Abstract
Bacteria can switch from a single-cell (planktonic) mode to a multicellular community (biofilm) mode via production of cell-cell aggregation and surface adhesion factors. In this report, we present evidence that the CsgD protein, a transcription regulator involved in biofilm formation in Escherichia coli, modulates the expression of the rpoS (σS) regulon. Protein pattern analysis of E. coli cells in stationary phase shows that CsgD affects the expression of several proteins encoded by σS-dependent genes. CsgD regulation of σS-dependent genes takes place at gene transcription level, does not bypass the need for rpoS, and is abolished in an rpoS-null mutant. Consistent with these results, we find that CsgD expression leads to an increase in σS intracellular concentration. Increase in σS cellular amount is mediated by CsgD-dependent transcription activation of iraP, encoding a factor involved in σS protein stabilization. Our results strongly suggest that the CsgD regulatory protein plays a major role as a relay between adhesion factors production and σS-dependent gene expression via σS protein stabilization. Direct coordination between biofilm formation and expression of the rpoS regulon could positively impact important biological processes, such as host colonization or response to environmental stresses.
Transition from the unicellular (planktonic) state to the multicellular community (biofilm) lifestyle can be considered one of the major developmental processes in bacteria. Indeed, most bacteria are capable of surface colonization and biofilm formation through the production of a large number of adhesion factors, whose expression is usually finely regulated in response to both environmental and physiological cues. Adaptation to growth as a biofilm affects cell morphology, physiology, and metabolism via major redirection of gene expression (4, 52, 54, 62); interestingly, biofilm formation appears to induce the expression of different stress responses (34, 54) and even programmed cell death (70).
In enterobacteria, curli fibers (also known as Tafi, thin aggregative fimbriae, in Salmonella) are an important factor in adhesion to surfaces, cell aggregation, and biofilm formation (19, 28, 68). Curli-encoding genes are clustered in two operons: csgBA encodes the structural components, while the divergently oriented csgDEFG operon encodes proteins involved in curli assembly and transport (38), as well as the CsgD transcription factor, which is necessary for csgBA transcription (28). Expression of the csg operons takes place in response to a combination of environmental conditions, such as low growth temperature (<32°C), low osmolarity, and slow growth (43). Such environmental signals are mediated at the gene expression level by a number of regulators, including, besides CsgD, global regulatory proteins such as OmpR, H-NS, CpxR, and the alternative σ factor σS (3, 25, 53, 58). However, this strict environmental control is lost in many bacterial strains due to mutations either in regulatory genes (3, 68) or in the csgDEFG promoter (57). Indeed, temperature-dependent regulation does not take place in several Salmonella and pathogenic Escherichia coli strains, in which curli are also expressed at 37°C and represent an important virulence factor (5, 6, 48). In contrast, curli operons are silent in a large number of laboratory strains, as well as in some clinical and environmental isolates, despite the presence of functional csg genes (57).
In addition to its role as activator of the csgBA operon, CsgD regulates a number of genes involved in biofilm formation and production of cell surface-associated structures, such as adrA, which promotes cellulose production (59, 75); the yhiU operon, encoding biosynthetic genes for the O antigen (26); and the bapA gene, which encodes a membrane protein acting as a positive determinant for biofilm formation (35). However, gene regulation by CsgD is not strictly limited to adhesion factors but also affects genes involved in transport, metabolism, and gene regulation, whose dependence on CsgD might possibly be linked to adaptation of cell physiology to the biofilm lifestyle (10, 16). Gene regulation by CsgD is tightly connected to production and sensing of cyclic di-GMP, a bacterial second messenger involved in various cellular processes, including biosynthesis of extracellular polysaccharides (63), biofilm formation (30), and virulence (50, 65), as well as morphological and physiological differentiation (47). Indeed, the CsgD-dependent adrA gene, involved in cellulose biosynthesis (75), encodes a cyclic di-GMP synthase (63). CsgD can also activate yoaD, whose gene product is a cyclic di-GMP phosphodiesterase, suggesting that CsgD is directly involved in feedback regulation of cyclic di-GMP intracellular levels and of cellulose biosynthesis (10). In turn, other proteins potentially involved in cyclic-di-GMP biosynthesis affect csgD expression in Salmonella (32).
The σS protein is an alternative sigma factor of RNA polymerase which directs transcription of genes involved in adaptation to slow growth and to cellular stresses (the rpoS regulon [37]). We show here that CsgD positively controls σS expression by activation of the iraP gene, a σS protein stabilization factor (8). Thus, CsgD can act as a relay between adhesion factor production and σS-dependent gene expression, coordinating the expression of biofilm determinants to stress response genes, and possibly virulence factors, belonging to the rpoS regulon.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
All strains used in the present study are listed in Table 1. Bacteria were grown in M9Glu/sup medium (M9 minimal medium supplemented with 0.5% glucose and 2.5% Luria broth) at 30°C. When needed, antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; tetracycline, 25 μg/ml; and kanamycin, 50 μg/ml.
TABLE 1.
Strains and plasmids used in the present study
| Strain or plasmid | Relevant genotype or characteristics | Source or reference |
|---|---|---|
| Strains | ||
| MG1655 | E. coli K-12 λ− F−, rph-1 | Reference strain |
| EB1.3 | MG1655 rpoS::Tn10 | 53 |
| LG03 | MG1655 iraP::kan | This study |
| LG05 | EB1.3 csgA::uidA-kan | This study |
| LG07 | EB1.3 csgD::uidA-kan | This study |
| PHL628 | MG1655 ompR234 | 53 |
| PHL856 | MG1655 csgA::uidA-kan | 53 |
| PHL1087 | PHL628 csgD::uidA-kan | 53 |
| PHL1088 | MG1655 csgD::uidA-kan | 53 |
| Plasmids | Ampicillin resistance, T7 RNA polymerase-dependent promoter | |
| pT7-7 | csgD gene open reading frame cloned into pT7-7 plasmid as an NdeI/PstI 651-bp fragment | S. Tabor, Institute of Cancer Research, London, United Kingdom |
| pT7-CsgD | E. coli K-12 λ− F−, rph-1 | 53 |
Biofilm formation assays.
Biofilm formation in microtiter plates was determined essentially as described previously (44). Cells were grown in liquid cultures in microtiter plates (0.2 ml) for 18 h in M9Glu/sup at 30°C. The liquid culture was removed, and the cell optical density at 600 nm (OD600) was determined spectrophotometrically. Cells attached to the microtiter plates were washed with 0.1 M phosphate buffer (pH 7.0) and then stained for 20 min with 1% crystal violet (CV) in ethanol. The stained biofilms were thoroughly washed with water and dried. CV staining was visually assessed, and the microtiter plates were scanned. For semiquantitative determination of biofilms, CV-stained cells were resuspended in 0.2 ml of 70% ethanol, and their absorbance was measured at 600 nm and normalized to the OD600 of the corresponding liquid culture.
Protein localization experiments.
Cell fractionation was performed as described previously (23). Portions (250 ml) of cultures grown in M9Glu/sup at 30°C for 18 h were centrifuged at 4,000 × g for 10 min at 4°C and washed with 5 ml of 0.1 M phosphate buffer pH 7.0 (PB). Cells were resuspended in 2 ml of PB with addition of 100 μg of lysozyme/ml and 1 mM EDTA (pH 8.0) and incubated at room temperature for 10 min. Cells were disintegrated by using a French press and centrifuged as described above to remove unbroken cells. The low-speed centrifugation supernatant was then centrifuged at 100,000 × g for 1 h at 4°C to separate the cytoplasm (supernatant) and the membrane fraction (pellet). The pellet was resuspended in 2 ml of 2% Sarkosyl in phosphate-buffered saline, left for 20 min at room temperature, and centrifuged at 40,000 × g at 10°C for 10 min to remove ribosomes and cytoplasmic proteins that were still associated with the membrane fraction. The pellet was resuspended in 1 ml of 1% Sarkosyl, precipitated again 20 min at room temperature, and centrifuged as described above. The supernatant, corresponding to inner membrane proteins, was collected, and the pellet, corresponding to outer membrane proteins, was resuspended in 0.5 ml of H2O. Protein concentrations were determined, and 20 μg of total proteins was loaded onto a 12% sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE). Specific bands were identified by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) analysis of the peptide products after in-gel trypsin digestion (15; performed by CRIBI, University of Padua, Padua,Italy [http://www.bio.unipd.it/cribi/]).
