Abstract
Mycobacterium tuberculosis utilizes the methylerythritol phosphate (MEP) pathway for biosynthesis of isopentenyl diphosphate and its isomer, dimethylallyl diphosphate, precursors of all isoprenoid compounds. This pathway is of interest as a source of new drug targets, as it is absent from humans and disruption of the responsible genes has shown a lethal phenotype for Escherichia coli. In the MEP pathway, 4-diphosphocytidyl-2-C-methyl-d-erythritol is formed from 2-C-methyl-d-erythritol 4-phosphate (MEP) and CTP in a reaction catalyzed by a 4-diphosphocytidyl-2-C-methyl-d-erythritol synthase (IspD). In the present work, we demonstrate that Rv3582c is essential for M. tuberculosis: Rv3582c has been cloned and expressed, and the encoded protein has been purified. The purified M. tuberculosis IspD protein was capable of catalyzing the formation of 4-diphosphocytidyl-2-C-methyl-d-erythritol in the presence of MEP and CTP. The enzyme was active over a broad pH range (pH 6.0 to 9.0), with peak activity at pH 8.0. The activity was absolutely dependent upon divalent cations, with 20 mM Mg2+ being optimal, and replacement of CTP with other nucleotide 5′-triphosphates did not support activity. Under the conditions tested, M. tuberculosis IspD had Km values of 58.5 μM for MEP and 53.2 μM for CTP. Calculated kcat and kcat/Km values were 0.72 min−1 and 12.3 mM−1 min−1 for MEP and 1.0 min−1 and 18.8 mM−1 min−1 for CTP, respectively.
Despite improvements in chemotherapeutics with supervised administration of an isoniazid, rifampin, and pyrazinamide drug regimen (10), tuberculosis (TB) remains one of the most prevalent infectious diseases worldwide, responsible for a total of 1.6 million deaths annually. The WHO recently reported that there are an estimated 8.8 million new TB cases per year, even though the TB incidence is stable or in decline globally. However, the total number of new TB cases is still rising slowly (41). In addition, present estimates indicate that 3.2% of the new cases of TB per annum are multidrug-resistant TB, showing resistance to both isoniazid and rifampin, the two first-line drugs (22).
In this context, isoprenoid synthesis is being studied with the goal of identifying new drug targets. In Mycobacterium tuberculosis, isoprenoids play diverse, essential roles. For example, polyprenyl phosphate acts as a carrier of activated sugar in the biosynthesis of the arabinan portion of arabinogalactan, arabinomannan, and lipoarabinomannan (40), the “linker unit” of mycobacterial arabinogalactan (21), and of lipid I and lipid II, which are fundamental to peptidoglycan synthesis (19). In addition, the side chain of menaquinone, the only lipoquinone in the electron transport chain in M. tuberculosis, is derived from a polyprenyl diphosphate molecule (32, 35). Recent reports have shown that both NADH type II dehydrogenase and F1F0 ATP synthase are effective drug targets in drug-resistant bacilli (1, 39), and it has been shown that the bactericidal effects of pyrazinamide are due to interference in the maintenance of a mycobacterial transmembrane proton gradient, resulting in depletion of energy (42). Therefore, it is reasonable to predict that the enzymes involved in the early steps of biosynthesis of essential isoprenoids provide new and valid drug targets.
Although they may be structurally and functionally complex, all isoprenoid compounds of eubacteria are generated from two simple five-carbon precursors, namely, isopentenyl diphosphate (IPP) and its isomer, dimethylallyl diphosphate (DMAPP) (4, 29). Until recently, these precursors were thought to be synthesized exclusively through the mevalonate pathway (11), as seen in animals. However, intensive work with Escherichia coli revealed the existence of an alternative means of synthesis, the methylerythritol phosphate (MEP) pathway (3, 13, 16, 18, 30), which is now accepted as the only source of IPP and DMAPP in many eubacteria, including M. tuberculosis (6). The hypothesis that this pathway contains attractive drug targets is supported by the observation that disruption of any step in the MEP pathway is lethal for E. coli (14, 37), but the essentiality of IspD has not been demonstrated previously for a gram-positive organism.
