Abstract
The use of whole-genome microarrays for monitoring mutagenized or otherwise engineered genetic derivatives is a potentially powerful tool for checking genomic integrity. Using comparative genomic hybridization of a number of unrelated, directed deletion mutants in Escherichia coli K-12 MG1655, we identified unintended secondary genomic deletions in the flhDC region in Δfnr, Δcrp, and ΔcreB mutants. These deletions were confirmed by PCR and phenotypic tests. Our findings show that nonmotile progeny are found in some MG1655 directed deletion mutants, and studies on the effects of gene knockouts should be viewed with caution when the mutants have not been screened for the presence of secondary deletions or confirmed by other methods.
Many studies on bacterial gene function or regulation are based on the comparison between a wild-type strain and a mutant strain, where the mutant has an inactivated or deleted gene but is otherwise assumed to be isogenic with the wild type. In Escherichia coli K-12 and other enteric bacteria, comparative studies between wild-type and mutant strains have been facilitated by the development of rapid methods for generating precise deletions, particularly λ-Red-mediated gene replacement techniques (8, 16, 27), but there has been no simple way of confirming that no unintended deletions had occurred during the mutagenesis procedure. Further, many mutants created in the past by a variety of methods remain in use, sometimes after extensive serial subculture but without verifying their genomic integrity by newer technologies.
Flagellum-mediated motility and its regulation have been well characterized in E. coli and other enteric bacteria and are regulated in E. coli K-12 by the activator complex encoded by flhDC (2, 15). The FlhDC heterotetramer regulates the synthesis and assembly of flagella, and motility is a phenotype that is crucial to fitness in many environments: it is characteristic of the vast majority of E. coli wild isolates, and yet it carries a heavy penalty in energy demands. Motility appears to be subject to counterselection in some biological contexts, for example, in the emergence of nonmotile Shigella spp. in more than one distinct lineage within the E. coli-Shigella phylogenetic cluster (12) and of nonmotile strains of enterohemorrhagic E. coli O157. A recent report also indicates that spontaneous flhDC mutants in K-12 MG1655 are advantaged in colonization of the mouse intestine (14). In contrast, an enterohemorrhagic E. coli strain with a defective flhC gene was impaired in colonization of the bovine digestive tract (9). The expression of flagella is important in avian colonization by O157 strains (3), while it is a hindrance in infections of pigs (4). These reports support the concept that the motility regulon may in some circumstances be unstable and subject to selective pressure and may be particularly liable to deletion when cells are stressed. Alternatively, spontaneous deletions of the flagellar regulon may be an advantage to cells under certain conditions. In addition to the flagellar regulon, FlhD is involved in the regulation of 29 operons of known function in E. coli K-12, including genes regulated by Aer involved in anaerobic metabolism and the Entner-Doudoroff pathway (22).
Comparative genomic hybridization (CGH) of E. coli strains using microarrays is a powerful tool to compare the gross genetic differences between strains and has been used to analyze evolutionary changes (26), determine the relationships between pathogenic E. coli and Shigella strains (10), characterize probiotic E. coli strains (11), and define the genome of the E. coli laboratory strain MC4100 in relation to the genome-sequenced K-12 strain MG1655 (19). CGH also has the potential to be a useful tool for characterizing bacterial gene knockout strains and bacterial subpopulations that may arise within cultures because of spontaneous deletions.
We report here the use of CGH to characterize genetically a series of regulatory gene deletion mutants we had made in E. coli K-12 MG1655 using the λ-Red method of Datsenko and Wanner (8). In each case the mutants had been verified by PCR using primers that anneal to DNA sequences on either side of the gene that had been replaced by an antibiotic resistance cassette. We describe the use of CGH to validate gene deletion mutants and describe the secondary gross deletions in the flagellar regulon that we detected in some knockout strains, which were confirmed by PCR and motility tests.
MATERIALS AND METHODS
Gene knockouts in E. coli K-12 MG1655 (CGSC 7740).
