Abstract
A total of 342 imipenem-resistant Acinetobacter baumannii isolates (IRABs) were collected from 16 Chinese cities. Six predominant clones had spread widely, and four clones were detected in distant hospitals. The majority of the IRABs contain blaOXA-23, with ISAba1 upstream of the gene. These results suggested that clonal spread played an important role in the outbreak of IRABs in China.
A. baumannii is recognized as an increasingly important opportunistic gram-negative pathogen that is frequently associated with nosocomial outbreaks worldwide (21). Carbapenems are useful agents that could combat many severe Acinetobacter infections. Resistance to carbapenems is now accumulating, largely through clonal spread (13). A few lineages even achieve an epidemic status, reaching multiple hospitals or countries (6). One mechanism of resistance to carbapenems is due to the carbapenem-hydrolyzing β-lactamases of molecular classes B and D (12). Four of the eight clusters of OXA-type carbapenemases were identified in A. baumannii, including clusters OXA-23, OXA-24, OXA-51, and OXA-58 (22). ISAba1 may have an important role in the expression and transfer of OXA-type carbapenemase genes (17, 20).
In this study, we investigated the prevalence of imipenem-resistant A. baumannii isolates (IRABs) in China and characterized the genes of the carbapenemases in IRABs. Three hundred forty-two nonduplicate IRABs were recovered from 16 cities through January 2005 to December 2005 in China (Table 1; Fig. 1). All isolates were identified by the Vitek GNI+ card (BioMérieux, Marcy-l'Etoile, France) as members of the Acinetobacter calcoaceticus-A. baumannii complex. Species identification was confirmed by sequence analyses of the 16S-23S rRNA gene spacer region (4).
TABLE 1.
The PFGE clone distribution of 342 clinical isolates among 16 cities
| City | Total no. of isolates | No. of isolates of PFGE clone
|
||||||
|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | Other | ||
| Beijing | 5 | 2 | 3 | |||||
| Shanghai | 30 | 17 | 10 | 3 | ||||
| Guangzhou | 17 | 10 | 7 | |||||
| Shenyang | 15 | 10 | 5 | |||||
| Chongqing | 4 | 1 | 3 | |||||
| Hangzhou | 149 | 17 | 5 | 97 | 24 | 6 | ||
| Ningbo | 23 | 21 | 1 | 1 | ||||
| Zhoushan | 1 | 1 | ||||||
| Jianxing | 7 | 6 | 1 | |||||
| Huzhou | 8 | 6 | 2 | |||||
| Quzhou | 8 | 6 | 2 | |||||
| Jinghua | 17 | 1 | 7 | 8 | 1 | |||
| Taizhou | 13 | 11 | 2 | |||||
| Lishui | 21 | 20 | 1 | |||||
| Shaoxing | 15 | 15 | ||||||
| Wenzhou | 9 | 4 | 3 | 2 | ||||
| Total | 342 | 62 | 32 | 160 | 31 | 10 | 8 | 39 |
FIG. 1.
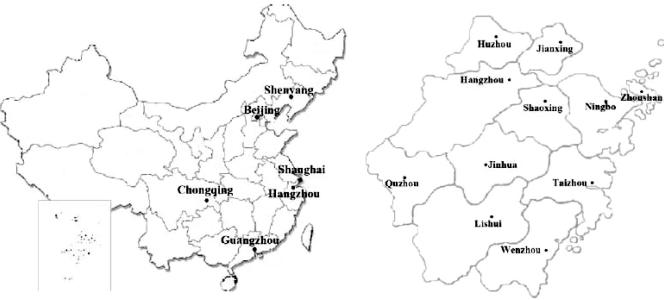
The geographic distribution of 16 cities in China. On the left, a map of China shows three autonomous cities (Beijing, Chongqing, and Shanghai) and three provincial cities (Hangzhou [Zhejiang], Guangzhou [Guangdong], and Shenyang [Liaoning]), in which Acinetobacter baumannii isolates were collected. On the right, a map shows the 11 cities, including Hangzhou, in Zhejiang Province.
