Abstract
The Amt proteins constitute a ubiquitous family of transmembrane ammonia channels that permit the net uptake of ammonium by cells. In many organisms, there is more than one amt gene, and these genes are subjected to nitrogen control. The mature Amt protein is a homo- or heterooligomer of three Amt subunits. We previously characterized an amt1 gene in the unicellular cyanobacterium Synechococcus elongatus strain PCC 7942. In this work, we describe the presence in this organism of a second amt gene, amtB, which encodes a protein more similar to the bacterial AmtB proteins than to any other characterized cyanobacterial Amt protein. The expression of amtB took place in response to nitrogen step-down, required the NtcA transcription factor, and occurred parallel to the expression of amt1. However, the transcript levels of amtB measured after 2 h of nitrogen deprivation were about 100-fold lower than those of amt1. An S. elongatus amtB insertional mutant exhibited an activity for uptake of [14C]methylammonium that was about 55% of that observed in the wild type, but inactivation of amtB had no noticeable effect on the uptake of ammonium when it was supplied at a concentration of 100 μM or more. Because an S. elongatus amt1 mutant is essentially devoid of [14C]methylammonium uptake activity, the mature Amt transporter is functional in the absence of AmtB subunits but not in the absence of Amt1 subunits. However, the S. elongatus amtB mutant could not concentrate [14C]methylammonium within the cells to the same extent as the wild type. Therefore, AmtB is necessary for full methylammonium uptake activity in S. elongatus.
The Amt proteins are transmembrane proteins that are ubiquitous in living organisms. These proteins are known as AMT proteins in higher plants and as MEP proteins in fungi, and the mammalian Rh proteins also belong to this protein family. A demonstrated function of these proteins is ammonia translocation across biological membranes. However, whether they mediate transport of the ammonium ion or translocation of the nonionic species ammonia (pKa [NH4+/NH3], 9.25) has been the subject of debate (36, 45). It has recently been shown that plant AMT proteins mediate electrogenic transport of ammonium, whereas the Rh proteins appear to mediate a nonelectrogenic translocation of ammonia (33). The crystal structures of the Amt proteins from Escherichia coli (23, 50) and Archaeoglobus fulgidus (1) have been determined. Inspection of these structures indicates that these proteins have a channel through which only ammonia can pass; however, the proteins initially bind ammonium, which is deprotonated before translocation. Coordinated transport of a proton could take place in some systems but not in others (25).
The available protein structures also show that the quaternary structure of Amt is a trimer in which each monomer provides a translocation pore. Many organisms carry more than one amt gene, and the formation of heterooligomers as well as of homooligomers has been shown for the yeast and plant proteins (26, 31). Interactions between the different polypeptides in the trimer could be deduced, but analysis of Saccharomyces cerevisiae mutants has shown that each MEP protein exhibits a different substrate affinity (30).
In many microbes, the amt genes are subjected to nitrogen control and are expressed mainly under nitrogen deficiency conditions. The Amt proteins appear, therefore, to have a role in uptake of ammonium when it is present at very low concentrations in the extracellular medium (30, 45). The Amt/MEP proteins are also responsible for a methylammonium uptake activity that was originally described in Penicillium chrysogenum (19) and is exhibited by many organisms. Uptake of [14C]methylammonium can be used to probe the activity of these proteins, although not all Amt/MEP proteins may recognize methylammonium as a substrate (34, 44). In many organisms, methylammonium is converted into methylglutamine by glutamine synthetase, and therefore measurements of [14C]methylammonium uptake involve both transport and metabolism of the substrate (5, 7).
The cyanobacteria are oxygenic photoautotrophs that can utilize for growth diverse inorganic sources of nitrogen, including nitrate and ammonium (14). Ammonium is a preferred nitrogen source that promotes repression of many nitrogen assimilation genes. When cyanobacterial cells are incubated in the absence of ammonium and with an adequate supply of CO2, the expression of these genes is activated by the transcriptional regulator NtcA (20). NtcA binds to a conserved sequence in the regulated promoters, GTAN8TAC (20, 27), and binding is most efficient in the presence of 2-oxoglutarate (48). The unicellular cyanobacterium Synechocystis sp. strain PCC 6803 has been shown to express, mainly under nitrogen deficiency conditions, three different amt genes, of which the amt1 gene is responsible for a major fraction of the methylammonium uptake activity (35). An amt1 gene has also been characterized in Synechococcus elongatus strain PCC 7942 (49). Its expression is NtcA dependent, and the encoded protein, Amt1, is essential for methylammonium uptake, for growth in the presence of low concentrations of ammonium, and for recapture of ammonium leaked out from the cells (49). In this work, a second S. elongatus amt gene is characterized, and the mechanism of ammonium uptake in this cyanobacterium is discussed.