The genome sequenced strain of MG1655, CGSC7740 (5) from the E. coli Stock Center (http://cgsc.biology.yale.edu/cgsc.html), was used throughout the present study. Gene knockouts in fnr (fumarate and nitrate reductase regulator) (7), crp (cyclic AMP receptor protein) (28), rpoS (general stress response sigma factor), creB (carbon source responsive response regulator) (1), fur (ferric uptake regulator), and gadA (glutamate decarboxylase) were made in MG1655 by using the λ-Red method of Datsenko and Wanner (8), wherein genes were replaced by chloramphenicol or kanamycin resistance cassettes from pKD3 or pKD4, respectively (8). Chloramphenicol- or kanamycin-resistant colonies were screened by PCR for replacement of the wild-type chromosomal gene by the antibiotic resistance cassette by using primers that flanked the gene that was replaced. Colonies were picked into 100 μl of sterile distilled H2O and heated to 100°C for 5 min. Cell lysates were centrifuged for 2 min at 13,000 × g, and 1 μl of the supernatant was added to 45 μl of ABgene 1.1x Reddymix PCR mix (ABgene, Epsom, United Kingdom) containing appropriate screening primers at a final amount of 20 pmol. The reaction volume was made up to 50 μl with sterile distilled water. The PCR cycling conditions used were 94°C for 4 min, followed by 30 cycles of 94°C for 1 min and 57°C for 1 min, followed by 72°C for 2 min. A final cycle of 72°C for 10 min completed any partial extension reactions. The deletion strains constructed and oligonucleotide primers used for mutagenesis and screening for gene loss are shown in Table 1.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides used in this studya
| E. coli strain, plasmid, or oligonucleotide | Sequence (5′-3′) | Source or reference |
|---|---|---|
| Strains | ||
| MG1655 (CGSC7740) | 5 | |
| MG1655 (Δcrp) | 28 | |
| MG1655 (Δcrp ΔflhDC) | This study | |
| MG1655 (Δfnr) | 7 | |
| MG1655 (Δfnr ΔflhDC) | This study | |
| MG1655 (Cet2) | Cariss and Avison, unpublished | |
| MG1655 (Cet2 ΔcreB) | This study | |
| MG1655 (Cet2 ΔcreB ΔflhDC) | This study | |
| MG1655 (ΔrpoS) | 18 | |
| MG1655 (ΔgadA) | This study | |
| MG1655 (Δfur) | This study | |
| Plasmids | ||
| pKD46 | 8 | |
| pKD3 | 8 | |
| pKD4 | 8 | |
| Oligonucleotides | ||
| Mutagenesis | ||
| CRP P1 | GCTCTGGAGAAAGCTTATAACAGAGGATAACCGCGCGTGTAGGCTGGAGCTGCTTC | 28 |
| CRP P2 | TGGCGCGCTACCAGGTAACGCGCCACTCCGACGGGACATATGAATATCCTCCTTAG | 28 |
| K12-FNR1 | AAATTGACAAATATCAATTACGGCTTGAGCAGACCTTGTAGGCTGGAGCTGCTTCG | 7 |
| K12-FNR2 | TGATATGACAGAAGGATAGTGAGTTATGCGGAAAAACATATGAATATCCTCCTTAG | 7 |
| K12-creB1 | CCTGTCATGCCGTGGCGGCAATAACAGAGGCGATTTGTGTAGGCTGGAGCTGCTTC | |
| K12-creB2 | AAAAATAGCCCAGCAACAACCGCATGCCGATACGCACATATGAATATCCTCCTTAG | |
| K12-rpoS2 | GGCCAGCCTCGCTTGAGACTGGCCTTTCTGACAGATGCTTACTTAGTGTAGGCTGGAGCTGCTTCGAA−3′ | |
| K12-rpoS1 | GGCTTTTGCTTGAATGTTCCGTCAAGGGATCACGGGTAGGAGCCACCTTCATATGAATATCCTCCTTAGTTCCT | |
| K12-gadA2 | GTTTTTTTTAAAGGCTGGGCATTCGGTTTTTACAACGTTATGTTATCAGTGTAGGCTGGAGCTGCTTCG−3′ | |
| K12-gadA1 | CTTCCATTGCGGATAAATCCTACTTTTTTATTGCCTTCAAATAAATTTAAGGAGTTCGAACATATGAATATCCTCCTTAGTTCC | |
| K12-fur1 | ATGAAGTGAACCGCTTAGTAACAGGACAGATTCCGCTGTAGGCTGGAGCTGCTTCG | |
| K12-fur2 | CCAACCCGCAGGTTGGCTTTTCTCGTTCAGGCTGGCCATATGAATATCCTCCTTAG | |