The MICs of 10 antimicrobial agents were determined by the agar dilution technique by following the standards of Clinical and Laboratory Standards Institute (5); these agents included imipenem (Merck KGaA, Darmstadt, Germany), meropenem (Sumitomo Pharmaceuticals Co., Ltd., Osaka, Japan), piperacillin (Sigma, Deisenhofen, Germany), ceftazidime (Sigma, Deisenhofen, Germany), cefepime (Sino-American Shanghai Squibb Pharmaceuticals Ltd., Shanghai, China), aztreonam (Sigma, Deisenhofen, Germany), amikacin (Sigma, Deisenhofen, Germany), gentamicin (Sigma, Deisenhofen, Germany), ciprofloxacin (Bayer, Leverkusen, Germany), and minocycline (Sigma, Deisenhofen, Germany). The MICs of four other agents, ampicillin-sulbactam, cefoperazone-sulbactam, piperacillin-tazobactam, and polymyxin E, were determined by Etest (AB Biodisk, Solna, Sweden) according to the manufacturer's instructions. The results were interpreted according to CLSI 2006 guidelines (5). Pseudomonas aeruginosa ATCC 27853 was used as a control. All isolates were found to be resistant to imipenem and meropenem. The rates of resistance to ampicillin-sulbactam, cefoperazone-sulbactam, and minocycline were 68.0%, 54.2%, and 75.9%, respectively. The rate of resistance to polymyxin E was 10.8%, the lowest among the tested agents. The rates of resistance to all other tested antimicrobial agents were more than 90%.
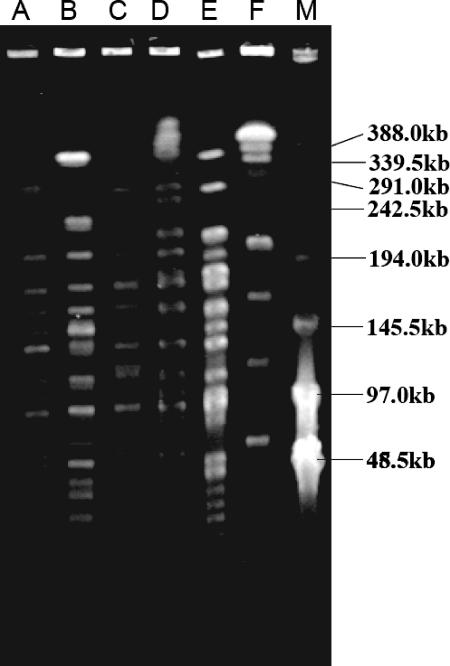
To characterize these isolates genetically, the genomic DNA of all the isolates was digested with ApaI and the resultant DNA fragments were subjected to pulsed-field gel electrophoresis (PFGE) as described previously (18). According to the datum-interpreting criteria described by Tenover et al. (19), these A. baumannii isolates belonged to 29 distinct clones (Fig. 2). Among them, six clones were dominant, consisting of 303 strains in total (Table 1).
FIG. 2.
PFGE analysis of the six major clones of A. baumannii. Genomic DNA isolated from the six major A. baumannii clones was digested with ApaI before PFGE analysis. Lane M, λ ladder used as molecular size marker; lanes A to F, clones A to F, respectively, of A. baumannii.
Crude β-lactamase preparations were extracted by the sonication method (25). pI values were determined by the PhastSystem electrophoresis system (Amersham Pharmacia Biotech, Piscataway, NJ) according to the manufacturer's instructions. pI reference proteins were stained with Coomassie brilliant blue R-250. The pattern was analyzed by Curve Expert software (version 1.3) developed by Daniel Hyams (free software). OXA-23-producing A. baumannii (a collection of our laboratory) was used as the positive control. Metallo-β-lactamase-producing isolates were screened by the imipenem-EDTA double-disk synergy test (10). IMP-4-producing A. baumannii and VIM-2-producing P. aeruginosa were used as the positive controls. All tested isolates had one band at pI 6.64 and were negative by the imipenem-EDTA double-disk synergy test.