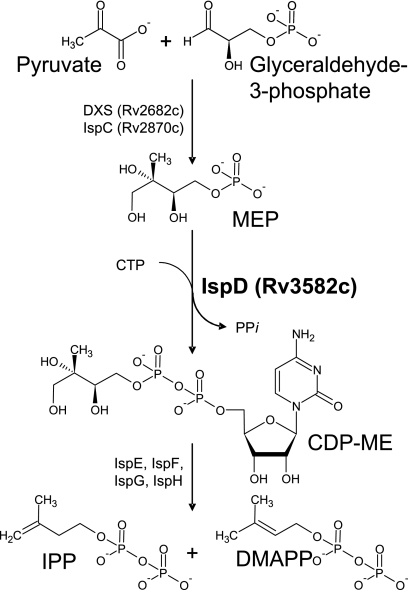
In the MEP pathway, IPP and DMAPP are produced by a series of catalytic reactions, starting with condensation of glyceraldehyde 3-phosphate and pyruvate (Fig. 1). The first two enzymes of this pathway in M. tuberculosis, 1-deoxy-d-xylulose 5-phosphate synthase (DXS) and 1-deoxy-d-xylulose 5-phosphate reductoisomerase (IspC), have been characterized previously (2, 5, 12). The product of IspC is 2-C-methyl-d-erythritol 4-phosphate (MEP), which provides the substrate for 4-diphosphocytidyl-2-C-methyl-d-erythritol (CDP-ME) synthase (IspD). In this step, IspD catalyzes transfer of the CMP moiety of CTP to MEP, producing CDP-ME, with the corresponding release of inorganic pyrophosphate (Fig. 1). Although IspD was predicted to be essential for M. tuberculosis, the preceding enzyme in the MEP pathway (IspC) was not (33). Therefore, the essentiality of this enzyme was determined for M. tuberculosis, the ispD gene was cloned and expressed, and the enzyme was characterized.
FIG. 1.
MEP pathway. The reaction catalyzed by CDP-ME synthase (IspD) is highlighted. In M. tuberculosis, IPP and DMAPP are synthesized through the activities of a cascade of enzymes (DXS through IspH). DXS, 1-deoxy-d-xylulose 5-phosphate synthase; IspC, 1-deoxy-d-xylulose 5-phosphate reductoisomerase; IspD, 4-diphosphocytidyl-2-C-methyl-d-erythritol synthase; IspE, 4-diphosphocytidyl-2-C-methyl-d-erythritol kinase; IspF, 2-C-methyl-d-erythritol 2,4-cyclodiphosphate synthase; IspG, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate synthase; IspH, 1-hydroxy-2-methyl-2-(E)-butenyl 4-diphosphate reductase. Identification numbers have been included for M. tuberculosis enzymes for which there is experimental evidence of activity.
MATERIALS AND METHODS
Materials.
H37Rv genomic DNA was provided by Colorado State University. All PCR reagents and cloning materials were purchased from Qiagen. [γ-32P]CTP (25 Ci/mmol) was purchased from ICN or synthesized enzymatically as previously described (9). MEP was purchased from Echelon Research Laboratories, Inc. (Salt Lake City, UT). All other chemicals used were at least analytical grade and were obtained from Sigma-Aldrich.
PCR amplification and cloning of the Rv3582c gene from M. tuberculosis.
The Rv3582c gene was amplified using oligonucleotide primers designed from sequences available in Tuberculist (http://genolist.pasteur.fr/TubercuList/). The oligonucleotides used were IspDFor and IspDRev, containing NdeI and XhoI restriction enzyme sites, respectively (Table 1). The PCR products were digested with NdeI and XhoI and ligated into pET28a(+) vector (EMD Biosciences, Inc.) that had been digested with the same enzymes and subsequently purified. Ligation mixtures were used to transform E. coli DH5α cells (Life Technologies), creating DH5α[pET28a(+)::Rv3582c], in which the target construct [pET28a(+)::Rv3582c] was propagated. The plasmid was isolated using Qiagen plasmid miniprep kits and sequenced by Macromolecular Resources, Colorado State University.
TABLE 1.
PCR primers used in this study
| Primer name | Sequencea | Restriction enzyme |
|---|---|---|
| IspDFor | CATATGAGGGAAGCGGGCGAAGTAG | NdeI |
| IspDRev | CTCGAGTCACCCGCGGAGTATAGCTTG | XhoI |
| IspDNFor | AAGCTTAGCATCCCGCATGCGGGCAGT | HindIII |
| IspDNRev | GGATCCGAGAGTCTGCCCGTCGAGCTG | BamHI |
| IspDCFor | GGATCCACCACCAAACTGGATCTGTTGC | BamHI |
| IspDCRev | GGTACCGGCGATTTCGTTCTCATGATCCG | KpnI |
| IspDint1 | CTCCGACCTTGAAAATCATCT | |
| IspDint2 | TACGACGTCGCGTTTATCC | |
| IspD/FCompPro | TTAATTAAGGCCGGCTGTTAGCATGGAGTAACG | PacI |
| IspD/FCompRev | TTAATTAAGCCAGCTTACCTGCCCAATTGCTG | PacI |
| IspD/FCompSh | TTAATTAAGACGCCAAAGCCGAGACCATCCTT | PacI |
| IspD US Probe F | GACGAGAATCAATGAGACCT | |
| IspD US Probe Rev | AGTGATATCGGCTCGGTGAC |
Restriction sites are underlined.
Expression and purification of recombinant Rv3582c.