| Screening | ||
| CRP screen 1 | GGATGCTACAGTAATACATTGATG | 28 |
| CRP screen 2 | GACCGAATCGTAATTCGCCAAG | 28 |
| FNR screen 1 | TGGTTATTGCGCCATGAAGG | 7 |
| FNR screen 2 | TGGTTGGTCGTCCTGGTTAG | 7 |
| creB screen 1 | ACGGCAAAGCTCAGGGCGAG | |
| creB screen 2 | GCAACGTTGCGGTGTCGATC | |
| rpoS screen 1 | GCCTGCACAAAATTCCAC | |
| rpoS screen 2 | CGGATTCTTAATTACCTGG | |
| gadA screen1 | GTTTGGGCGATTTTTATTACG | |
| gadA screen2 | GCACTGTAATTTCCATTAGCG | |
| yecM forward | GCGGGAGTATACTGCAAATG | |
| yecM reverse | CGTTAGTGCACATTCCATGC | |
| yecT forward | GCGGGAGTATACTGCAAATG | |
| yecT reverse | CGTTAGTGCACATTCCATGC | |
| flhB forward | CGACAAACGCACACTATGCT | |
| flhB reverse | AGCAACACCATGATCGACAA | |
| cheZ forward | GGCTATGTGGTGAAGCCATT | |
| cheZ reverse | AGCATAGTGTGCGTTTGTCG | |
| tap forward | TCACCAATAAACCGCAAACA | |
| tap reverse | CATGTCGCGTTTATGGTCAG | |
| tar forward | AAATGAACCCGATGATCTGC | |
| tar reverse | AAATGCCCAAAAAGACATGC | |
| cheW forward | ATTTCTGCTGCGACCATTCT | |
| cheW reverse | GCCGCGAAATTGAAATAAAA | |
| cheA forward | CATCAGCCTGCTGGTACTGA | |
| cheA reverse | GTTCGCAATCCGTGTTACCT | |
| motA forward | AACATTCCAGCAGCGGTAAC | |
| motA reverse | TCAGCCACATCACCAGAAAA | |
| motB forward | GCTCTATTCCAGCGAACGTC | |
| motB reverse | GCTGAAGCCAAAAGTTCCTG | |
| motA forward | AACATTCCAGCAGCGGTAAC | |
| motA reverse | TCAGCCACATCACCAGAAAA | |
| flhC forward | GGTTAAGCTGGCAGAAACCA | |
| flhC reverse | CCACTGTTGACCATGACAGG | |
| flhD forward | CCAAAAAGGTGGTTCTGCTT | |
| flhD reverse | AACCAGTCGGTTGAGAATGG | |
| insB_5 forward | GCTCCAGTGGCTTCTGTTTC | |
| insB_5 reverse | TCGACGCAACTGTACTCGTC | |
| insA_5 forward | GCTTGCTAGCGTAGCGAAAA | |
| insA_5 reverse | CGTCTTCCGGAGACTGTCAT | |
| yecG forward | GATTGCGTCGTTTCTCCATT | |
| yecG reverse | GGCAGGAGTGCTTCATTAGC | |
| otsA forward | GGATGTCTGGAGCTGGCTTG | |
| otsA reverse | CAAGTGAGGTCGATGTGCTG | |
| otsB forward | CCATAACGGTTGGCTGTTC | |
| otsB reverse | TCCTCATTCCCTGTTTCACC |
The strains, plasmids, and PCR primers used in this study are listed. Regions underlined in the mutagenesis primers correspond to homology to genomic DNA sequences flanking genes that were deleted; nonunderlined regions are homologous to the pKD3 and pKD4 antibiotic resistance cassettes (8).
Genomic DNA preparations.
Cultures (20 ml) of MG1655 wild-type and mutant strains were grown in LB broth (23) with appropriate antibiotic selection at 37°C to an optical density at 600 nm (OD600) of 1.7 to 1.8. Ten milliliters of the cultures were harvested by centrifugation at 6,000 × g for 10 min. Total DNA was prepared by using the QIAGEN Genomic DNA buffer set (QIAGEN, Crawley, United Kingdom) and Genomic Tip 500/G columns according to the manufacturer's protocols, except that the total DNA precipitated by isopropanol was washed three times with 70% (vol/vol) of cold ethanol to remove any residual salt before the material was dried and resuspended in QIAGEN EB buffer. Total DNA was sheared by repeated passage through a 19G sterile needle and quantified by using a Nanodrop ND1000 microspectrophotometer (Nanodrop Technologies, Inc.). DNA purity was tested by digestion using the NaCl-sensitive restriction endonuclease HindIII.
CGH microarray experiments.
