone; Meridian Biosciences, Cincinnati, OH). Neonates were considered to be infected with RV if at least one of the stool samples was positive for the virus. RV double-stranded RNA was extracted from stools or laboratory strains by the TRIzol (Invitrogen Corp., Carlsbad, CA) method, and the RNA was used for polyacrylamide gel electrophoresis and the determination of the G and P types of the strains by reverse transcription-PCR (6). Genotyping was initially performed using common G (G1 to G4 and G9) and P (P4, P6, P8, and P11) type primers in a multiplex assay (3). After initially detecting numerous nontypeable strains, we subsequently included a G12-specific primer in the oligonucleotide pool for the G-typing PCR assay (18). Nucleotide sequencing of a fragment of the VP7 gene of strains that could not be typed with primers for common strains (G1 to G4 and G9) was performed using a previously described method (18). Multiple sequence alignments of the VP7 gene sequences (spanning nucleotides [nt] 68 to 580) of these strains with the equivalent regions of genotype G12 sequences from the GenBank database were carried out using CLUSTAL W 1.81 as described previously (19). Distances were calculated using the Kimura two-parameter method, and a dendrogram was constructed using the neighbor-joining method with MEGA software, version 2.1 (19).
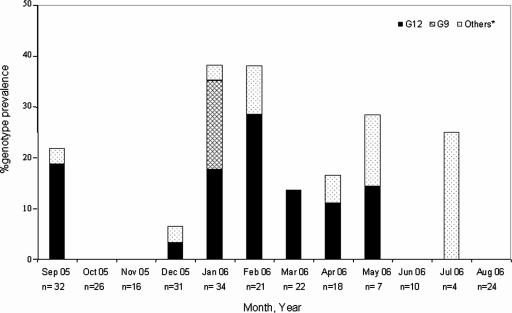
In this survey, RV antigens were detected in 16% of neonates (39 of 245) born during this period. Although surveillance was conducted for 1 year, the dates of isolation of the majority of the RV isolates (21 of 39) clustered in January and February 2006 (Fig. 1). Most RVs (34 of 39) were detected in newborns between 5 and 9 days of age.
FIG. 1.
Seasonal distribution and percent prevalence of VP7 genotypes of neonatal RV strains detected during the period from 2005 to 2006 at the AIIMS nursery. *, includes G2 strains, strains involved in mixed infections, and nontypeable strains.
Our analysis of the G and P genotypes of strains from all 39 RV-infected neonates yielded the following results: 28 of 39 strains (72%) were originally nontypeable, 6 of 39 (15%) were G9, 4 of 39 (10%) had mixed G types, and 1 was identified as a G2 strain. All of the strains had P[6] VP4 specificity. Nucleotide sequencing of 513-bp VP7 gene fragments (spanning nt 68 to 580) from four nontypeable strains identified them as genotype G12 based on the high degree of homology of the sequences to those of G12 strains listed in the GenBank database. Subsequently, using reverse transcription-PCR with G12 primers, we found that 21 of 24 remaining nontypeable strains were also G12, and samples from the remaining 3 newborns were exhausted. Interestingly, the G12P[6] strain was also detected in all four cases of mixed infections, making the total number of G12 strains 29 (74% of the total).
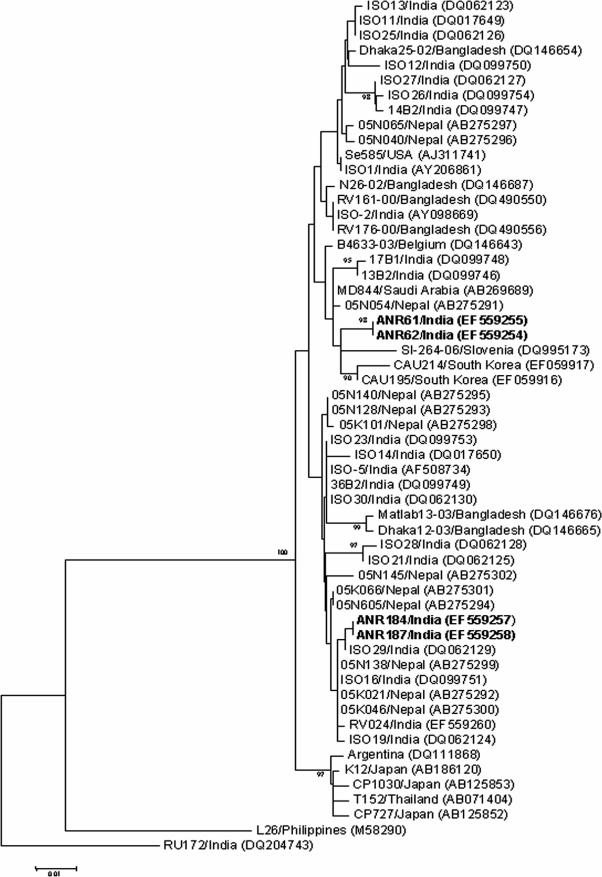
An analysis of the VP7 gene nucleotides of G12 strains (n = 4) by using a distance matrix demonstrated ∼89 to 99% homology to those of other human G12 strains reported worldwide. The maximum homology was observed with G12 strains from Calcutta and strain Se585 from the United States (data not shown). A phylogenetic analysis using MEGA version 2.1 confirmed the close relationship to recent G12 isolates from many countries (Fig. 2). Polyacrylamide gel electrophoresis analysis of RV genomic double-stranded RNA identified a single RNA migration pattern (electropherotype) for 30 of the 39 strains examined (data not shown). The amount of RNA in the remaining strains was insufficient for visualization by silver staining.