Recombinant Rv3582c was expressed and purified as previously described (5, 17). Briefly, transformation of BL21(DE3) (Novagen) with pET28a(+)::Rv3582c afforded the recombinant strain BL21(DE3)[pET28a(+)::Rv3582c]. Protein expression was induced in the presence of 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 20°C for 10 h. The recombinant protein, carrying a His6 tag, was purified by immobilized metal-affinity chromatography, using a linear gradient of 50 to 200 mM imidazole in washing buffer (50 mM 4-morpholinepropanesulfonic acid [MOPS] [pH 7.9], 1 mM MgCl2, 10% glycerol, and 1 mM β-mercaptoethanol). Eluted fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting and visualized with Coomassie brilliant blue 250R and an anti-His antibody (Sigma-Aldrich), respectively. Fractions containing recombinant Rv3582c estimated to be at least 95% pure by Coomassie staining of sodium dodecyl sulfate-polyacrylamide gels were pooled, desalted on a PD-10 column (Millipore), and stored at −70°C.
Assay for IspD activity.
IspD activity was monitored by 32PPi release, employing liquid scintillation spectrometry. Reaction mixtures contained 50 mM Tris-HCl (pH 7.9), 20 mM sodium fluoride, 10 mM MgCl2, 1 mM dithiothreitol, 38.5 pmol of purified IspD enzyme, and MEP and [γ-32P]CTP (10 dpm/pmol) at the indicated final concentrations in a final volume of 100 μl. Incubations were carried out at 37°C for 30 min. Reactions were terminated by adding 200 μl of a slurry of activated charcoal in 10 mM Tris-HCl buffer (pH 8.0). The slurry was then loaded into an empty spin column and centrifuged at 5,000 × g for 3 min to elute 32PPi, and residual [γ-32P]CTP was retained on the charcoal.
Determination of enzymatic properties of Rv3582c.
To determine the optimal pH for enzyme activity, reaction mixtures at various pH values containing pH-appropriate buffers (morpholineethanesulfonic acid [MES], MOPS, Tris, or [(2-hydroxy-1,1-bis(hydroxymethyl)ethyl)amine]-1-propanesulfonic acid [TAPS]) were used. Optimal concentrations for divalent cations were determined in assay mixtures containing MgCl2, MnCl2, CaCl2, or ZnCl2 at the indicated concentrations. The effect of CTP concentration on activity was determined using a constant concentration of MEP (100 μM) and various concentrations of CTP. The effect of MEP concentration was determined using a constant concentration of CTP (100 μM) and various concentrations of MEP. The Km and Vmax values of the enzyme for different substrates were calculated by nonlinear regression analysis (GraFit 5.0.13; Erithacus Software Ltd.).
Determination of Rv3582c essentiality by gene switching analysis.
Previously published methods were employed to determine the essentiality of the Rv3582c gene (23, 25, 36). A deletion delivery vector was constructed. The primer pairs IspDNFor-IspDNRev and IspDCFor-IspDCRev were used to amplify the regions to either side of Rv3582c and to introduce relevant restriction enzyme sites (Table 1). PCR products were cloned into pCR-Blunt II-TOPO (Invitrogen). The fragments were then cloned as HindIII-BamHI and BamHI-KpnI fragments into p2NIL (25), and the marker gene from pGOAL19 was cloned into the PacI site to generate the final delivery vector, in which 537 bp of Rv3582c was deleted. Single crossovers were generated by electroporation into M. tuberculosis and selection on 100 μg/ml hygromycin, 20 μg/ml kanamycin, and 50 μg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). A single colony was selected and streaked out in the absence of any antibiotics to allow the second crossover event to occur. Double-crossover colonies were selected on 2% (wt/vol) sucrose and 50 μg/ml X-Gal; white colonies were patch tested for kanamycin and hygromycin sensitivity to ensure that they had lost the plasmid by recombination. PCR was then used to determine whether each double-crossover product had the wild-type or deletion allele. Primers used were IspDint1 and IspDint2 (Table 1), which amplify 2.0-kbp and 1.3-kbp fragments from the wild-type and deletion alleles, respectively. Two merodiploid strains carrying an extra copy of Rv3582c were constructed using an L5 mycobacteriophage-derived vector, which integrates into the attB site via site-specific recombination with attP mediated by the L5 integrase, as follows. The primer pairs IspD/FCompPro-IspD/FCompRev and IspD/FCompSh-IspD/FCompRev were used to amplify Rv3582c and the regions to either side and to introduce PacI sites (Table 1). PCR products were ligated into pCR-Blunt II-TOPO. The products were then cloned as PacI fragments into the integrating vector pAPA3 to generate pIspD-PRO and pIspD-SH, respectively. The integrity and direction of the constructs were confirmed by DNA sequencing. Both vectors were electroporated into the single-crossover strain carrying the delivery vector, in which 537 bp of Rv3582c was deleted, and recombinants were isolated on 10 μg/ml gentamicin, 100 μg/ml hygromycin, 20 μg/ml kanamycin, and 50 μg/ml X-gal. The merodiploids were streaked out on plates without any antibiotics, and double-crossover products were isolated as described above, except that gentamicin was included at all stages. Deletion of the chromosomal copy of Rv3582c was confirmed by Southern blotting. To generate a probe for Southern analysis, the region upstream of ispD was PCR amplified using primers IspD US Probe F and IspD US Probe Rev (Table 1), and the isolated fragment was labeled with an AlkPhos Direct system (GE Healthcare). Genomic M. tuberculosis DNA (2 μg) was digested with BamHI, and the digestion products were separated in an agarose gel and transferred by vacuum blotter to a Hybond N+ membrane (GE Healthcare). The membrane was hybridized for 16 h in AlkPhos Direct hybridization buffer, with a blocking agent (GE Healthcare) added at 65°C with the labeled probe. Primary and secondary posthybridization washes were carried out (two primary washes for 10 min each at 55°C and two secondary washes for 5 min each at room temperature, per the manufacturer's instructions), and the probe was detected by CDP-Star (GE Healthcare). The requirement of the integrated gene copy was established via gene switching using pUC-Hyg-Int (20), as previously described (26).