Microarrays were printed using an Operon Array Ready 70-mer E. coli oligonucleotide set version 1.0 (Operon Biotechnologies, Cologne, Germany) and processed as previously described (7). Total DNA from each strain (5 μg) was labeled in a 50-μl reaction mixture with either Fluorolink Cy3 or Cy5 d-CTP (GE-Amersham, Little Chalfont, United Kingdom) in a reaction containing 60 ng of random hexamers (Invitrogen, Paisley, United Kingdom)/μl; 0.1 mM dA, dG, and dT NTPs; 0.04 mM dCTP (Bioline, London, United Kingdom); 50 U of Klenow exo− fragment of DNA polymerase I; and 10× EcoPol buffer (New England Biolabs, Hitchin, United Kingdom). Total DNA was mixed with random hexamers and sterile filtered high-pressure liquid chromatography-grade water (VWR, Lutterworth, United Kingdom) to a volume of 41.5 μl and then heated to 95°C for 5 min before rapid cooling on ice and brief centrifugation at 13,000 × g in a microcentrifuge. The remaining components were added, and the reaction was incubated overnight in the dark at 37°C. Cy dye-labeled total DNA was purified by using a QIAGEN QIAquick PCR purification kit according to the manufacturer's protocol and eluted in sterile high-pressure liquid chromatography-grade water before quantification of the DNA concentration and Cy dye incorporation using a Nanodrop microspectrophotometer.
Next, 80 pmol of Cy3-labeled total DNA from wild-type MG1655 was cohybridized with 80 pmol of Cy5-labeled total DNA from a mutant to the oligonucleotide arrays. Microarray slides were prehybridized, hybridized, washed, and scanned as previously described (7). The signal intensity from each printed feature on the array was quantified by using Genepix v5.0 software (Molecular Devices Corp. Sunnyvale, CA), and microarray data were analyzed by using Genespring software (v6.1; Agilent Instruments, South Queensferry, West Lothian, United Kingdom). Intensity-dependent LOWESS normalization was used to transform raw signal data in order to eliminate Cy dye bias, and spots with an intensity value lower than the cutoff value for the error model were filtered out. Transcriptomics experiments and analysis were carried out as previously described (7).
Motility tests.
The Δfnr, Δcrp, and ΔcreB mutants identified by CGH as having deletions in the flagellar regulon were tested for motility by using soft agar motility tests with point inoculation of a colony from an overnight plate culture of the strains into LB agar plates containing 0.35% (wt/vol) agar. The motility test plates were incubated at 37°C for 24 h, and the zone of bacterial growth was measured.
PCR confirmation of CGH data.
Secondary gross deletions identified by microarray CGH of the MG1655 mutants were confirmed by PCR of each individual gene, as described above. The primers shown in Table 1 were used to amplify the genomic DNA flanking the gross gene deletion in the Δfnr, Δcrp, and ΔcreB secondary deletion strains.
Data deposition.
The CGH data from these experiments is deposited under accession number GSE7695, and transcriptomics data for the nonmotile Δfnr mutant are available under accession number GSE3591, in the gene expression omnibus (GEO) at the National Center for Biotechnology Information (http:www.ncbi.nlm.nih.gov/geo).
RESULTS AND DISCUSSION
CGH of the MG1655 fnr mutants.
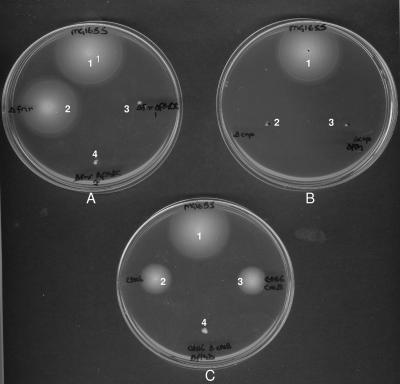
Three putative fnr deletion mutants were characterized by PCR using FNR primers A and B (Table 1). PCR confirmed that fnr had been replaced by the kanamycin resistance cassette from pKD4 in all three mutant strains (data not shown). The Δfnr mutants were each characterized by CGH against the MG1655 (CGSC7740) parental strain total DNA. The operon 70-mer oligonucleotide array set we used was designed to contain one oligonucleotide per gene and lacks the coverage to detect small changes in the genome, but it showed that in two of the mutants fnr and a cluster of eight other genes at a different location on the genome were deleted (Fig. 1), suggesting that these were possible siblings, while in the third mutant only fnr was missing. CGH data showed that the unintended deletion was contiguous and included the insB5 component of IS1, the flhDC master regulators of the flagella biosynthesis operon, motA, motB, cheA, cheW, and tar, indicating that regulation of the flagellar regulon had been lost. Soft agar motility tests of the MG1655 Δfnr mutants carrying the motility gene deletion confirmed that the strains were nonmotile, whereas wild-type MG1655 and the Δfnr mutant with no other deletions in it were motile (Fig. 2A).