σS determination by Western blotting.
Protein amounts in cytoplasmic samples were determined by the Bradford method, and 20-μg portions of total proteins were loaded onto an SDS-PAGE gel (12% acrylamide). Proteins were transferred on Hybond P membranes (Amersham Life Sciences) and incubated with the polyclonal rabbit antibodies against the σS protein (55). The anti-σS antibodies were detected by using a secondary anti-rabbit antibody conjugated with fluorescein. For σS turnover experiments, 50 μg of cell extracts (cytoplasmic fractions) was incubated at 37°C for different times (0, 5, 10, and 20 min). Reactions were stopped by the addition of an equal amount of SDS-PAGE loading buffer, and the samples were used for Western blotting. Bands were quantified by using the ImageQuant 5.2 software (Molecular Dynamics).
RNA isolation, cDNA synthesis, and real-time-PCR analysis.
For RNA isolation, strains were grown in M9Glu/sup at 30°C to stationary phase (OD600 = 2). The cells were harvested by centrifugation at 13,000 rpm for 5 min at 4°C, and total RNA was extracted by using an RNeasy minikit (QIAGEN). RNA samples were checked by agarose gel electrophoresis to assess the lack of degradation and then quantified spectrophotometrically. Genomic DNA was removed by DNase I treatment. Reverse transcription was performed on 1 μg of total RNA, along with negative control samples incubated without reverse transcriptase. cDNA synthesis efficiency was verified by electrophoresis on agarose gel in comparison to negative controls. Real-time PCR was performed by using the SYBR green PCR master mixture, and the results were determined with an iCycle iQ real-time detection system (Bio-Rad). Reaction mixtures (25 μl) included 0.1 μg of cDNA and 300 nM concentrations of primers in the reaction buffer and enzyme supplied by the manufacturer. The sequences of the primers used are listed (see Table S1 in the supplemental material). All reactions were performed in triplicate, including negative control samples, which never showed significant threshold cycles (CT). The relative amounts of the transcripts were determined by using 16S rRNA as the reference gene ([CT(gene of interest) − CT(16S)] = ΔCT).
Other methods.
β-Glucuronidase assays were performed as described previously (10). Bacteriophage P1vir transductions were carried out as described previously (40). The correctness of transduction was checked by PCR verification of the presence of the antibiotic cassette used for selection in the gene of interest, except for the rpoS mutants, which were checked by catalase activity assays as previously described (69).
RESULTS
Ectopic CsgD expression in MG1655.
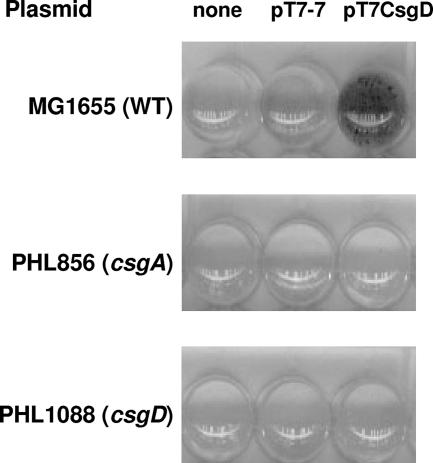
Several E. coli laboratory strains, including MG1655, as well as some clinical and environmental isolates, can only produce curli fibers in very limited amounts, insufficient to promote biofilm formation in standard laboratory assays (44), even though the genes necessary for curli biosynthesis and assembly are fully functional (57). However, transformation of MG1655 with the pT7CsgD plasmid results in an ability to form biofilm (Fig. 1). In the pT7CsgD plasmid, the csgD gene is under the control of a T7 RNA polymerase-dependent promoter. In E. coli strains such as MG1655, which do not carry the T7 RNA polymerase-encoding gene, low-level, constitutive transcription of genes cloned under the control of T7 promoters can still take place due to weak promoter recognition by bacterial RNA polymerase (12). Surface adhesion properties conferred by the pT7CsgD plasmid were totally abolished by inactivation of either the csgA gene, whose gene product is the curli major subunit, or the chromosomal csgD gene (Fig. 1); inactivation of chromosomal csgD has a polar effect on the whole csgDEFG operon, thus preventing expression of genes involved in curli assembly and export. These results indicate that, as expected, stimulation of biofilm formation by pT7CsgD is mediated by curli production.
FIG. 1.
Surface adhesion by MG1655 (WT), PHL856 (csgA), and PHL1088 (csgD) transformed with either the pT7-7 or the pT7CsgD plasmids. Surface adhesion was performed in polypropylene microtiter plates as described in Materials and Methods; bacterial biofilm was revealed by CV staining. The images shown are direct scans of microtiter wells.
CsgD effects on MG1655 protein production.
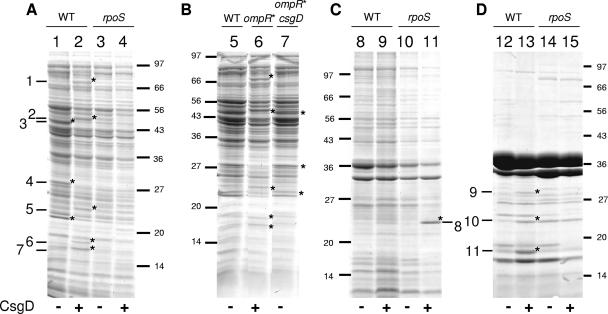
In order to investigate the possible effects of CsgD on global gene expression in the MG1655 strain, we carried out protein analysis of fractionated cell extracts on monodimensional SDS-PAGE, comparing MG1655 transformed either with pT7CsgD or with the pT7-7 control vector. We have performed a similar approach in another laboratory strain, PHL565 (10), which, however, is impaired in expression of the rpoS gene (P. Landini, unpublished observation), a feature common to several laboratory isolates (31). Unlike PHL565, in the rpoS-proficient MG1655 strain CsgD expression from pT7CsgD significantly affects protein production pattern (Fig. 2). We excised the bands differentially expressed in MG1655/pT7CsgD and identified the corresponding proteins by MALDI-TOF after in-gel trypsin digestion (Table 2). We found that CsgD positively affects expression of the PflB, GadA, WrbA, and Dps proteins in the cytoplasmic fraction and of the Dps, CsgG, and OmpW proteins in the outer membrane fraction. The CsgB and CsgA proteins, i.e., the structural components of curli fibers, which are expressed in a CsgD-dependent fashion, cannot be visualized by SDS-PAGE since, once assembled into curli fibers, they form an extremely tight structure that cannot enter the polyacrylamide gel (28). However, evidence of curli production in the MG1655/pT7CsgD strain comes from functional assays (Fig. 1) and from csg gene expression experiments (Fig. 4). Interestingly, four of the six proteins produced at higher level in the presence of CsgD, namely, GadA (band 2 in Fig. 2), WrbA (band 5), Dps (bands 6, 7, and 11), and CsgG (band 9), are encoded by genes known to belong to the rpoS regulon, i.e., their transcription is dependent upon the alternative sigma factor σS. The GadA, WrbA, and Dps proteins are, respectively, a glutamate decarboxylase involved in resistance to acid stress (21, 64), a quinone reductase part of a response to oxidative stress (41, 45), and a bacterial ferritin able to protect DNA from iron-mediated hydroxyl-radical formation (39, 73). Interestingly, two bands corresponding to the Dps protein were found in the cytoplasm fraction of MG1655/pT7CsgD (bands 6 and 7, Fig. 2A); this would suggest the existence of different Dps isoforms, as also observed in recent two-dimensional gel analysis of the MG1655 proteome (36). Despite Dps being a cytoplasmic protein, we also detected its presence in the outer membrane fraction (band 11, Fig. 2C), as already reported for other biofilm-forming E. coli strains (34), thus suggesting that a fraction of the Dps protein might be associated with the outer membrane. Unlike Dps, CsgG and OmpW are bona fide outer membrane proteins (38, 49). The CsgG protein is a component of the curli transport system and is encoded by a gene belonging to the csgDEFG operon, which also encodes CsgD (28, 38). Consistent with their being part of the rpoS regulon, no CsgD-dependent increase in protein expression of GadA, WrbA, Dps, and CsgG could be detected in EB1.3/pT7CsgD, an rpoS mutant of MG1655 transformed with the pT7CsgD plasmid (Fig. 2, lanes 4 and 10), thus suggesting that regulation by CsgD does not bypass the need for σS. In addition to known members of the rpoS regulon, we find that expression of the PflB protein (band 1), encoded by a gene thus far never described as σS dependent, is stimulated by CsgD in the MG1655 background only, suggesting that its CsgD-dependent expression requires a functional rpoS gene (Fig. 2A). In contrast, expression of the OmpW protein (band 10), although stimulated by CsgD, appears to be negatively regulated by σS (Fig. 2, compare lanes 11 and 13).