Other procedures.
The enzyme reactions were performed under conditions that were linear for both time and concentration of purified recombinant Rv3582c, and all reported values are averages for triplicate assays. Protein concentrations were estimated using a bicinchoninic acid protein assay kit (Pierce). Radioactive samples were analyzed using a Beckman LS6500 liquid scintillation spectrometer. Blast searches were performed using the National Center for Biotechnology Information (NCBI) website and the Tuberculist website (http://bioweb.pasteur.fr/GenoList/TubercuList/). Multiple sequence alignments were performed using the Multalin interface (8) and were formatted using the sequence manipulation suite available at the University of Alberta website.
RESULTS
Identification and expression of M. tuberculosis IspD.
Rv3582c from M. tuberculosis showed 31% identity with the amino acid sequence of E. coli IspD, suggesting an analogous function in M. tuberculosis. Rv3582c is 696 bp in length, encoding a polypeptide of 234 amino acids with a molecular mass of 26 kDa, which is predicted to be cytosolic. Rv3582c was amplified by PCR from the M. tuberculosis H37Rv genome and cloned into the pET28a(+) vector for overexpression in E. coli BL21(DE3). The cytosolic fractions from the control and recombinant BL21(DE3) strains were tested for IspD activity. The specific activity in the cytosol from the recombinant strain was about 1.5-fold higher than that of the control strain, indicating that Rv3582c encodes functional IspD in M. tuberculosis. Overexpression of the predicted 26-kDa polypeptide was confirmed by Western blot hybridization using an anti-His antibody.
Determination of M. tuberculosis Rv3582c gene essentiality.
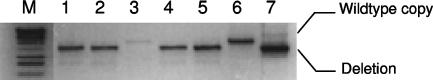
All attempts to generate an Rv3582c deletion mutant in the wild-type background failed. Forty double-crossover strains were tested, and all were wild type (see Materials and Methods). Two different merodiploid strains were used to demonstrate that the Rv3582c gene was essential; the pIspD-SH strain carried the Rv3582c gene expressed from the antigen 85A promoter, and the pIspD-PRO strain carried a larger DNA fragment containing not only the Rv3582c gene but also the surrounding genes predicted to be in the same operon, including the probable native promoter. Using this method, double-crossover strains with both the wild-type and deletion alleles were isolated for both merodiploid strains. The PCR primers IspDint1 and IspDint2 were used to verify the presence of the wild-type (2.0 kbp) or deletion (1.3 kbp) allele of ispD in genomic DNA (Fig. 2). Three of eight mutant double-crossover colonies for the strain carrying pIspD-SH and 7 of 32 for the strain carrying pIspD-PRO were isolated (P = 0.003 and P = 0.002, respectively). One colony of each strain was selected for further study (named AS1 and AS2, respectively). A previously described “gene switching” method (26), which relies on the fact that an integrated vector can be replaced efficiently by an alternative version by transformation (24), was employed to show that the integrated wild-type allele cannot be removed when it is the only functional copy present in the cell. Replacement of the integrated Rv3582c-containing vectors (gentamicin resistance) in strains AS1 and AS2 with an “empty” vector (pUC-Hyg-Int), which did not have the Rv3582c gene but carried an alternative marker gene (hygromycin resistance), was attempted. In both cases, no viable hygromycin-resistant colonies were obtained after electroporation, indicating that the cells cannot survive without a functional copy of Rv3582c.
FIG. 2.
Demonstration of ispD essentiality for M. tuberculosis H37Rv survival. Double-crossover strains were isolated in the wild-type and merodiploid backgrounds and analyzed using PCR primers IspDint1 and IspDint2 to amplify ispD. All strains in the wild-type background were wild type. Strains with either a wild-type or deletion allele were isolated in the merodiploid background. Lane M, lamdba HindIII marker; lanes 1 to 5, double-crossover strains generated from the merodiploid background; lane 6, wild-type genomic control; lane 7, delivery vector control. Lanes 1, 2, 4, and 5 show the product expected for a deletion strain, and lane 3 shows the product expected for a wild-type strain.
In vitro reaction requirements for Rv3582c.