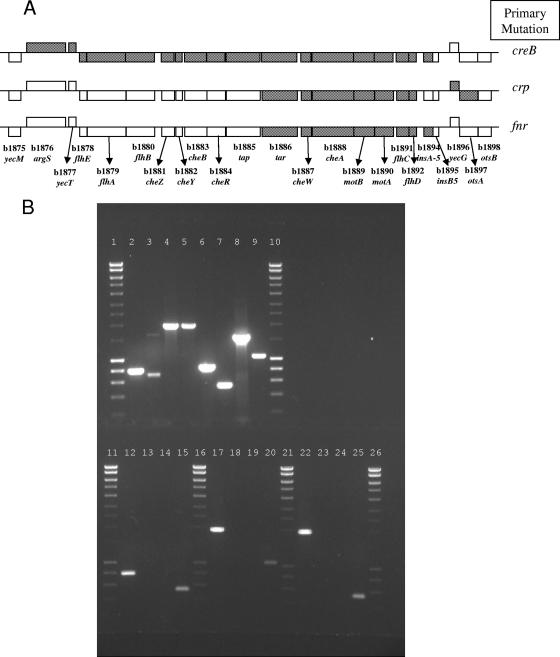
FIG. 1.
(A) Graphical representation of CGH data showing gene loss in the flhDC region for the nonmotile Δfnr, Δcrp, and ΔcreB mutants. Genes are denoted by their “b number” and name. Genes are denoted by boxes. Genes encoded on the top strand of the genome are shown above the line; those on the bottom strand are shown below the line. Gray-shaded boxes indicate a loss of signal in the mutant strain as determined by the CGH array. (B) Demonstration PCRs confirming gene presence or loss in the flhDC region for each mutant. No PCR product indicates a loss of all or part of the gene in the nonmotile mutants. Top row of the gel (MG1655; CGSC7740): lanes 1 and 10, DNA marker (ABgene Hyperladder I); lane 2, yecM; lane 3, yecT; lane 4, tap; lane 5, tar; lane 6, insB5; lane 7, insA5; lane 8, otsA; lane 9, otsB. Bottom row of the gel: lanes 11, 16, 21, and 26, DNA marker (ABgene Hyperladder I); lanes 12 to 15, nonmotile MG1655 ΔcreB mutant; lane 12, yecM; lane 13, yecT; lane 14, insB5; lane 15, insA5; lanes 17 to 20, nonmotile MG1655 Δcrp mutant; lane 17, tap; lane 18, tar; lane 19, otsA; lane 20, otsB; lanes 22 to 25, nonmotile MG1655 Δfnr mutant; lane 22, tap; lane 23, tar; lane 24, insB5; lane 25, insA5. For each mutant, a PCR for each deleted gene, as well as for genes flanking the deletion, was carried out to confirm the CGH data (data not shown).
FIG. 2.
Motility tests of wild-type MG1655 and deletion mutants where CGH had confirmed that flhDC were deleted. (A) Spots: 1, MG1655 (CGSC7740; wild type); 2, MG1655 (Δfnr); 3, MG1655 (Δfnr ΔflhDC clone 1); 4, MG1655 (Δfnr ΔflhDC clone 2). (B) Spots: 1, MG1655 (CGSC7740) (wild type); 2, MG1655 (Δcrp clone 1); 3, MG1655 (Δcrp ΔflhDC clone 1). (C) Spots: 1, MG1655 (CGSC7740, wild type); 2, MG1655 (Cet2); 3, MG1655 (Cet2 ΔcreB); 4, MG1655 (Cet2 ΔcreB ΔflhDC).
CGH and motility test data were confirmed by transcriptomics experiments (GEO accession number GSE3591), which showed that expression of the flagellar regulon was ablated in the mutant, and by attempting PCR amplification of the region identified by CGH as being deleted and of genes upstream and downstream of this region (Fig. 1A and B). PCR was used to attempt to amplify yecM, yecT, flhB, cheZ, cheY, tap, tar, cheA, cheW, motA, motB, flhC, flhD, insA5, insB5, yecG, otsA, and otsB from the chromosome of wild-type MG1655 and from the Δfnr mutant using primers detailed in Table 1. PCR products for each of the genes detailed above were detected in the wild-type MG1655 strain, as would be expected, but not in the Δfnr mutants where CGH had identified a deletion in the flagellum genes (Fig. 1A and B). In the Δfnr strains with the CGH detected secondary mutations, insB5, flhD, flhC, motA, motB, cheA, cheW, and tar did not amplify as determined by PCR, confirming the CGH results. The Δfnr mutant where only fnr had been deleted was used in transcriptomics experiments reported elsewhere (7).