FIG. 2.
SDS-PAGE of fractionated cell extracts. (A) Cytoplasmic proteins. Lane 1, MG1655/pT7-7; lane 2, MG1655/pT7CsgD; lane 3, EB1.3/pT7-7; lane 4, EB1.3/pT7CsgD. (B) Cytoplasmic proteins. Lane 5, MG1655; lane 6, PHL628; lane 7, PHL1087. (C) Inner membrane proteins. Lane 8, MG1655/pT7-7; lane 9, MG1655/pT7CsgD; lane 10, EB1.3/pT7-7; lane 11, EB1.3/pT7CsgD. (D) Outer membrane proteins. Lane 12, MG1655/pT7-7; lane 13, MG1655/pT7CsgD; lane 14, EB1.3/pT7-7; lane 15, EB1.3/pT7CsgD. The relevant genotype of the different bacterial strains is indicated in the figure. ompR* stands for the ompR234 mutation resulting in increased csgD expression (68). The position of molecular mass markers is shown (numbers indicate molecular masses in kilodaltons). Asterisks indicate bands differentially expressed in a CsgD-dependent manner that were excised and identified by MALDI-TOF (numbered from 1 to 11).
TABLE 2.
Gene characteristics
| Band no.a | Gene productb | Predicted molecular mass (Da)c | Gene function (reference) | Regulation of corresponding gene (reference) |
|---|---|---|---|---|
| 1 | PflB* | 85,357 | Pyruvate formate lyase I (56) | Induced by ArcA and FNR (61) |
| 2 | GadA* | 52,685 | Glutamate decarboxylase (64) | Positively regulated by rpoS, CRP, gadE, gadW, and gadX; negatively regulated by H-NS (21, 66) |
| 3 | TnaA† | 52,773 | Tryptophan deaminase (tryptophanase) (22) | Positively activated by rpoS and by CRP (33, 74) |
| 3 | GatZ† | 47,109 | Subunit of tagatose-1,6-bisphosphate aldolase 2 (42) | Positively activated by CRP (74); repressed by ArcA (60) |
| 4 | GatY† | 30,812 | Subunit of tagatose-1,6-bisphosphate aldolase 2 (42) | Same transcription unit as gatZ |
| 5 | WrbA* | 20,846 | Quinone reductase; response to oxidative stress (45) | Positively regulated by rpoS (33, 46, 71) |
| 6, 7, and 11 | Dps* | 18,695 | Bacterial ferritin, DNA protection during stationary phase (39, 73) | Positively regulated by rpoS (2, 33, 46, 71) |
| 8 | CsgD* | 24,935 | Transcriptional regulator (28) | Positively regulated by rpoS, hns, and ompR (25, 58) |
| 9 | CsgG* | 30,557 | Outer membrane protein; curli transport component (38) | Same transcription unit as csgD |
| 10 | OmpW* | 22,928 | Outer membrane protein; receptor for colicin S4 (49) | Positively regulated by FNR (18) |
As in Fig. 2. Band 3 was identified as a mixture of two proteins and thus appears twice, once for each protein.
*, Increased expression in MG1655/pT7CsgD; †, decreased expression in MG1655/pT7CsgD.
Predicted molecular masses were obtained from the EcoCyc database (http://www.ecocyc.org/).
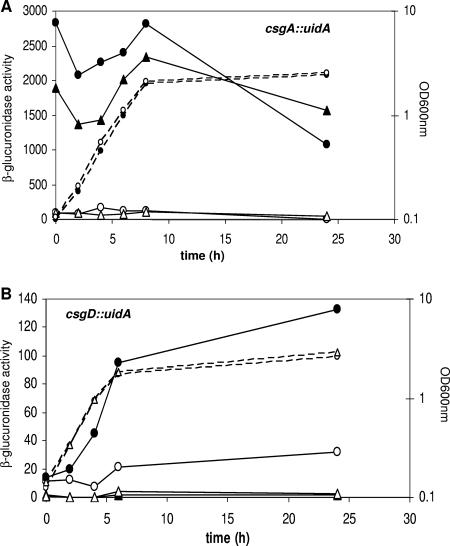
FIG. 4.
β-Glucuronidase activity measured from either csgA::uidA (A) or from csgD::uidA (B) chromosomal fusions. Cultures were grown in M9Glu/sup at 30°C. (A) Reporter gene expression from the csgB promoter was measured either in PHL856 (MG1655 csgA::uidA; circles) or in LG05 (MG1655 rpoS csgA::uidA, triangles) transformed either with pT7-7 (open symbols) or pT7CsgD (closed symbols). Dashed lines indicate growth curves of MG1655/pT7-7 (○) and MG1655/pT7CsgD (•) but are representative for all of the strains tested. (B) Reporter gene expression from the csgD promoter was measured either in PHL1088 (MG1655 csgD::uidA; circles) or in LG07 (MG1655 rpoS csgD::uidA, triangles) transformed either with pT7-7 (open symbols) or with pT7CsgD (closed symbols). Dashed lines indicate growth curves of MG1655/pT7-7 (○) and EB1.3/pT7-7 (•) but are representative for all of the strains tested. The data are an average of four independent experiments. Standard deviations were always lower than 15% and are not shown for clarity.
Among the proteins negatively regulated by CsgD, we could identify band 5 as a mixture of TnaA (tryptophanase [22]) and GatZ (involved in galactitol catabolism). Identification of band 4, probably a complex of different proteins, was inconclusive, although GatY, also involved in galactitol catabolism, appears to be present. The gatY gene, together with gatZ, is part of the gatZYABC operon (42), which, as we already showed, is negatively regulated by CsgD (10). Neither TnaA nor the GatZ/GatY complex could be detected in cell extracts from the rpoS mutant EB1.3 strain (Fig. 2A), suggesting that expression of the corresponding genes is σS dependent. Indeed, at least for the tnaA gene, dependence on rpoS has already been reported (33, 36). Negative regulation of rpoS-dependent genes by the CsgD protein might be due to direct interaction with their promoters: indeed, at least for the tnaA gene, an imperfect inverted repeat with high similarity to the putative CsgD binding site (9) overlaps the start site of the tnaLAB transcript, suggesting possible direct control of the tnaA gene by CsgD.