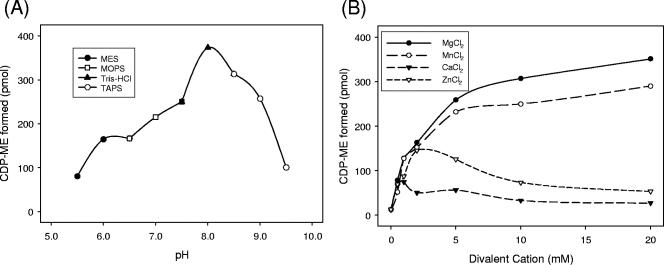
Recombinant Rv3582c was purified by immobilized nickel-affinity chromatography (Fig. 3). The reaction catalyzed by purified recombinant Rv3582c was tested for optimal pH, divalent cation specificity, and nucleotide 5-triphosphate specificity. The enzyme was active over a broad pH range (6.0 to 9.0), with optimal activity at pH 8.0 (Fig. 4A), and showed an absolute dependence on divalent cations (Fig. 4B). The activity was optimal in the presence of 20 mM Mg2+. Mn2+ also supported activity at a comparable level. Zn2+ supported activity, with an optimal concentration of 2.5 mM. Ca2+ was less effective at supporting catalytic activity at all concentrations tested (Fig. 4B). The addition of 10 mM EDTA to the reaction mixture completely abolished activity. In addition, the enzyme is specific for CTP, as other nucleotide 5-triphosphates were poor substrates (with less than 6% of the activity seen with CTP under optimal conditions) (data not shown).
FIG. 3.
Purification of recombinant IspD. Analysis of protein fractions from E. coli transformed with pET28a(+)::Rv3582c is shown. Lane 1, cell lysates prior to IPTG treatment; lane 2, cell lysates after IPTG treatment; lane 3, purified His6-IspD fraction after immobilized metal-affinity chromatography; lane 4, Western blot hybridization analysis of purified IspD using an anti-His antibody; lane M, molecular size markers (kDa). In lane 3, Rv3582c expression was visualized with Coomassie brilliant blue 250R.
FIG. 4.
Effects of pH and divalent cation concentration on M. tuberculosis IspD activity. (A) The optimal pH for catalytic activity was determined using MES (pH 5.5 to 6.5), MOPS (pH 6.5 to 7.5), Tris-HCl (pH 7.5 to 8.5), and TAPS (pH 8.5 to 9.5). (B) Divalent cations (Mg2+, Mn2+, Zn2+, or Ca2+) were added to the reaction mixtures at the indicated concentrations. Reaction mixtures containing 38.5 pmol of purified IspD enzyme were prepared as described in Materials and Methods.
Kinetic characterization of Rv3582c.
Purified IspD activity was linear with increasing protein levels up to a level of 192.5 pmol, and reactions were also linear with time, for up to 30 min. The effects of the MEP or CTP concentration on reaction rates were determined by varying the concentration of one substrate while keeping the other fixed, and the KmMEP and KmCTP were calculated to be 58.5 μM and 53.2 μM, respectively (Table 2).
TABLE 2.
Calculated kinetic parameters for M. tuberculosis IspDa
| Substrate | Km (μM) | Vmax (pmol/min) | kcat (min−1) | kcat/Km (mM−1 min−1) |
|---|---|---|---|---|
| MEP | 58.5 ± 5.4 | 28.0 ± 0.73 | 0.72 | 12.3 |
| CTP | 53.2 ± 4.5 | 38.6 ± 0.87 | 1.0 | 18.8 |
Each reaction mix contained 38.5 pmol of M. tuberculosis IspD. The Km and Vmax values were calculated from averaged data from three independent experiments, using nonlinear regression analysis (GraFit 5.0.13).
DISCUSSION
The enzyme encoded by Rv3582c is clearly a functional CDP-ME synthase. IspD was predicted to be essential in M. tuberculosis, but the preceding enzyme in the MEP pathway (IspC) was not (33), providing impetus to determine the essentiality of IspD in M. tuberculosis. The inability to isolate a nonfunctional mutant using the two-step disruption strategy indicates that Rv3582c is essential for M. tuberculosis survival in vivo, as predicted by Himar1-based transposon mutagenesis experiments (33). The essentiality of the IspD enzyme for the growth of two gram-negative organisms, i.e., Salmonella enterica and E. coli, has previously been reported (7, 14, 34). Thus, the data indicate that IspD is essential for both gram-negative and gram-positive organisms and likely represents a valid drug target for both classes of bacilli.
M. tuberculosis IspD was active over a broad pH range, as previously reported for the E. coli enzyme (28). The addition of EDTA or BioRex 70 to reaction mixtures showed that divalent cations are absolutely required for its cytidylyltransferase activity. A concentration of 20 mM Mg2+ or Mn2+ supported optimal or nearly optimal activity; however, Zn2+ could also support M. tuberculosis IspD activity at low concentrations. Interestingly, E. coli IspD catalytic activity was not supported by Zn2+ at all (31), suggesting subtle differences in the active sites of the enzymes from these organisms. In E. coli IspD, Mg2+, which is essential for activity, forms coordination bonds with the α-, β-, and γ-phosphate oxygens of CTP or the α-phosphate oxygen of CDP-ME, as shown for the glycerol 3-phosphate cytidyltransferase of Bacillus subtilis (27, 38). It is assumed that the divalent cations play a similar role in IspD from M. tuberculosis. Not surprisingly, M. tuberculosis IspD showed a high degree of specificity for CTP, which is likely achieved from the pyrimidine base forming hydrogen bonds with the backbone amides of conserved amino acids and stacking interactions inside the catalytic domain (27, 28). In fact, mutation of these conserved amino acids abolished or reduced the catalytic activity of E. coli IspD (28).