FIG. 2.
Neighbor-joining dendrogram of the VP7 gene (nt 68 to 580) of neonatal G12 RV strains from AIIMS (shown in boldface) and other G12 strains. Reference sequences were obtained from the GenBank and EMBL databases; accession numbers are given in parentheses.
We previously reported that asymptomatic RV infection in neonates at AIIMS conferred protection against severe diarrhea upon reinfection (2). Based on this observation, the vaccine candidate 116E (type G9P[11]) has been developed and is presently being evaluated in clinical trials (3). After a decade, we reinitiated surveillance with the aim of determining whether closely related strains were still present among neonates in the nursery. During the period from September 2005 to August 2006, a large number of neonates born in the AIIMS nursery were found to be infected with RV. However, the observed rate of RV detection was much lower than that in our previous study (16 versus 73%) (2). Unlike the previous study, this survey did not detect any G9P[11] (116E-like) strains (2). Instead, G12P[6], a recently emerging strain among children with diarrhea worldwide, was detected for the first time in a nursery outbreak among asymptomatic newborns in New Delhi (3, 14, 16, 18, 20, 21). This finding reinforces the hypothesis that some novel strains of RV may emerge in newborns whose high titers of maternal antibody to common strains select for strains that contain novel serotype antigens. In a recent study, Ray and Kelkar observed that low titers of neutralizing antibodies to common strains (e.g., G1, G2, G4, and G9) among the mothers predisposed the children to infection with those serotypes (17). In another study, Ramachandran et al. noted that low titers of neutralizing antibodies to strain 116E in cord blood predisposed newborns to infection with 116E-like strains (15). In the present study, we detected G12P[6] infections concomitantly with a low incidence of G9P[6] strains. Since G9P[6] strains have been circulating in the Delhi community for the past several years, we hypothesize that preexisting immunity among neonates in the form of maternal antibodies to these strains and lower titers of antibodies to G12 may explain these results. In the future, it will be interesting to test titers of G12-specific neutralizing antibodies in the mothers or in the cord blood of the babies infected with these strains to further examine this hypothesis.
The original source of the G12 RV and the mechanism of its introduction into the AIIMS nursery are unknown. The unusual G9P[11] strains previously introduced at AIIMS had a probable bovine origin (8), as indicated by a close relationship with the VP4 gene of a bovine P[11] RV. Since P[11] strains of various serotypes are common in India, we speculated that introduction by members of the hospital staff or mothers admitted to the nursery or by other mechanisms was possible. G12 strains have been found in animals from India (9), so a mechanism of introduction into the nursery similar to that of the G9P[11] strains may be operative. For both the earlier P[11] strains and the present G12 isolates, introduction may have been followed by reassortment with other nursery strains. In addition, since the animal P[11] and G12 gene homologues are significantly divergent from those of G9P[11] and G12P[6] isolates at AIIMS, either the evolution of the VP7 or VP4 gene to its present-day form occurred or other progenitors are yet to be discovered. Further analysis and comparison of these strains both from the nursery and from the community may reveal their relatedness and may help in understanding the evolution and origin of G12 RVs.