The only major difference observed in the inner membrane compartment was the presence of a rather large band of ca. 25 kDa in EB1.3/pT7CsgD, identified as the CsgD protein. This result suggests that the rpoS mutation might either stimulate ectopic CsgD expression from the pT7CsgD plasmid or promote CsgD protein association to the cytoplasmic membrane.
The induction of proteins belonging to the rpoS regulon in MG1655/pT7CsgD is not due to changes in growth rate (see Fig. 4), nor does it depend on a stress response elicited by nonphysiological protein overexpression from the pT7CsgD plasmid. Indeed, ectopic expression of the AdrA protein (a cyclic-di-GMP synthase), as well as truncated (inactive) forms of CsgD, did not lead to any detectable changes in protein expression (data not shown). In addition, the same pattern of protein regulation was observed when we compared MG1655/pT7CsgD to PHL628, an ompR234 mutant of MG1655 showing increased transcription of the csgD gene (53), both in the cytoplasm (Fig. 2B) and in the membrane compartments (data not shown). Proteins whose production is increased in the cytoplasmic fraction of PHL628 correspond to those identified in MG1655/pT7CsgD, as determined MALDI-TOF analysis, and their expression was totally abolished in PHL1087, a csgD::kan derivative of PHL628 (Fig. 2B).
Transcription activation of rpoS-dependent genes by CsgD.
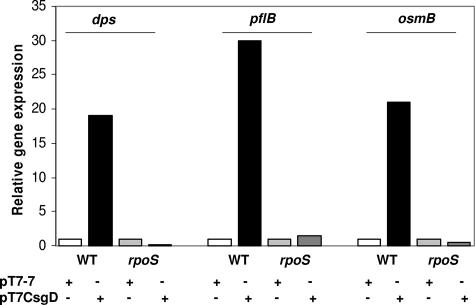
It is likely that the CsgD-dependent effects on protein expression in MG1655 take place via transcription regulation of the corresponding genes. Although monodimensional SDS-PAGE can only provide an incomplete view of CsgD-mediated effects on global protein production in MG1655, our results (Fig. 2 and Table 2) suggest that CsgD might somehow affect the expression of the rpoS regulon. In order to confirm this possibility, we determined CsgD effects on transcription of the dps, pflB, and osmB genes by real-time PCR, both in the MG1655 strain and in its rpoS derivative EB1.3. These genes were chosen as representatives of known rpoS-dependent genes encoding proteins whose production is stimulated by CsgD (dps), genes encoding proteins stimulated by CsgD not assigned to the rpoS regulon (pflB), and rpoS-dependent genes encoding proteins for which no stimulation by CsgD could be detected in our SDS-PAGE experiments (osmB). As shown in Fig. 3, CsgD activates transcription of dps, pflB, and osmB genes by a similar extent (19- to 30-fold); however, CsgD transcription activation can only occur in the MG1655 strain, and it is totally abolished in its rpoS derivative EB1.3. These results confirm that CsgD-mediated effects on protein expression take place at gene transcription level and suggest that CsgD can activate expression of the rpoS regulon, or at least of a subset of σS-dependent promoters. Interestingly, both the CsgD-dependent csgB and the adrA promoters, as well as the csgD promoter itself, have been proposed to be under the control of σS and regulated by the Crl protein, a specific modulator of σS activity (7, 51, 55, 57). Thus, it might be possible that CsgD could act as a specific activator for the σS-associated form of RNA polymerase (EσS). In order to better understand the interplay between σS and CsgD, we compared the effect of rpoS inactivation on CsgD-dependent transcription at either the csgB or the csgD promoter. Activation of its own promoter by CsgD is suggested by increased production of the CsgG protein (Fig. 2), encoded by a gene which is part of the csgDEFG operon. Thus, we transformed with either the pT7CsgD or the pT7-7 control vector two derivatives of the MG1655 strain carrying, respectively, a csgA::uidA (transcription of the csgA gene is directed by the csgB promoter) or a csgD::uidA chromosomal transcriptional fusion. The results shown in Fig. 4 indicate that CsgD expression results in increased transcription levels for both the csgB and the csgD promoters. However, CsgD-dependent transcription activation at the csgB and csgD promoters greatly differs both in timing and in the extent of dependence on the rpoS gene: transcription from the csgB promoter (Fig. 4A) is activated by CsgD regardless of growth phase, and rpoS inactivation only results in a slight reduction (down to ca. 65%) in promoter activity. Thus, although σS is required for optimal transcription levels, the csgB promoter is activated by CsgD in a manner largely independent of σS, strongly suggesting that σS is not directly involved in protein-protein interaction between CsgD and RNA polymerase leading to transcription activation. CsgD activation of csgB transcription in the EB1.3 strain also indicates that the CsgD protein is fully competent in transcription activation even in an rpoS mutant, despite its localization in the inner membrane compartment (Fig. 2B), which is unusual for a transcription regulator. In contrast to csgB, CsgD-mediated stimulation of its own promoter only takes place in late-log and stationary phase and is totally abolished by inactivation of the rpoS gene (Fig. 4B).
FIG. 3.
Relative transcription of the dps, pflB, and osmB genes in MG1655 (WT) and EB1.3 (isogenic rpoS mutant), as determined by real-time PCR. Bars: □, strains transformed with pT7-7; ▪, strains transformed with pT7CsgD. Relative transcription values were set to 1 for both MG1655/pT7-7 and EB1.3/pT7-7 for better comparison of the CsgD-dependent effects in either strain.
Effect of CsgD on σS intracellular concentrations.
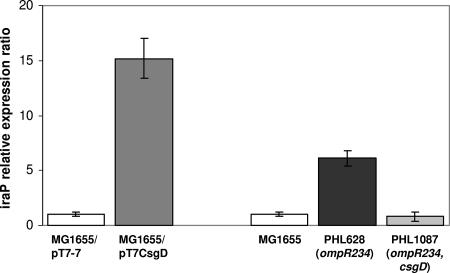
Results shown in Fig. 4, indicating that CsgD does not function as an EσS-specific transcription activator, and the observation that rpoS-dependent promoters stimulated by CsgD (Fig. 3) lack a putative CsgD binding site would suggest that the CsgD protein might affect expression of the rpoS regulon by altering either σS activity or its intracellular concentration. Indeed, in a previous report, we have shown by gene array experiments that CsgD can activate the yaiB gene (10). The product of the yaiB (now iraP) gene has recently been shown to be a stabilization factor for the σS protein, which acts by inhibiting RssB-mediated degradation of σS in response to phosphate starvation (8). Thus, CsgD activation of the iraP gene should result in improved σS stability and possibly increase σS intracellular concentrations, resulting in increased transcription of σS-dependent genes. In order to confirm our previous results, we compared iraP expression in both the MG1655/pT7-7 and the MG1655/pT7CsgD strains by real-time PCR experiments. As shown in Fig. 5, CsgD can increase transcription levels of the iraP gene by ∼15-fold. Stimulation of iraP transcription by ∼6-fold was also observed when we compared MG1655 to its ompR234 mutant derivative PHL628 (Fig. 5), which expresses the csgDEFG operon at higher levels than MG1655 (53). Activation of iraP transcription in the ompR234 mutant strain was totally abolished by csgD inactivation (Fig. 5), thus confirming that iraP activation observed in the PHL628 strain completely depends on CsgD. CsgD-induced iraP expression in either MG1655/pT7CsgD or PHL628 is consistent with increased production of proteins encoded by rpoS-dependent genes observed in both strains (Fig. 2).
FIG. 5.
Expression levels of the iraP genes, normalized to 16S rRNA, are shown as relative values in MG1655/pT7CsgD in comparison to MG1655/pT7-7, as well as in PHL628 (ompR234) and PHL1087 (ompR234 csgD::kan) in comparison to MG1655. Samples were taken from cultures grown overnight in M9Glu/sup at 30°C. Values shown are the average of at least three experiments, and standard deviations are shown.