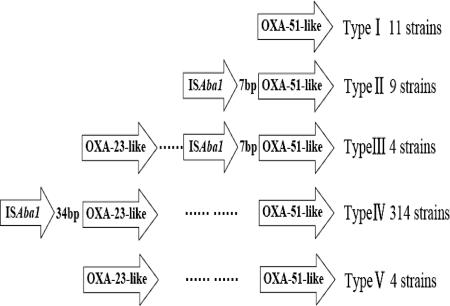
blaIMP-type, blaVIM-type, blaSIM-1, blaOXA-23-like, blaOXA-24-like, blaOXA-58-like, and blaOXA-51-like were analyzed by PCR using primers as described previously (9, 11, 20). PCR mapping was performed using the ISAba1-OXA-23-like primers or the ISAba1-OXA-51-like primers as reported previously (26). The results showed that all the isolates contained blaOXA-51-like but were negative for the OXA-24-like, OXA-58-like, SIM-1, IMP-type, or VIM-type gene. Three hundred twenty-two isolates contained the blaOXA-23-like gene. PCR with the ISAba1-OXA-23-like primers generated a PCR product in 314 IRABs, and sequencing analysis showed that the PCR product contained the OXA-23 gene (the GenBank accession number AJ132105), suggesting that a majority of the clones contain the OXA-23 gene with ISAba1 upstream of it. PCR with the ISAba1-OXA-51-like primers generated a PCR product, which contained the OXA-66 gene (the GenBank accession number AY750909) in 13 strains, as revealed by sequencing analysis. There was a 34-bp nucleotide sequence between ISAba1 and blaOXA-23 and a 7-bp nucleotide sequence between ISAba1 and blaOXA-66. According to the PCR mapping, the 342 strains belonged to five types (Fig. 3).
FIG. 3.
PCR mapping of blaOXA-51-like, blaOXA-23-like, and ISAba1 in IRABs. According to the PCR mapping results, 342 strains were divided into five types: type I, consisting of 11 strains, contains blaOXA-51-like without the upstream ISAba1; type II, consisting of 9 strains, contains blaOXA-51-like with the upstream ISAba1; type III, consisting of 4 strains, contains both blaOXA-51-like with the upstream ISAba1 and blaOXA-23-like without the upstream ISAba1; type IV, consisting of 314 strains, contains blaOXA-51-like without ISAba1 and blaOXA-23-like with ISAba1; and type V, consisting of 4 strains, contains blaOXA-51-like and blaOXA-23-like without ISAba1 upstream of either gene.
Attempts to transfer blaOXA-23 by conjugation using the rifampin-resistant Escherichia coli 600 as the recipient failed. Repeated attempts to detect the presence of plasmids capable of hybridizing with the blaOXA-23 and blaOXA-66 probes also failed. ApaI-digested PFGE DNAs were transferred to nylon membranes and were hybridized with a v-dCTP (DuPont Corporation)-labeled blaOXA-23 gene probe. The fragments of the ApaI-digested PFGE DNA of pulsotypes A, B, and C showed hybridization bands at about >220 kb in clone A, about >300 kb in clone B, and about 220 kb in clone C, respectively (data not shown). These results are indicative of a chromosomal location for the blaOXA-23 gene.
Our study revealed that IRABs have resistance to most antimicrobial agents. Some studies have indicated that colistin may be useful for treating infections caused by carbapenem-resistant pathogens (14). We confirmed that in vitro, the most active agent against these resistant isolates was polymyxin E.
This is the first report to investigate IRABs in China on such a large scale. We found that clones A, B, C, and D were the dominant isolates and had spread widely among hospitals, cities, and even remote areas. Some isolates with the same genetic basis spread in many hospitals in China, suggesting that the spread of isolates plays an important role in the increase of IRABs. So, it is important to monitor and control the spread of A. baumannii having resistance to most β-lactams.
In China, blaOXA-23 and blaOXA-51-like were the most popular carbapenemase genotypes. OXA-23-producing IRABs spread widely in the world (1, 2, 7, 9, 16, 23, 24). blaOXA-51-like genes are the endogenous genes of A. baumannii, and their protein products have weak carbapenem-hydrolyzing activities (3, 8). An insertion of ISAba1 may provide a promoter for the OXA-type carbapenemase genes to increase their expression (20). There were only 15 isolates (4.39%) that lacked ISAba1 upstream of OXA-type carbapenemase coding genes, suggesting that an insertion of ISAbal may be a major factor responsible for the carbapenem resistance.
All isolates belonging to clones B, D, E, and F had a PCR mapping of type IV. In contrast, isolates of clones A and C and sporadic isolates had complex PCR mapping results (Table 2). In the process of the dissemination, the strains may have acquired or lost different gene elements under different antimicrobial pressures (15).