The specificity constants (kcat/Km) of M. tuberculosis IspD for both substrates were lower than those previously determined for E. coli IspD (28), suggesting that IspD of M. tuberculosis is less efficient. However, the KmMEP and KmCTP values used for calculating specificity constants for the E. coli enzyme were 0.37 mM and 0.76 mM, respectively, which are much higher than the KmMEP (3.14 μM) and KmCTP (131 μM) values determined for the enzyme from the same source, but using a different assay system (31). The latter values are more similar to the values reported here for IspD from M. tuberculosis and the values of 3 μM (KmCTP) and 20 μM (KmMEP) reported for the enzyme from Campylobacter jejuni (15).
To achieve minimal toxicity for clinical trials, any potential drug therapy for TB should ideally target factors that are absent in the host but essential for bacillus survival. In humans, the mevalonate pathway is exclusively employed for IPP biosynthesis, and no ortholog of M. tuberculosis IspD exists. The essential nature of IspD in M. tuberculosis indicates that the enzyme is a valid drug target per se. However, inhibition of IspD will result in an inhibition of the synthesis of both IPP and DMAPP, precursors of all isoprenoid compounds synthesized downstream in the biosynthetic pathway, resulting in multiple adverse effects caused by the depletion of isoprenoid compounds that play crucial roles in cell wall biosynthesis and energy production.
Previous enzymatic characterization of IspD orthologs from other organisms were done using 1H- or 13C-nuclear magnetic resonance spectroscopy (15, 30, 31) or thin-layer chromatography-based assays (27, 28), which either require expensive equipment or are tedious. In the present study, the PPi released during the reaction catalyzed by IspD was utilized to monitor M. tuberculosis IspD activity. This assay is facile and relatively inexpensive. It is anticipated that the release of PPi can be coupled to commercially available enzymes to facilitate high-throughput screening for novel inhibitors of IspD. Thus, the results presented here allow the design and implementation of high-throughput screens to attempt to identify new classes of inhibitors and lead compounds, which may lead to the development of new antituberculosis drugs.
Acknowledgments
This work was supported by NIH/NIAID grant AI-065357, EU project QLRT-2001-00887, and the Biotechnology and Biological Sciences Research Council project BB/D000181. H37Rv genomic DNA was provided by Colorado State University under NIH/NIAID contract N01-AI-75320 for tuberculosis research material and vaccine testing.
We thank Dina Robertson for technical assistance.
Footnotes
Published ahead of print on 5 October 2007.
REFERENCES
- 1.Andries, K., P. Verhasselt, J. Guillemont, H. W. Gohlmann, J. M. Neefs, H. Winkler, J. Van Gestel, P. Timmerman, M. Zhu, E. Lee, P. Williams, D. de Chaffoy, E. Huitric, S. Hoffner, E. Cambau, C. Truffot-Pernot, N. Lounis, and V. Jarlier. 2005. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307:223-227. [DOI] [PubMed] [Google Scholar]
- 2.Argyrou, A., and J. S. Blanchard. 2004. Kinetic and chemical mechanism of Mycobacterium tuberculosis 1-deoxy-d-xylulose-5-phosphate isomeroreductase. Biochemistry 43:4375-4384. [DOI] [PubMed] [Google Scholar]
- 3.Arigoni, D., S. Sagner, C. Latzel, W. Eisenreich, A. Bacher, and M. H. Zenk. 1997. Terpenoid biosynthesis from 1-deoxy-d-xylulose in higher plants by intramolecular skeletal rearrangement. Proc. Natl. Acad. Sci. USA 94:10600-10605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bach, T. J. 1995. Some new aspects of isoprenoid biosynthesis in plants—a review. Lipids 30:191-202. [DOI] [PubMed] [Google Scholar]
- 5.Bailey, A. M., S. Mahapatra, P. J. Brennan, and D. C. Crick. 2002. Identification, cloning, purification, and enzymatic characterization of Mycobacterium tuberculosis 1-deoxy-d-xylulose 5-phosphate synthase. Glycobiology 12:813-820. [DOI] [PubMed] [Google Scholar]
- 6.Brennan, P. J., and D. C. Crick. 2007. The cell-wall core of Mycobacterium tuberculosis in the context of drug discovery. Curr. Top. Med. Chem. 7:475-488. [DOI] [PubMed] [Google Scholar]