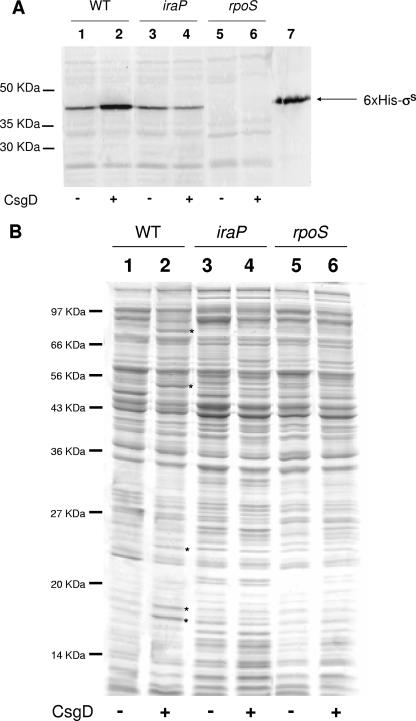
In order to prove the possibility that CsgD expression might indeed stimulate the rpoS regulon by IraP-dependent stabilization of σS, we compared the levels of σS intracellular amounts in both MG1655/pT7-7 and MG1655/pT7CsgD by Western blotting experiments (Fig. 6A). Expression of CsgD from the pT7CsgD plasmid resulted in a clearly detectable increase in σS intracellular amounts in the MG1655 strain. CsgD-dependent stimulation of σS intracellular amounts was also observed in E. coli strains with different genetic backgrounds and at different antibody dilutions (data not shown). As expected, no bands reacting with the anti-σS antibody were detected in the rpoS mutant EB1.3, regardless of the presence of pT7CsgD (lanes 5 and 6). Inactivation of the iraP gene completely abolishes CsgD-dependent increase in σS intracellular concentration (Fig. 6A, lane 4), strongly suggesting that this effect indeed takes place through IraP-mediated stabilization of the σS protein. In the growth conditions used in our experiments, σS cellular levels do not differ significantly in MG1655 compared to its iraP mutant derivative (Fig. 6A, compare lanes 1 and 3), a finding in agreement with previous results indicating that the IraP protein is only essential for σS stability under phosphate starvation conditions (8). In order to correlate σS intracellular concentration and expression of σS-dependent genes, cell extracts from the MG1655 strain and in its EB1.3 (rpoS) and LG03 (iraP) derivatives were compared by SDS-PAGE. As shown in Fig. 6B, a strict correlation exists between σS intracellular concentration and expression of proteins encoded by σS-dependent genes. Consistent with the lack of CsgD-dependent increase in σS intracellular concentration in LG03 (iraP mutant; Fig. 6A, lane 4), the CsgD protein failed to stimulate expression of the GadA, WrbA and Dps proteins in this strain (Fig. 6B, lane 4). The band running with electrophoretic mobility similar to WrbA in cell extracts of the EB1.3 and LG03 was identified as adenosine phosphoribosyltransferase (data not shown), and its expression appears to be CsgD independent (Fig. 6B).
FIG. 6.
(A) Western blotting with anti-σS antibodies. Cell extracts were prepared from overnight cultures grown in M9Glu/sup at 30°C, and 20 μg of total proteins was loaded onto a SDS-polyacrylamide gel. Lane 1, MG1655/pT7-7; lane 2, MG1655/pT7CsgD; lane 3, LG03/pT7-7; lane 4, LG03/pT7CsgD; lane 5, EB1.3/pT7-7; lane 6, EB1.3/pT7CsgD; lane 7, purified His6-tagged σS. The positions of the 50-, 35-, and 30-kDa molecular mass markers are shown. (B) SDS-PAGE analysis of cytoplasmic proteins (20 μg of total proteins). Lane order is as described in panel A. Differently expressed proteins are indicated by an asterisk.
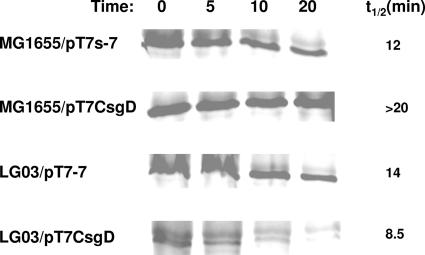
To confirm that CsgD-dependent accumulation of σS is indeed mediated by σS stabilization via the IraP protein, we performed σS protein turnover assays in cell extracts of either MG1655 or its iraP mutant derivative (LG03), grown both in the presence or in the absence of the pT7CsgD plasmid. As shown in Fig. 7, the stability of the σS protein is increased in cell extracts of MG1655/pT7CsgD compared to MG1655/pT7-7; however, no CsgD-dependent σS stabilization could be detected in cell extracts of the LG03/pT7CsgD strain, a finding consistent with a direct role of the IraP protein in CsgD-mediated stabilization of σS. In the absence of CsgD expression, the σS half-life in cell extracts of the iraP mutant is similar to MG1655 (14 versus 12 min, respectively), as detected by image analysis.
FIG. 7.
σS turnover experiments. Cell extracts of MG1655/pT7-7, MG1655/pT7CsgD, LG03/pT7-7, and LG03/pT7CsgD were incubated at 37°C. Samples (50 μg of proteins) were taken at the indicated times and loaded onto an SDS-polyacrylamide gel. The amount of σS protein was determined by Western blotting. σS half-life values were obtained by quantification of the σS-corresponding bands using the ImageQuant 5.2 image analysis program. The experiment was performed twice with very similar results.
DISCUSSION
The CsgD protein, a transcription regulator responsible for the production of adhesion factors (curli fibers) and biofilm formation in enterobacteria, can activate expression of the iraP gene (10) (Fig. 5). The IraP protein acts as a stabilization factor for the alternative σ factor σS by binding to the RssB protein and thus preventing σS proteolysis by the RssB-ClpXP protein complex. IraP-dependent stabilization of σS takes place in response to phosphate starvation (8). We report here that CsgD transcription activation of the iraP gene does indeed result in a significant increase of σS intracellular concentration by positively affecting σS protein stability (Fig. 6 and 7), thus leading to altered expression of σS-dependent genes (Fig. 2, 3, 4B, and 6B and Table 1). CsgD modulation of the rpoS regulon (or at least of a subset of σS-dependent genes) via iraP activation was observed both in MG1655 transformed with the pT7CsgD plasmid, allowing low-level, constitutive expression of the CsgD protein, and in the curli-proficient PHL628 strain (53), in which the csgD gene is expressed from its own promoter (Fig. 2). Thus, CsgD-mediated stabilization of σS, and its consequent effect on the expression of the rpoS regulon, do not depend on nonphysiological CsgD expression from the pT7CsgD plasmid. Our experiments were performed in phosphate-rich medium, suggesting that IraP-mediated σS stabilization might take place in response to environmental cues other than phosphate starvation; this would be in agreement with a recent report that, in Salmonella, iraP transcription is activated during growth at low Mg2+ concentration, resulting in IraP-mediated σS stabilization (67). In the csgD-expressing PHL628 strain of E. coli, CsgD-dependent iraP transcription can only take place in response to environmental signals leading to csgD expression and curli production, i.e., low temperature and osmolarity (data not shown). These observations would be consistent with a role for IraP in σS stabilization in a broad range of physiological conditions. From our results, we cannot establish whether CsgD activates iraP expression via direct interaction with the iraP promoter or through additional factors: we could not find in the iraP promoter region any sequence closely resembling the putative binding site for CsgD identified in the CsgD-dependent csgB, adrA, and pepD promoters (9). However, this does not rule out the possibility of CsgD binding to less conserved sites in the iraP promoter; despite various attempts, we have never succeeded in purifying active CsgD protein for in vitro DNA-binding assays.