TABLE 2.
The distribution of carbapenemase genes in the major PFGE clones of IRABs
| Type | No. of strains of clone
|
No. of strains | ||||||
|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | Other | ||
| I | 1 | 2 | 8 | 11 | ||||
| II | 3 | 4 | 2 | 9 | ||||
| III | 4 | 4 | ||||||
| IV | 57 | 32 | 152 | 31 | 10 | 8 | 24 | 314 |
| V | 1 | 2 | 1 | 4 | ||||
| Total (no. of affected cities) | 62 (8) | 32 (3) | 160 (9) | 31 (2) | 10 (1) | 8 (1) | 39 (14) | 342 (16) |
Nucleotide sequence accession numbers.
Sequences for the 34 bp between ISAba1 and blaOXA-23 and the 7 bp between ISAba1 and blaOXA-66 have been submitted to GenBank under accession numbers DQ923478 (26) and DQ923479, respectively.
Acknowledgments
This work was supported by a research grant from the National Natural Science Foundation of China (no. NSFC30370073), a grant from High Technology Research and Development Program of China (863 program, no. 2006AA02Z413), and a grant from the Program for New Century Excellent Talents in University (no. NCET-04-0552).
Footnotes
Published ahead of print on 17 October 2007.
REFERENCES
- 1.Akalin, H., C. Ozakin, and S. Gedikoglu. 2006. Epidemiology of Acinetobacter baumannii in a university hospital in Turkey. Infect. Control Hosp. Epidemiol. 27:404-408. [DOI] [PubMed] [Google Scholar]
- 2.Boo, T. W., F. Walsh, and B. Crowley. 2006. First report of OXA-23 carbapenemase in clinical isolates of Acinetobacter species in the Irish Republic. J. Antimicrob. Chemother. 58:1101-1102. [DOI] [PubMed] [Google Scholar]
- 3.Brown, S., H. K. Young, and S. G. Amyes. 2005. Characterisation of OXA-51, a novel class D carbapenemase found in genetically unrelated clinical strains of Acinetobacter baumannii from Argentina. Clin. Microbiol. Infect. 11:15-23. [DOI] [PubMed] [Google Scholar]
- 4.Chang, H. C., Y. F. Wei, L. Dijkshoorn, M. Vaneechoutte, C. T. Tang, and T. C. Chang. 2005. Species-level identification of isolates of the Acinetobacter calcoaceticus-Acinetobacter baumannii complex by sequence analysis of the 16S-23S rRNA gene spacer region. J. Clin. Microbiol. 43:1632-1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2006. Performance standards for antimicrobial susceptibility testing; 16th informational supplement. CLSI document M100-S16. Clinical and Laboratory Standards Institute, Wayne, PA.
- 6.Coelho, J. M., J. F. Turton, M. E. Kaufmann, J. Glover, N. Woodford, M. Warner, M. F. Palepou, R. Pike, T. L. Pitt, B. C. Patel, and D. M. Livermore. 2006. Occurrence of carbapenem-resistant Acinetobacter baumannii clones at multiple hospitals in London and southeast England. J. Clin. Microbiol. 44:3623-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalla-Costa, L. M., J. M. Coelho, H. A. Souza, M. E. Castro, C. J. Stier, K. L. Bragagnolo, A. Rea-Neto, S. R. Penteado-Filho, D. M. Livermore, and N. Woodford. 2003. Outbreak of carbapenem-resistant Acinetobacter baumannii producing the OXA-23 enzyme in Curitiba, Brazil. J. Clin. Microbiol. 41:3403-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Héritier, C., L. Poirel, P. E. Fournier, J. M. Claverie, D. Raoult, and P. Nordmann. 2005. Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob. Agents Chemother. 49:4174-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeon, B. C., S. H. Jeong, I. K. Bae, S. B. Kwon, K. Lee, D. Young, J. H. Lee, J. S. Song, and S. H. Lee. 2005. Investigation of a nosocomial outbreak of imipenem-resistant Acinetobacter baumannii producing the OXA-23 beta-lactamase in Korea. J. Clin. Microbiol. 43:2241-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee, K., Y. S. Lim, D. Yong, J. H. Yum, and Y. Chong. 