- 7.Cornish, R. M., J. R. Roth, and C. D. Poulter. 2006. Lethal mutations in the isoprenoid pathway of Salmonella enterica. J. Bacteriol. 188:1444-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crick, D. C., J. R. Scocca, J. S. Rush, D. W. Frank, S. S. Krag, and C. J. Waechter. 1994. Induction of dolichyl-saccharide intermediate biosynthesis corresponds to increased long chain cis-isoprenyltransferase activity during the mitogenic response in mouse B cells. J. Biol. Chem. 269:10559-10565. [PubMed] [Google Scholar]
- 10.Davies, P. D. 2003. The role of DOTS in tuberculosis treatment and control. Am. J. Respir. Med. 2:203-209. [DOI] [PubMed] [Google Scholar]
- 11.Dhe-Paganon, S., J. Magrath, and R. H. Abeles. 1994. Mechanism of mevalonate pyrophosphate decarboxylase: evidence for a carbocationic transition state. Biochemistry 33:13355-13362. [DOI] [PubMed] [Google Scholar]
- 12.Dhiman, R. K., M. L. Schaeffer, A. M. Bailey, C. A. Testa, H. Scherman, and D. C. Crick. 2005. 1-Deoxy-d-xylulose 5-phosphate reductoisomerase (IspC) from Mycobacterium tuberculosis: towards understanding mycobacterial resistance to fosmidomycin. J. Bacteriol. 187:8395-8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flesch, G., and M. Rohmer. 1988. Prokaryotic hopanoids: the biosynthesis of the bacteriohopane skeleton. Formation of isoprenic units from two distinct acetate pools and a novel type of carbon/carbon linkage between a triterpene and d-ribose. Eur. J. Biochem. 175:405-411. [DOI] [PubMed] [Google Scholar]
- 14.Freiberg, C., B. Wieland, F. Spaltmann, K. Ehlert, H. Brotz, and H. Labischinski. 2001. Identification of novel essential Escherichia coli genes conserved among pathogenic bacteria. J. Mol. Microbiol. Biotechnol. 3:483-489. [PubMed] [Google Scholar]
- 15.Gabrielsen, M., F. Rohdich, W. Eisenreich, T. Grawert, S. Hecht, A. Bacher, and W. N. Hunter. 2004. Biosynthesis of isoprenoids: a bifunctional IspDF enzyme from Campylobacter jejuni. Eur. J. Biochem. 271:3028-3035. [DOI] [PubMed] [Google Scholar]
- 16.Horbach, S., H. Sahm, and R. Welle. 1993. Isoprenoid biosynthesis in bacteria: two different pathways? FEMS Microbiol. Lett. 111:135-140. [DOI] [PubMed] [Google Scholar]
- 17.Kaur, D., P. J. Brennan, and D. C. Crick. 2004. Decaprenyl diphosphate synthesis in Mycobacterium tuberculosis. J. Bacteriol. 186:7564-7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lois, L. M., N. Campos, S. R. Putra, K. Danielsen, M. Rohmer, and A. Boronat. 1998. Cloning and characterization of a gene from Escherichia coli encoding a transketolase-like enzyme that catalyzes the synthesis of d-1-deoxyxylulose 5-phosphate, a common precursor for isoprenoid, thiamin, and pyridoxol biosynthesis. Proc. Natl. Acad. Sci. USA 95:2105-2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahapatra, S., T. Yagi, J. T. Belisle, B. J. Espinosa, P. J. Hill, M. R. McNeil, P. J. Brennan, and D. C. Crick. 2005. Mycobacterial lipid II is composed of a complex mixture of modified muramyl and peptide moieties linked to decaprenyl phosphate. J. Bacteriol. 187:2747-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mahenthiralingam, E., B. I. Marklund, L. A. Brooks, D. A. Smith, G. J. Bancroft, and R. W. Stokes. 1998. Site-directed mutagenesis of the 19-kilodalton lipoprotein antigen reveals no essential role for the protein in the growth and virulence of Mycobacterium intracellulare. Infect. Immun. 66:3626-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mikusova, K., M. Mikus, G. S. Besra, I. Hancock, and P. J. Brennan. 1996. Biosynthesis of the linkage region of the mycobacterial cell wall. J. Biol. Chem. 271:7820-7828. [DOI] [PubMed] [Google Scholar]
- 22.Ormerod, L. P. 2005. Multidrug-resistant tuberculosis (MDR-TB): epidemiology, prevention and treatment. Br. Med. Bull. 73-74:17-24. [DOI] [PubMed] [Google Scholar]
- 23.Parish, T., J. Lewis, and N. G. Stoker. 2001. Use of the mycobacteriophage L5 excisionase in Mycobacterium tuberculosis to demonstrate gene essentiality. Tuberculosis (Edinburgh) 81:359-364. [DOI] [PubMed] [Google Scholar]
- 24.Parish, T., M. Schaeffer, G. Roberts, and K. Duncan. 2005. HemZ is essential for heme biosynthesis in Mycobacterium tuberculosis. Tuberculosis (Edinburgh) 85:197-204. [DOI] [PubMed] [Google Scholar]