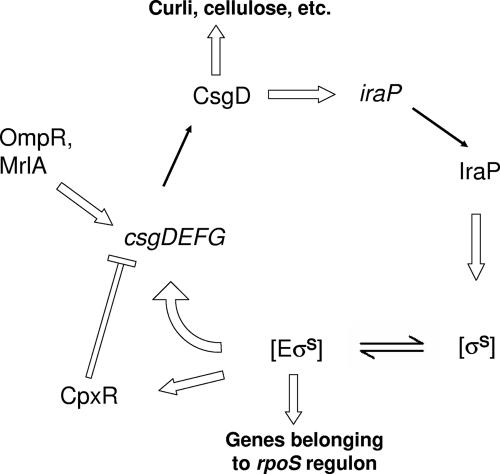
CsgD-mediated increase of σS cellular concentrations via the iraP gene would trigger an autoactivation loop, as summarized in Fig. 8, leading to an increased production of CsgD-dependent adhesion determinants such as curli fibers and cellulose. Indeed, in most curli-producing strains, transcription of the csgDEFG operon requires a functional rpoS gene (58), although it is not clear whether dependence on the rpoS gene is due to direct recognition of the csgD promoter by the EσS form of RNA polymerase or whether it is mediated by rpoS-dependent expression of the mrlA gene, encoding a transcription factor which, in turn, activates csgDEFG transcription (11). Either way, an increase in the cellular σS pool in conditions permitting csg gene expression (i.e., low osmolarity) leads to increased transcription from the csgD promoter; in turn, production of the CsgD protein results in iraP activation and consequent further stabilization of σS. Evidence for this autoregulatory mechanism in the MG1655 strain is provided by σS-dependent activation of the csgD promoter in the presence of pT7CsgD (Fig. 4B) and by consequent production of proteins encoded by the csgDEFG operon, such as CsgG (Fig. 2C), in response to CsgD-dependent accumulation of σS. Indeed, σS-dependent activation of the csgDEFG operon observed in Fig. 4B is impaired in an iraP mutant strain (data not shown). This autoregulatory circuitry might be further fueled by σS-dependent induction of genes encoding di-guanylate cyclases, i.e., proteins able to synthesize the second messenger di-cyclic-GMP, which, in turn, can positively affect csg gene expression (32, 72). A possible feedback control for this regulation loop could be provided by the CpxA/CpxR two-component regulatory system, whose activity is triggered by curli overexpression and results in repression of both the csgD and the csgB promoters (53). Consistent with this model, transcription of the cpxRA operon is itself positively controlled by σS (24).
FIG. 8.
Model for the CsgD/σS autoactivation loop. Thin black arrows indicate gene expression; block white arrows indicate positive regulatory interactions; white lines indicate negative interactions. See the text for details.
Besides more efficient expression of the csgDEFG operon, however, it is likely that CsgD-σS interaction plays a more general role, i.e., to coordinate the production of adhesion and cell aggregation factor (curli fibers and cellulose) to σS-dependent gene expression. Such coordination would relay the transition from single cell to biofilm to expression of the rpoS regulon, i.e., one of the main stress responses in bacteria (29). Interestingly, biofilm formation in E. coli, even when independent of CsgD and of curli production, appears to be tightly connected to σS-dependent gene expression (1, 4, 17, 34, 62) and to general induction of stress responses (4, 54), possibly suggesting that biofilm formation might represent a switch to a “resistance form” better equipped to withstand environmental stresses. However, to our knowledge, our work is the first demonstration of a mechanism for activation of a stress response (the rpoS regulon) by a positive regulator of biofilm formation.
In addition to resistance to environmental stresses, production of adhesion factors (curli and fimbriae) and σS-dependent gene expression play a role in pathogenic enterobacteria in adaptation and survival in the host and even in virulence: the σS-dependent gad genes allow survival of bacteria to acid stress in the gastric tract (20), whereas curli fibers are involved in bacterial internalization in epithelial host cells (27). Finally, σS controls the spvR virulence system in Salmonella (14). Thus, correlation between adhesion factor production and the rpoS regulon could constitute an important regulation mechanism of virulence in enterobacteria.
Other than its effect on the rpoS regulon, CsgD expression appears to regulate a subset of anaerobiosis-dependent genes, such as ompW and pflB, both induced in anaerobic conditions by FNR, either alone or in concert with ArcA (18, 61). In contrast, the gatZYABC operon, negatively controlled by CsgD (10) (Table 2) is repressed by ArcA in anaerobic conditions (60). It is thus possible that CsgD-mediated cell aggregation might, to some extent, mimic a switch to anaerobic conditions. Finally, CsgD expression also appears to affect expression of the tnaA gene (Fig. 2A), whose product catalyzes tryptophan degradation to indole and pyruvate. Indole acts as a signal molecule, and its accumulation results in inhibition of cell division (13), and repression of tnaA might prevent untimely indole accumulation leading to growth arrest. The impact of CsgD on expression of genes not related to adhesion factors' production, such as tnaA and anaerobic genes, as well as the rpoS regulon, is likely to reflect the cell's need to reprogram its global gene expression in response to transition from planktonic cells to biofilm.
Supplementary Material
Acknowledgments
We thank Françoise Norel for the gift of the anti-σS antibody, Corinne Dorel for endless supply of strains and plasmids, Eva Brombacher and Andrea Baratto for help with strain construction, Alexandre Bougdour for the gift of the iraP::kan strain, and Stefano Bertagnoli for performing the β-glucuronidase assays.
This study was supported in part by CARIPLO Foundation grant N. 2005.1076/10.4878.