2003. Evaluation of the Hodge test and the imipenem-EDTA double-disk synergy test for differentiating metallo-β-lactamase-producing isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 41:4623-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee, K., J. H. Yum, D. Yong, H. M. Lee, H. D. Kim, J. D. Docquier, G. M. Rossolini, and Y. Chong. 2005. Novel acquired metallo-β-lactamase gene, blaSIM-1, in a class 1 integron from Acinetobacter baumannii clinical isolates from Korea. Antimicrob. Agents Chemother. 49:4485-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livermore, D. M. 2002. The impact of carbapenemases on antimicrobial development and therapy. Curr. Opin. Investig. Drugs 3:218-224. [PubMed] [Google Scholar]
- 13.Livermore, D. M., and N. Woodford. 2006. The beta-lactamase threat in Enterobacteriaceae, Pseudomonas and Acinetobacter. Trends Microbiol. 14:413-420. [DOI] [PubMed] [Google Scholar]
- 14.Nordmann, P., and L. Poirel. 2002. Emerging carbapenemases in gram-negative aerobes. Clin. Microbiol. Infect. 8:321-331. [DOI] [PubMed] [Google Scholar]
- 15.Peleg, A. Y., C. Franklin, J. M. Bell, and D. W. Spelman. 2006. Emergence of carbapenem resistance in Acinetobacter baumannii recovered from blood cultures in Australia. Infect. Control Hosp. Epidemiol. 27:759-761. [DOI] [PubMed] [Google Scholar]
- 16.Poirel, L., and P. Nordmann. 2006. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin. Microbiol. Infect. 12:826-836. [DOI] [PubMed] [Google Scholar]
- 17.Poirel, L., and P. Nordmann. 2006. Genetic structures at the origin of acquisition and expression of the carbapenem-hydrolyzing oxacillinase gene blaOXA-58 in Acinetobacter baumannii. Antimicrob. Agents Chemother. 50:1442-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seifert, H., and P. Gerner-Smidt. 1995. Comparison of ribotyping and pulsed-field gel electrophoresis for molecular typing of Acinetobacter isolates. J. Clin. Microbiol. 33:1402-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turton, J. F., M. E. Ward, N. Woodford, M. E. Kaufmann, R. Pike, D. M. Livermore, and T. L. Pitt. 2006. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol. Lett. 258:72-77. [DOI] [PubMed] [Google Scholar]
- 21.Villegas, M. V., and A. I. Hartstein. 2003. Acinetobacter outbreaks, 1977-2000. Infect. Control Hosp. Epidemiol. 24:284-295. [DOI] [PubMed] [Google Scholar]
- 22.Walther-Rasmussen, J., and N. Hoiby. 2006. OXA-type carbapenemases. J. Antimicrob. Chemother. 57:373-383. [DOI] [PubMed] [Google Scholar]
- 23.Wilks, M., A. Wilson, S. Warwick, E. Price, D. Kennedy, A. Ely, and M. R. Millar. 2006. Control of an outbreak of multidrug-resistant Acinetobacter baumannii-calcoaceticus colonization and infection in an intensive care unit (ICU) without closing the ICU or placing patients in isolation. Infect. Control Hosp. Epidemiol. 27:654-658. [DOI] [PubMed] [Google Scholar]
- 24.Ying, C. M., T. K. Ling, C. C. Lee, and J. M. Ling. 2006. Characterization of carbapenem-resistant Acinetobacter baumannii in Shanghai and Hong Kong. J. Med. Microbiol. 55:799-802. [DOI] [PubMed] [Google Scholar]
- 25.Yu, Y. S., Q. Yang, X. W. Xu, H. S. Kong, G. Y. Xu, and B. Y. Zhong. 2004. Typing and characterization of carbapenem-resistant Acinetobacter calcoaceticus-baumannii complex in a Chinese hospital. J. Med. Microbiol. 53:653-656. [DOI] [PubMed] [Google Scholar]
- 26.Zhou, H., B. R. Pi, Q. Yang, Y. S. Yu, Y. G. Chen, L. J. Li, and S. S. Zheng. 2007. Dissemination of imipenem-resistant Acinetobacter baumannii strains carrying the ISAba1-blaOXA-23 genes in a Chinese hospital. J. Med. Microbiol. 56:1076-1080. [DOI] [PubMed] [Google Scholar]