The identical electropherotypes of all G12 RV strains infecting neonates in the present study suggested a common source of nosocomial infection. A similar observation was also made in our earlier study on neonatal RV infections, which demonstrated a point-source nosocomial infection with a single strain (G9P[11]) that persisted for several years at AIIMS. RV strains infecting neonates have been hypothesized to represent a possible source of emergence of new strains that subsequently spread in the local community. Previously, G8 or G9 strains that were found to be prevalent in hospital nurseries have also been identified in the community (5, 6, 10, 12).
Globally, P[6] strains were distinctly labeled as neonatal strains because they were identified in newborns in Sweden, the United Kingdom, Venezuela, and Australia (4, 7). Sequence analyses demonstrated that the corresponding community and neonatal strains were virtually identical; in the case of G9, the newborn strains were identified 2 years earlier than the community strains, and P[6] strains are now commonly found in older children with diarrhea. The high degree of sequence homology among G12 strains from India and those worldwide also indicates the recent emergence of these strains in the populations or alternatively suggests that we should next expect to find G12 strains in the Delhi population of older children with RV diarrhea.
In conclusion, G9P[11], a strain previously prevalent in neonates, is no longer present in the AIIMS nursery, while G12P[6], which has been recently detected at a high frequency in diarrheal children worldwide, was detected for the first time among asymptomatic neonates, also at a high frequency. This finding supports previous observations that newborns are sensitive hosts for novel strains of RV whose outer capsid proteins, VP4 and VP7, are not neutralized by high titers of circulating maternal antibodies. Selected surveillance of newborns for RV in settings where nosocomial infections are common may provide an early discovery of new strains that may later enter routine circulation in older children.
Acknowledgments
Financial support for this study was obtained from the Department of Biotechnology and the Department of Science and Technology, Government of India.
Technical assistance from Mary Wilson, Seema Verma, and Jaseem Khan and the participation of the parents of the children are gratefully acknowledged.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the CDC.
Footnotes
Published ahead of print on 29 August 2007.
REFERENCES
- 1.Angel, J., M. A. Franco, and H. B. Greenberg. 2007. Rotavirus vaccines: recent developments and future considerations. Nat. Rev. Microbiol. 5:529-539. [DOI] [PubMed] [Google Scholar]
- 2.Bhan, M. K., F. W. Lew, S. Sazawal, B. K. Das, J. R. Gentsch, and R. I. Glass. 1993. Protection conferred by neonatal rotavirus infection against subsequent rotavirus diarrhea. J. Infect. Dis. 168:282-287. [DOI] [PubMed] [Google Scholar]
- 3.Bhandari, N., P. Sharma, R. I. Glass, P. Ray, H. Greenberg, S. Taneja, M. Saksena, C. D. Rao, J. R. Gentsch, U. Parashar, Y. Maldonado, R. L. Ward, and M. K. Bhan. 2006. Safety and immunogenecity of two live attenuated human rotavirus vaccine candidates, 116E and 1321, in infants: results of a randomized controlled trial. Vaccine 24:5817-5823. [DOI] [PubMed] [Google Scholar]
- 4.Bishop, R. F., G. L. Barnes, E. Cipriani, and J. S. Lund. 1983. Clinical immunity after neonatal rotavirus infection: a prospective longitudinal study in young children. N. Engl. J. Med. 309:72-76. [DOI] [PubMed] [Google Scholar]
- 5.Cunliffe, N. A., S. Rogerson, W. Dove, B. D. Thindwa, J. Greensill, C. D. Kirkwood, R. L. Broadhead, and C. A. Hart. 2002. Detection and characterization of rotaviruses in hospitalized neonates in Blantyre, Malawi. J. Clin. Microbiol. 40:1534-1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das, B. K., J. R. Gentsch, H. G. Cicirello, P. A. Woods, A. Gupta, M. Ramachandran, R. Kumar, M. K. Bhan, and R. I. Glass. 1994. Characterization of rotavirus strains from newborns in New Delhi, India. J. Clin. Microbiol. 32:1820-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flores, J., K. Midthun, Y. Hoshino, K. Green, M. Gorziglia, A. Z. Kapikian, and R. M. Chanock. 