Footnotes
Published ahead of print on 14 September 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Adams, J. L., and R. J. McLean. 1999. Impact of rpoS deletion on Escherichia coli biofilms. Appl. Environ. Microbiol. 65:4285-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altuvia, S., M. Almiron, G. Huisman, R. Kolter, and G. Storz. 1994. The dps promoter is activated by OxyR during growth and by IHF and sigma S in stationary phase. Mol. Microbiol. 13:265-272. [DOI] [PubMed] [Google Scholar]
- 3.Arnqvist, A., A. Olsen, and S. Normark. 1994. Sigma S-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by sigma 70 in the absence of the nucleoid-associated protein H-NS. Mol. Microbiol. 13:1021-1032. [DOI] [PubMed] [Google Scholar]
- 4.Beloin, C., J. Valle, P. Latour-Lambert, P. Faure, M. Kzreminski, D. Balestrino, J. A. Haagensen, S. Molin, G. Prensier, B. Arbeille, and J. M. Ghigo. 2004. Global impact of mature biofilm lifestyle on Escherichia coli K-12 gene expression. Mol. Microbiol. 51:659-674. [DOI] [PubMed] [Google Scholar]
- 5.Ben Nasr, A., A. Olsen, U. Sjobring, W. Muller-Esterl, and L. Bjorck. 1996. Assembly of human contact phase proteins and release of bradykinin at the surface of curli-expressing Escherichia coli. Mol. Microbiol. 20:927-935. [DOI] [PubMed] [Google Scholar]
- 6.Bian, Z., A. Brauner, Y. Li, and S. Normark. 2000. Expression of and cytokine activation by Escherichia coli curli fibers in human sepsis. J. Infect. Dis. 181:602-612. [DOI] [PubMed] [Google Scholar]
- 7.Bougdour, A., C. Lelong, and J. Geiselmann. 2004. Crl, a low temperature-induced protein in Escherichia coli that binds directly to the stationary phase sigma subunit of RNA polymerase. J. Biol. Chem. 279:19540-19550. [DOI] [PubMed] [Google Scholar]
- 8.Bougdour, A., S. Wickner, and S. Gottesman. 2006. Modulating RssB activity: IraP, a novel regulator of sigma(S) stability in Escherichia coli. Genes Dev. 20:884-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brombacher, E., C. Dorel, A. J. Zehnder, and P. Landini. 2003. The curli biosynthesis regulator CsgD co-ordinates the expression of both positive and negative determinants for biofilm formation in Escherichia coli. Microbiology 149:2847-2857. [DOI] [PubMed] [Google Scholar]
- 10.Brombacher, E., A. Baratto, C. Dorel, and P. Landini. 2006. Gene expression regulation by the curli activator CsgD protein: modulation of cellulose biosynthesis and control of negative determinants for microbial adhesion. J. Bacteriol. 188:2027-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown, P. K., C. M. Dozois, C. A. Nickerson, A. Zuppardo, J. Terlonge, and R. Curtiss. 2001. MlrA, a novel regulator of curli (AgF) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium. Mol. Microbiol. 41:349-363. [DOI] [PubMed] [Google Scholar]
- 12.Brown, W. C., and J. L. Campbell. 1993. A new cloning vector and expression strategy for genes encoding proteins toxic to Escherichia coli. Gene 127:99-103. [DOI] [PubMed] [Google Scholar]
- 13.Chant, E. L., and D. K. Summers. 2007. Indole signalling contributes to the stable maintenance of Escherichia coli multicopy plasmids. Mol. Microbiol. 63:35-43. [DOI] [PubMed] [Google Scholar]
- 14.Chen, C. Y., N. A. Buchmeier, S. Libby, F. C. Fang, M. Krause, and D. G. Guiney. 1995. Central regulatory role for the RpoS sigma factor in expression of Salmonella Dublin plasmid virulence genes. J. Bacteriol. 177:5303-5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, X., L. M. Smith, and E. M. Bradbury. 2000. Site-specific mass tagging with stable isotopes in proteins for accurate and efficient protein identification. Anal. Chem. 72:1134-1143. [DOI] [PubMed] [Google Scholar]
- 16.Chirwa, N. T., and M. B. Herrington. 2003. CsgD, a regulator of curli and cellulose synthesis, also regulates serine hydroxymethyltransferase synthesis in Escherichia coli K-12. Microbiology 149:525-535. [DOI] [PubMed] [Google Scholar]
- 17.Collet, A., S. Vilain, P. Cosette, G. A. Junter, T. Jouenne, R. S. Phillips, and P. Di Martino. 2007. Protein expression in Escherichia coli S17-1 biofilms: impact of indole. Antonie Leeuwenhoek 91:71-85. [DOI] [PubMed] [Google Scholar]
- 18.Constantinidou, C., J. L. Hobman, L. Griffiths, M. D. Patel, C. W. Penn, J. A. Cole, and T. W. Overton. 2006. A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K-12 adapts from aerobic to anaerobic growth. J. Biol. Chem. 281:4802-4815. [DOI] [PubMed] [Google Scholar]
- 19.Cookson, A. L., W. A. Cooley, and M. J. Woodward. 2002. The role of type 1 and curli fimbriae of Shiga toxin-producing Escherichia coli in adherence to abiotic surfaces. Int. J. Med. Microbiol. 292:195-205. [DOI] [PubMed] [Google Scholar]
- 20.Cotter, P. D., C. G. Gahan, and C. Hill. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40:465-475. [DOI] [PubMed] [Google Scholar]
- 21.De Biase, D., A. Tramonti, F. Bossa, and P. Visca. 1999. The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 32:1198-1211. [DOI] [PubMed] [Google Scholar]
- 22.Deeley, M. C., and C. Yanofsky. 1981. Nucleotide sequence of the structural gene for tryptophanase of Escherichia coli K-12. J. Bacteriol. 147:787-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deflaun, M. F., B. M. Marshall, E. P. Kulle, and S. B. Levy. 1994. Tn5 insertion mutants of Pseudomonas fluorescens defective in adhesion to soil and seeds. Appl. Environ. Microbiol. 60:2637-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Wulf, P., O. Kwon, and E. C. Lin. 1999. The CpxRA signal transduction system of Escherichia coli: growth-related autoactivation and control of unanticipated target operons. J. Bacteriol. 181:6772-6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gerstel, U., C. Park, and U. Romling. 2003. Complex regulation of csgD promoter activity by global regulatory proteins. Mol. Microbiol. 49:639-654. [DOI] [PubMed] [Google Scholar]
- 26.Gibson, D. L., A. P. White, S. D. Snyder, S. Martin, C. Heiss, P. Azadi, M. Surette, and W. W. Kay. 2006. Salmonella produces an O-antigen capsule regulated by AgfD and important for environmental persistence. J. Bacteriol. 188:7722-7730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gophna, U., M. Barlev, R. Seijffers, T. A. Oelschlager, J. Hacker, and E. Z. Ron. 2001. Curli fibers mediate internalization of Escherichia coli by eukaryotic cells. Infect. Immun. 69:2659-2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hammar, M., A. Arnqvist, Z. Bian, A. Olsen, and S. Normark. 1995. Expression of two csg operons is required for production of fibronectin- and Congo red-binding curli polymers in Escherichia coli K-12. Mol. Microbiol. 18:661-670. [DOI] [PubMed] [Google Scholar]
- 29.Hengge-Aronis, R. 2002. Recent insights into the general stress response regulatory network in Escherichia coli. J. Mol. Microbiol. Biotechnol. 4:341-346. [PubMed] [Google Scholar]
- 30.Hickman, J. W., D. F. Tifrea, and C. S. Harwood. 2005. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. USA 102:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jishage, M., and A. Ishihama. 1997. Variation in RNA polymerase sigma subunit composition within different stocks of Escherichia coli W3110. J. Bacteriol. 179:959-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kader, A., R. Simm, U. Gerstel, M. Morr, and U. Romling. 2006. Hierarchical involvement of various GGDEF domain proteins in rdar morphotype development of Salmonella enterica serovar Typhimurium. Mol. Microbiol. 60:602-616. [DOI] [PubMed] [Google Scholar]
- 33.Lacour, S., and P. Landini. 2004. σS-dependent gene expression at the onset of stationary phase in Escherichia coli: function of σS-dependent genes and identification of their promoter sequences. J. Bacteriol. 186:7186-7195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lacqua, A., O. Wanner, T. Colangelo, M. G. Martinotti, and P. Landini. 2006. Emergence of biofilm-forming subpopulations upon exposure of Escherichia coli to environmental bacteriophages. Appl. Environ. Microbiol. 72:956-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Latasa, C., A. Roux, A. Toledo-Arana, J. M. Ghigo, C. Gamazo, J. R. Penades, and I. Lasa. 2005. BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol. Microbiol. 58:1322-1339. [DOI] [PubMed] [Google Scholar]
- 36.Lelong, C., K. Aguiluz, S. Luche, L. Kuhn, J. Garin, T. Rabilloud, and J. Geiselmann. 2007. The Crl-RpoS regulon of Escherichia coli. Mol. Cell Proteomics 6:648-659. [DOI] [PubMed] [Google Scholar]