Screening other gene knockouts in MG1655.
CGH of Δfur, ΔrpoS, and ΔgadA mutants made by using the Datsenko and Wanner method (8) showed that only the required gene had been deleted. However, in two other mutants we made, the Δcrp and ΔcreBmutants, CGH identified gross unintended secondary deletions. One of two Δcrp mutants had only crp deleted, while the other also contained additional deletions similar to those in the nonmotile fnr mutants, despite being made in different experiments and by different researchers. The secondary deletions in this Δcrp mutant were also centered on a contiguous tract of the chromosome from otsA to tar inclusive, amounting to 9.52 kb in total (Fig. 1A). PCR confirmed the CGH results except that CGH of the crp mutant indicated that insA5 and insB5 were present in the mutant, but PCR showed that they were absent. This inconsistency is probably due to cross-hybridization between multiple copies of Is1 found in the MG1655 genome and the insA5 and insB5 oligonucleotides in the operon array, although it is unclear why this phenomenon was not seen in the creB and fnr mutants. Neither of the Δcrp mutants (Fig. 2B) was motile, a finding in agreement with previous reports that CRP mutants are nonmotile (25), and this shows that in certain cases motility tests alone cannot be used to determine the loss of flagellar genes. The Δcrp mutant strain that did not contain secondary deletions was used for experiments published elsewhere (28).
A ΔcreB mutant, in a Cet2 (creC point mutant [see reference 1]) background (which was constructed independently by S. J. L. Cariss and M. B. Avison in Bristol [unpublished data]) was also shown by CGH to contain a secondary genome deletion. Although this secondary deletion was in approximately the same region of the chromosome as in the Δcrp and Δfnr mutants, it was more extensive, covering the region between insB5 and yecT/argS (approximately 15.5 to 17.2 kb) (Fig. 1A), and PCR confirmed the CGH result (Fig. 1B). In the ΔcreB mutant, the antibiotic resistance cassette replacing creB had been excised using FLP recombinase to negate polar effects on expression of the downstream creC observed if the cassette remained. flhD PCR analysis of mutants retained at each stage of the creB deletion procedure showed that deletion of the flagellar regulator had occurred during excision of the antibiotic resistance cassette rather than during λ-Red-mediated recombination (data not shown). This finding was in contrast to the Δfnr and Δcrp secondary deletion mutants which had both lost flhDC and contiguous genes at some point during λ-Red-mediated gene replacement: the antibiotic resistance cassette had not been excised from either of these strains. Another ΔcreB mutant that had not lost the flh region was made in the MG1655 Cet2 mutant background. Although the Cet2 mutant and this ΔcreB derivative were both reduced in motility compared to wild-type MG1655, motility was totally abolished in the ΔcreB mutant with the secondary deletion in the flhDC region (Fig. 2C).
The similarity of the secondary deletions in the Δfnr, Δcrp, and ΔcreB mutants may be a feature of an unstable genomic region in MG1655 centered around IS1 upstream of flhD (2) rather than a problem specifically associated with the mutagenesis technique we used and could be due to a low level of spontaneous deletions in the flhDC region in MG1655 cultures. The deletions we detected are highly similar to those reported by Leatham et al., who found that selection of spontaneous nonmotile MG1655 flhDC deletion mutants occurred under nutrient-limited conditions in the mouse intestine, and these mutants grew faster than wild-type MG1655 on several sole carbon sources, including d-gluconate, l-fucose, d-glucuronate, and d-mannose (14). All of the flagellar regulon mutants we detected were in directed mutants where genes involved in central metabolism control had been deleted and occurred before or during the λ-Red mutagenesis procedure (Δfnr and Δcrp) or after it (ΔcreB). Loss of motility may be an advantage to cells that are metabolically challenged by the loss of important metabolic regulators, since there is an energy penalty in flagellar production (14), and reports of additional regulation of metabolism by flhD (14, 22) have shown that the loss of this gene allows E. coli K-12 MG1655 cells to utilize a wider range of carbon sources than the wild-type strain can. The secondary gene deletions we detected could be spontaneous, but there is evidence that the λ-Red mutagenesis procedure causes unintentional genomic deletions or mutations (6, 17, 20, 21), and other gene deletion methods such as suicide vector-driven allelic exchange mutagenesis have resulted in a high frequency of secondary mutations in extraintestinal pathogenic E. coli strains (13), so there is a real need to validate mutants as thoroughly as possible. A commonly used strategy to avoid problems of unintended deletions or mutations in E. coli K-12 has been to transduce mutations into a wild-type strain using bacteriophage P1, but unless the original mutant genome has no other mutations in the region that will be transduced, this can result in the transduction of undetected secondary mutations into the new host, such as when a secondary mutation in the astC gene was cotransduced with a ynjB lesion and was detected by using phenotype arrays (6).