- 25.Parish, T., and N. G. Stoker. 2000. glnE is an essential gene in Mycobacterium tuberculosis. J. Bacteriol. 182:5715-5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pashley, C. A., and T. Parish. 2003. Efficient switching of mycobacteriophage L5-based integrating plasmids in Mycobacterium tuberculosis. FEMS Microbiol. Lett. 229:211-215. [DOI] [PubMed] [Google Scholar]
- 27.Richard, S. B., M. E. Bowman, W. Kwiatkowski, I. Kang, C. Chow, A. M. Lillo, D. E. Cane, and J. P. Noel. 2001. Structure of 4-diphosphocytidyl-2-C-methylerythritol synthetase involved in mevalonate-independent isoprenoid biosynthesis. Nat. Struct. Biol. 8:641-648. [DOI] [PubMed] [Google Scholar]
- 28.Richard, S. B., A. M. Lillo, C. N. Tetzlaff, M. E. Bowman, J. P. Noel, and D. E. Cane. 2004. Kinetic analysis of Escherichia coli 2-C-methyl-d-erythritol-4-phosphate cytidyltransferase, wild type and mutants, reveals roles of active site amino acids. Biochemistry 43:12189-12197. [DOI] [PubMed] [Google Scholar]
- 29.Rohdich, F., A. Bacher, and W. Eisenreich. 2005. Isoprenoid biosynthetic pathways as anti-infective drug targets. Biochem. Soc. Trans. 33:785-791. [DOI] [PubMed] [Google Scholar]
- 30.Rohdich, F., J. Wungsintaweekul, W. Eisenreich, G. Richter, C. A. Schuhr, S. Hecht, M. H. Zenk, and A. Bacher. 2000. Biosynthesis of terpenoids: 4-diphosphocytidyl-2C-methyl-d-erythritol synthase of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 97:6451-6456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rohdich, F., J. Wungsintaweekul, M. Fellermeier, S. Sagner, S. Herz, K. Kis, W. Eisenreich, A. Bacher, and M. H. Zenk. 1999. Cytidine 5′-triphosphate-dependent biosynthesis of isoprenoids: YgbP protein of Escherichia coli catalyzes the formation of 4-diphosphocytidyl-2-C-methylerythritol. Proc. Natl. Acad. Sci. USA 96:11758-11763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Saito, Y., and K. Ogura. 1981. Biosynthesis of menaquinones. Enzymatic prenylation of 1,4-dihydroxy-2-naphthoate by Micrococcus luteus membrane fractions. J. Biochem. (Tokyo) 89:1445-1452. [DOI] [PubMed] [Google Scholar]
- 33.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77-84. [DOI] [PubMed] [Google Scholar]
- 34.Sauret-Gueto, S., A. Ramos-Valdivia, E. Ibanez, A. Boronat, and M. Rodriguez-Concepcion. 2003. Identification of lethal mutations in Escherichia coli genes encoding enzymes of the methylerythritol phosphate pathway. Biochem. Biophys. Res. Commun. 307:408-415. [DOI] [PubMed] [Google Scholar]
- 35.Sharma, V., M. E. Hudspeth, and R. Meganathan. 1996. Menaquinone (vitamin K2) biosynthesis: localization and characterization of the menE gene from Escherichia coli. Gene 168:43-48. [DOI] [PubMed] [Google Scholar]
- 36.Smith, D. A., T. Parish, N. G. Stoker, and G. J. Bancroft. 2001. Characterization of auxotrophic mutants of Mycobacterium tuberculosis and their potential as vaccine candidates. Infect. Immun. 69:1142-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Testa, C. A., and M. J. Brown. 2003. The methylerythritol phosphate pathway and its significance as a novel drug target. Curr. Pharm. Biotechnol. 4:248-259. [DOI] [PubMed] [Google Scholar]
- 38.Weber, C. H., Y. S. Park, S. Sanker, C. Kent, and M. L. Ludwig. 1999. A prototypical cytidylyltransferase: CTP:glycerol-3-phosphate cytidylyltransferase from Bacillus subtilis. Structure 7:1113-1124. [DOI] [PubMed] [Google Scholar]
- 39.Weinstein, E. A., T. Yano, L. S. Li, D. Avarbock, A. Avarbock, D. Helm, A. A. McColm, K. Duncan, J. T. Lonsdale, and H. Rubin. 2005. Inhibitors of type II NADH:menaquinone oxidoreductase represent a class of antitubercular drugs. Proc. Natl. Acad. Sci. USA 102:4548-4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolucka, B. A., M. R. McNeil, E. de Hoffmann, T. Chojnacki, and P. J. Brennan. 1994. Recognition of the lipid intermediate for arabinogalactan/arabinomannan biosynthesis and its relation to the mode of action of ethambutol on mycobacteria. J. Biol. Chem. 269:23328-23335. [PubMed] [Google Scholar]
- 41.World Health Organization. 2007. Global tuberculosis control: surveillance, planning, financing. WHO report 2007. WHO/HTM/TB/2007.376. World Health Organization, Geneva, Switzerland.
- 42.Zhang, Y., M. M. Wade, A. Scorpio, H. Zhang, and Z. Sun. 2003. Mode of action of pyrazinamide: disruption of Mycobacterium tuberculosis membrane transport and energetics by pyrazinoic acid. J. Antimicrob. Chemother. 52:790-795. [DOI] [PubMed] [Google Scholar]