- 37.Loewen, P. C., and R. Hengge-Aronis. 1994. The role of the sigma factor sigma S (KatF) in bacterial global regulation. Annu. Rev. Microbiol. 48:53-80. [DOI] [PubMed] [Google Scholar]
- 38.Loferer, H., M. Hammar, and S. Normark. 1997. Availability of the fibre subunit CsgA and the nucleator protein CsgB during assembly of fibronectin-binding curli is limited by the intracellular concentration of the novel lipoprotein CsgG. Mol. Microbiol. 26:11-23. [DOI] [PubMed] [Google Scholar]
- 39.Martinez, A., and R. Kolter. 1997. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J. Bacteriol. 179:5188-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, J. H. (ed.). 1972. Experiments in molecular genetics, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 41.Natalello, A., S. M. Doglia, J. Carey, and R. Grandori. 2007. Role of flavin mononucleotide in the thermostability and oligomerization of Escherichia coli stress-defense protein WrbA. Biochemistry 46:543-553. [DOI] [PubMed] [Google Scholar]
- 42.Nobelmann, B., and J. W. Lengeler. 1996. Molecular analysis of the gat genes from Escherichia coli and of their roles in galactitol transport and metabolism. J. Bacteriol. 178:6790-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olsen, A., A. Arnqvist, M. Hammar, and S. Normark. 1993. Environmental regulation of curli production in Escherichia coli. Infect. Agents Dis. 2:272-274. [PubMed] [Google Scholar]
- 44.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 45.Patridge, E. V., and J. G. Ferry. 2006. WrbA from Escherichia coli and Archaeoglobus fulgidus is an NAD(P)H:quinone oxidoreductase. J. Bacteriol. 188:3498-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patten, C. L., M. G. Kirchhof, M. R. Schertzberg, R. A. Morton, and H. E. Schellhorn. 2004. Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol. Genet. Genomics 272:580-591. [DOI] [PubMed] [Google Scholar]
- 47.Paul, R., S. Weiser, N. C. Amiot, C. Chan, T. Schirmer, B. Giese, and U. Jenal. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Persson, K., W. Russell, M. Morgelin, and H. Herwald. 2003. The conversion of fibrinogen to fibrin at the surface of curliated Escherichia coli bacteria leads to the generation of proinflammatory fibrinopeptides. J. Biol. Chem. 278:31884-31890. [DOI] [PubMed] [Google Scholar]
- 49.Pilsl, H., D. Smajs, and V. Braun. 1999. Characterization of colicin S4 and its receptor, OmpW, a minor protein of the Escherichia coli outer membrane. J. Bacteriol. 181:3578-3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pratt, J. T., R. Tamayo, A. D. Tischler, and A. Camilli. 2007. PilZ domain proteins bind cyclic diguanylate and regulate diverse processes in Vibrio cholerae. J. Biol. Chem. 282:12860-12870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pratt, L. A., and T. J. Silhavy. 1998. Crl stimulates RpoS activity during stationary phase. Mol. Microbiol. 29:1225-1236. [DOI] [PubMed] [Google Scholar]
- 52.Prigent-Combaret, C., O. Vidal, C. Dorel, and P. Lejeune. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 181:5993-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Prigent-Combaret, C., E. Brombacher, O. Vidal, A. Ambert, P. Lejeune, P. Landini, and C. Dorel. 2001. Complex regulatory network controls initial adhesion and biofilm formation in Escherichia coli via regulation of the csgD gene. J. Bacteriol. 183:7213-7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ren, D., L. A. Bedzyk, S. M. Thomas, R. W. Ye, and T. K. Wood. 2004. Gene expression in Escherichia coli biofilms. Appl. Microbiol. Biotechnol. 64:515-524. [DOI] [PubMed] [Google Scholar]
- 55.Robbe-Saule, V., L. M. Dias, A. Kolb, and F. Norel. 2007. Physiological effects of Crl in Salmonella are modulated by σS level and promoter specificity. J. Bacteriol. 189:2976-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodel, W., W. Plaga, R. Frank, and J. Knappe. 1988. Primary structures of Escherichia coli pyruvate formate-lyase and pyruvate-formate-lyase-activating enzyme deduced from the DNA nucleotide sequences. Eur. J. Biochem. 177:153-158. [DOI] [PubMed] [Google Scholar]
- 57.Romling, U., W. D. Sierralta, K. Eriksson, and S. Normark. 1998. Multicellular and aggregative behavior of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol. Microbiol. 28:249-264. [DOI] [PubMed] [Google Scholar]
- 58.Romling, U., Z. Bian, M. Hammar, W. D. Sierralta, and S. Normark. 1998. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180:722-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Romling, U., M. Rohde, A. Olsen, S. Normark, and J. Reinkoster. 2000. AgfD, the checkpoint of multicellular and aggregative behavior in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 36:10-23. [DOI] [PubMed] [Google Scholar]
- 60.Salmon, K. A., S. P. Hung, N. R. Steffen, R. Krupp, P. Baldi, G. W. Hatfield, and R. P. Gunsalus. 2005. Global gene expression profiling in Escherichia coli K-12: effects of oxygen availability and ArcA. J. Biol. Chem. 280:15084-15096. [DOI] [PubMed] [Google Scholar]
- 61.Sawers, G. 1993. Specific transcriptional requirements for positive regulation of the anaerobically inducible pfl operon by ArcA and FNR. Mol. Microbiol. 10:737-747. [DOI] [PubMed] [Google Scholar]
- 62.Schembri, M. A., K. Kjaergaard, and P. Klemm. 2003. Global gene expression in Escherichia coli biofilms. Mol. Microbiol. 48:253-267. [DOI] [PubMed] [Google Scholar]
- 63.Simm, R., M. Morr, A. Kader, M. Nimtz, and U. Romling. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123-1134. [DOI] [PubMed] [Google Scholar]
- 64.Smith, D. K., T. Kassam, B. Singh, and J. F. Elliott. 1992. Escherichia coli has two homologous glutamate decarboxylase genes that map to distinct loci. J. Bacteriol. 174:5820-5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tischler, A. D., and A. Camilli. 2005. Cyclic diguanylate regulates Vibrio cholerae virulence gene expression. Infect. Immun. 73:5873-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tramonti, A., M. De Canio, I. Delany, V. Scarlato, and D. De Biase. 2006. Mechanisms of transcription activation exerted by GadX and GadW at the gadA and gadBC gene promoters of the glutamate-based acid resistance system in Escherichia coli. J. Bacteriol. 188:8118-8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tu, X., T. Latifi, A. Bougdour, S. Gottesman, and E. A. Groisman. 2006. The PhoP/PhoQ two-component system stabilizes the alternative sigma factor RpoS in Salmonella enterica. Proc. Natl. Acad. Sci. USA 103:13503-13508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vidal, O., R. Longin, C. Prigent-Combaret, C. Dorel, M. Hooreman, and P. Lejeune. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 180:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Visick, J. E., and S. Clarke. 1997. RpoS- and OxyR-independent induction of HPI catalase at stationary phase in Escherichia coli and identification of rpoS mutations in common laboratory strains. J. Bacteriol. 179:4158-4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Webb, J. S., L. S. Thompson, S. James, T. Charlton, T. Tolker-Nielsen, B. Koch, M. Givskov, and S. Kjelleberg. 2003. Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185:4585-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weber, H., T. Polen, J. Heuveling, V. F. Wendisch, and R. Hengge. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: sigmaS-dependent genes, promoters, and sigma factor selectivity. J. Bacteriol. 187:1591-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weber, H., C. Pesavento, A. Possling, G. Tischendorf, and R. Hengge. 2006. Cyclic-di-GMP-mediated signalling within the sigma network of Escherichia coli. Mol. Microbiol. 62:1014-1034. [DOI] [PubMed] [Google Scholar]
- 73.Zhao, G., P. Ceci, A. Ilari, L. Giangiacomo, T. M. Laue, E. Chiancone, and N. D. Chasteen. 2002. Iron and hydrogen peroxide detoxification properties of DNA-binding protein from starved cells: a ferritin-like DNA-binding protein of Escherichia coli. J. Biol. Chem. 277:27689-27696. [DOI] [PubMed] [Google Scholar]
- 74.Zheng, D., C. Constantinidou, J. L. Hobman, and S. D. Minchin. 2004. Identification of the CRP regulon using in vitro and in vivo transcriptional profiling. Nucleic Acids Res. 32:5874-5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zogaj, X., M. Nimtz, M. Rohde, W. Bokranz, and U. Romling. 2001. The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol. Microbiol. 39:1452-1463. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.