1986. Conservation of the fourth gene among rotaviruses recovered from asymptomatic newborn infants and its possible role in attenuation. J. Virol. 60:972-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gentsch, J., B. K. Das, B. Jiang, M. K. Bhan, and R. I. Glass. 1993. Similarity of the VP4 protein of human rotavirus strain 116E to that of the bovine B223 strain. Virology 194:424-430. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh, S., V. Varghese, S. Samajdar, S. K. Bhattacharya, N. Kobayashi, T. N. Naik, S. Ghosh, V. Varghese, S. Samajdar, S. K. Bhattacharya, N. Kobayashi, and T. N. Naik. 2006. Molecular characterization of a porcine group A rotavirus strain with G12 genotype specificity. Arch. Virol. 151:1329-1344. [DOI] [PubMed] [Google Scholar]
- 10.Glass, R. I., M. K. Bhan, P. Ray, R. Bahl, U. D. Parashar, H. Greenberg, C. D. Rao, N. Bhandari, Y. Maldonado, R. L. Ward, D. I. Bernstein, and J. R. Gentsch. 2005. Development of candidate rotavirus vaccines derived from neonatal strains in India. J. Infect. Dis. 192:S30-S35. [DOI] [PubMed] [Google Scholar]
- 11.Hoshino, Y., R. W. Jones, J. Ross, N. Santos, and A. Z. Kapikian. 2003. Human rotavirus strains bearing VP4 gene P[6] allele recovered from asymptomatic or symptomatic infections share similar, if not identical, VP4 neutralization specificities. Virology 316:1-8. [DOI] [PubMed] [Google Scholar]
- 12.Laird, A. R., J. R. Gentsch, T. Nakagomi, O. Nakagomi, and R. I. Glass. 2003. Characterization of serotype G9 rotavirus strains isolated in the United States and India from 1993 to 2001. J. Clin. Microbiol. 41:3100-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parashar, U. D., C. J. Gibson, J. S. Bresee, and R. I. Glass. 2006. Rotavirus and severe childhood diarrhea. Emerg. Infect. Dis. 12:304-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rahman, M., J. Matthijnssens, X. Yang, T. Delbeke, I. Arijs, K. Taniguchi, M. Iturriza-Gomara, N. Iftekharuddin, T. Azim, and M. Van Ranst. 2007. Evolutionary history and global spread of the emerging g12 human rotaviruses. J. Virol. 81:2382-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramachandran, M., A. Vij, R. Kumar, B. K. Das, J. R. Gentsch, M. K. Bhan, and R. I. Glass. 1998. Lack of maternal antibodies to P serotypes may predispose neonates to infections with unusual rotavirus strains. Clin. Diagn. Lab. Immunol. 5:527-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramani, S., I. Banerjee, B. P. Gladstone, R. Sarkar, D. Selvapandian, A. M. Le Fevre, S. Jaffar, M. Iturriza-Gomara, J. J. Gray, M. K. Estes, D. W. Brown, and G. Kang. 2007. Geographic information systems and genotyping in identification of rotavirus G12 infections in residents of an urban slum with subsequent detection in hospitalized children: emergence of G12 genotype in South India. J. Clin. Microbiol. 45:432-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ray, P. G., and S. D. Kelkar. 2004. Prevalence of neutralizing antibodies against different rotavirus serotypes in children with severe rotavirus-induced diarrhea and their mothers. Clin. Diagn. Lab. Immunol. 11:186-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Samajdar, S., V. Varghese, P. Barman, S. Ghosh, U. Mitra, P. Dutta, S. K. Bhattacharya, M. V. Narasimham, P. Panda, T. Krishnan, N. Kobayashi, and T. N. Naik. 2006. Changing pattern of human group A rotaviruses: emergence of G12 as an important pathogen among children in eastern India. J. Clin. Virol. 36:183-188. [DOI] [PubMed] [Google Scholar]
- 19.Subodh, S., M. K. Bhan, and P. Ray. 2006. Genetic characterization of VP3 gene of group A rotaviruses. Virus Genes 33:143-145. [DOI] [PubMed] [Google Scholar]
- 20.Taniguchi, K., T. Urasawa, N. Kobayashi, M. Gorziglia, and S. Urasawa. 1990. Nucleotide sequence of VP4 and VP7 genes of human rotaviruses with subgroup I specificity and long RNA pattern: implication for new G serotype specificity. J. Virol. 64:5640-5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uchida, R., B. D. Pandey, J. B. Sherchand, K. Ahmed, M. Yokoo, T. Nakagomi, L. E. Cuevas, N. A. Cunliffe, C. A. Hart, and O. Nakagomi. 2006. Molecular epidemiology of rotavirus diarrhea among children and adults in Nepal: detection of G12 strains with P[6] or P[8] and G11P[25] strain. J. Microbiol. 44:3499-3505. [DOI] [PMC free article] [PubMed] [Google Scholar]