Use of single gene deletion mutants has been crucial to understanding gene function in E. coli and other bacterial strains because, provided there are no polar or other secondary effects, any phenotypic, proteomic, or transcriptomic differences in the mutants compared to the wild-type are specifically attributable to the targeted deletion (13). CGH shows that confirmation of a mutation by PCR screening or Southern hybridization is not sufficient for the validation of recombineered deletion mutants because both methods are used to detect whether the directed mutation has occurred and not whether there have been any other genomic changes. Oligonucleotide microarray CGH of deletion mutants is a powerful tool for detecting secondary gross deletions and has the advantage that it interrogates the whole genome, with a higher resolution, than do macrorestriction profiles and pulsed-field gel electrophoresis (13). Oligonucleotide arrays, like PCR arrays, are designed based on the genome sequence of the strain, but unlike PCR arrays oligonucleotide arrays are not dependent on amplifying the targets for printing on the array from the parental strain, which itself may contain unknown deletions (24). Oligonucleotide microarray CGH is therefore also a useful tool for confirming that the parental strain has not accumulated gross deletions during storage or growth. Although the oligonucleotide array we used lacks the coverage to detect small changes in the genome, higher-resolution “tiling” arrays, or complete genome resequencing, should allow researchers to identify small deletions or point mutations that are not detectable using the present lower-resolution methods.
We confirm here that there is genomic instability in the flhDC region of the MG1655 genome (14) and show that there were significant secondary deletions in several mutants we constructed, which if undetected could have led to incorrect assignment of regulator function in subsequent experiments.
Acknowledgments
This study was supported by grants EGA16107 and JIF13209 to Birmingham University and BB/C514266 to M.B.A. at Bristol University from the UK Biotechnology and Biological Sciences Research Council. G.A.H.-A. was supported by a Ph.D. scholarship from CONACYT (Mexico).
We thank Antony Jones from the School of Biosciences Functional Genomics laboratory for help in printing the microarrays.
Footnotes
Published ahead of print on 5 October 2007.
REFERENCES
- 1.Avison, M. B., R. E. Horton, T. R. Walsh, and P. M. Bennett. 2001. Escherichia coli CreBC is a global regulator of gene expression that responds to growth in minimal media. J. Biol. Chem. 276:26955-26961. [DOI] [PubMed] [Google Scholar]
- 2.Barker, C. S., B. M. Prüss, and P. Matsumura. 2004. Increased motility of Escherichia coli by insertion sequence element integration into the regulatory region of the flhD operon. J. Bacteriol. 186:7529-7537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Best, A., R. M. La Reggione, D. Clifford, W. A. Cooley, A. R. Sayers, and M. J. Woodward. 2006. A comparison of Shiga-toxin negative Escherichia coli O157 aflagellate and intimin deficient mutants in porcine in vitro and in vivo models of infection. Infect. Immun. 113:67-72. [DOI] [PubMed] [Google Scholar]
- 4.Best, A., R. M. La Reggione, A. R. Sayers, and M. J. Woodward. 2005. Role for flagella but not intimin in the persistent infection of the gastrointestinal tissues of specific-pathogen-free chicks by Shiga toxin-negative Escherichia coli O157:H7. Infect. Immun. 73:1836-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 6.Bochner, B. R., P. Gadzinski, and E. Panomitros. 2001. Phenotype microarrays for high-throughput phenotypic testing and assay of gene function. Genome Res. 11:1246-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Constantinidou, C., J. L. Hobman, L. Griffiths, M. D. Patel, C. W. Penn, J. A. Cole, and T. W. Overton. 2006. A reassessment of the FNR regulon and transcriptomics analysis of the effects of nitrate, nitrite, NarXL, and NarQP, as Escherichia coli K-12 adapts from aerobic to anaerobic growth. J. Biol. Chem. 281:4802-4815. [DOI] [PubMed] [Google Scholar]
- 8.Datsenko, K., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobbin, H. S., C. J. Hovde, C. J. Williams, and S. A. Minnich. 2006. The Escherichia coli O157 flagellar regulatory gene flhC and not the flagellin gene fliC impacts colonization of cattle. Infect. Immun. 74:2894-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukiya, S., H. Mizoguchi, T. Tobe, and H. Mori. 2004. Extensive genomic diversity in pathogenic Escherichia coli and Shigella strains revealed by comparative genomic hybridization microarray. J. Bacteriol. 186:3911-3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grozdanov, L., C. Raasch, J. Schulze, U. Sonnenborn, G. Gottschalk, J. Hacker, and U. Dobrindt. 2004. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J. Bacteriol. 186:5432-5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson, J. 2002. Evolution of pathogenic Escherichia coli, p. 55-77. In M. S. Donnenberg (ed.), Escherichia coli: virulence mechanisms of a versatile pathogen. Academic Press, Inc., San Diego, CA.
- 13.Johnson, J. R., H. A. Lockman, K. Owens, S. Jelacic, and P. I. Tarr. 2003. High-frequency secondary mutations after suicide-driven allelic exchange mutagenesis in extraintestinal pathogenic Escherichia coli. J. Bacteriol. 185:5301-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leatham, M. P., S. J. Stevenson, E. J. Gauger, K. A. Krogfelt, J. J. Lins, T. L. Haddock, S. M. Autieri, T. Conway, and P. S. Cohen. 2005. Mouse intestine selects nonmotile flhDC mutants of Escherichia coli MG1655 with increased colonizing ability and better utilization of carbon sources. Infect. Immun. 73:8039-8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu, X., and P. Matsumura. 1994. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J. Bacteriol. 176:7345-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy, K. C. 1998. Use of bacteriophage λ recombination functions to promote gene replacement in Escherichia coli. J. Bacteriol. 180:2063-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy, K. C., and K. G. Campellone. 2003. Lambda Red-mediated recombinogenic engineering of enterohemorrhagic and enteropathogenic Escherichia coli. BMC Mol. Biol. 4:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perni, S., G. Sharma, J. L. Hobman, P. A. Lund, C. J. Kershaw, G. A. Hidalgo-Arroya, C. W. Penn, X. T. Deng, J. L. Walsh, and M. G. Kong. 2007. Probing bactericidal mechanisms induced by cold atmospheric plasmas with Escherichia coli mutants. Appl. Phys. Lett. 90:7392. [Google Scholar]
- 19.Peters, J. E., T. E. Thate, and N. L. Craig. 2003. Definition of the Escherichia coli MC4100 genome by use of a DNA array. J. Bacteriol. 185:2017-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poteete, A. R., A. C. Fenton, and A. Nadkarni. 2004. Chromosomal duplications and cointegrates generated by the bacteriophage lambda Red system in Escherichia coli K-12. BMC Mol. Biol. 5:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poteete, A. R., H. R. Wang, and P. L. Foster. 2002. Phage λ Red-mediated adaptive mutation. J. Bacteriol. 184:3753-3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prüss, B. M., J. W. Campbell, T. K. Van Dyck, C. Zhu, Y. Kogan, and P. Matsumura. 2003. FlhD/FlhC is a regulator of anaerobic respiration and the Entner-Doudoroff pathway through induction of the methyl-accepting chemotaxis protein Aer. J. Bacteriol. 185:534-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.Soupene, E., W. C. van Heeswijk, J. Plumbridge, V. Stewart, D. Bertenthal, H. Lee, G. Prasad, O. Paliy, P. Charernnoppakul, and S. Kustu. 2003. Physiological studies of Escherichia coli strain MG1655: growth defects and apparent cross-regulation of gene expression. J. Bacteriol. 185:5611-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soutourina, O., A. Kolb, E. Krin, C. Laurent-Winter, S. Rimsky, A. Danchin, and P. Bertin. 1999. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J. Bacteriol. 181:7500-7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wick, L. M., W. Qi, D. W. Lacher, and T. S. Whittam. 2005. Evolution of genomic content in the stepwise emergence of Escherichia coli O157:H7. J. Bacteriol. 187:1783-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu, D., H. M. Ellis, E.-C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng, D., C. Constantinidou, J. L. Hobman, and S. D. Minchin. 2004. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 32:5874-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]