MATERIALS AND METHODS
Strains and growth conditions.
S. elongatus strain PCC 7942 was grown axenically at 30°C in the light (85 microeinsteins m−2 s−1) in BG11 (nitrate-containing) medium (41) or in BG110 (BG11 medium lacking NaNO3) supplemented with 4 mM NH4Cl and 8 mM N-tris(hydroxymethyl)methyl-2-aminoethanesulfonic acid (TES)-NaOH buffer (pH 7.5). For mutants, the medium was supplemented with 7 μg chloramphenicol ml−1, 2 μg spectinomycin ml−1, or 2 μg streptomycin ml−1. For plates, the medium was solidified with separately sterilized 1% agar (Difco). Cultures used for RNA isolation and for uptake assays were grown in BG110C (BG110 supplemented with 10 mM NaHCO3) supplemented with 8 mM NH4Cl and 16 mM TES-NaOH buffer (pH 7.5) and bubbled with a mixture of CO2 (1%, vol/vol) and air. At the mid-exponential phase of growth (4 to 5 μg of chlorophyll a [Chl] ml−1), the cells were harvested at room temperature, washed twice with BG110C, resuspended in BG110C or BG11C (BG11 supplemented with 10 mM NaHCO3), and incubated for the indicated times under culture conditions with CO2-enriched air. Cyanobacterial cell mass was estimated by measuring the concentration of Chl in the cultures, as determined using methanolic extracts of the cells (29). The concentration of proteins was determined by a modified Lowry assay (32), using bovine serum albumin as a standard.
E. coli strain DH5α was grown in LB medium to which antibiotics were added, when necessary, at the following concentrations: ampicillin, 50 μg ml−1; spectinomycin, 25 μg ml−1; streptomycin, 25 μg ml−1; and chloramphenicol, 30 μg ml−1.
Inactivation of amtB.
A DNA fragment was amplified by PCR using primers amtB-7942-1 and amtB-7942-2 (Table 1). PCR amplification was carried out in a 50-μl mixture containing 2 ng of genomic DNA from strain PCC 7942, each deoxynucleoside triphosphate at a concentration of 0.2 mM, 50 pmol of each primer, 2.5 U of Taq polymerase (EcoTaq; Ecogen S.R.L.), and the reaction buffer supplied with this enzyme (Ecogen S.R.L.). The program used for amplification was 95°C for 2 min, followed by 35 cycles of denaturation for 1 min at 95°C, annealing for 1 min at 60°C, and polymerization for 1.5 min at 72°C and then by incubation at 72°C for 10 min. The PCR product was cloned in the pGEM-T vector (Promega), and the plasmid generated was designated pCSP30. The 2-kb Smr Spr gene cassette C.S3 from pRL463 (10), excised with SalI, was inserted into a unique XhoI site that was present in the pCSP30 insert in order to generate plasmid pCSP31.
TABLE 1.
Some deoxyoligonucleotide primers used in this work
| Primer | Sequence |
|---|---|
| amtB-7942-1 | 5′ TCA GGC ATG TTC GAC AAA CG 3′ |
| amtB-7942-2 | 5′ CCT AGC GCT CTT TGC TGT AGG C 3′ |
| amt1-qPCR-1 | 5′ GGG CGG CTC CTT CAT CGT C 3′ |
| amt1-qPCR-2 | 5′ TCC ATC CCG TGT TCG CCA ATA 3′ |
| amtB-qPCR-1 | 5′ GTC GCC GCC GCA CAA TG 3′ |
| amtB-qPCR-2 | 5′ GCC GCC CAA ACA GGA ATG AA 3′ |
| glnA-qPCR-1 | 5′ ACC GGC GGC CAG TGT GAG 3′ |
| glnA-qPCR-2 | 5′ CGG CGC GTG CTT GAG GAT A 3′ |
| glnB-qPCR-1 | 5′ CAG AAG TGC GCG GGT TTT GGT C 3′ |
| glnB-qPCR-2 | 5′ TGC GTC GGC GTT TTT CTC G 3′ |
| rpoD1-qPCR-1 | 5′ CGT AAG CCC ACC GAG GAA GAG A 3′ |
| rpoD1-qPCR-2 | 5′ GCG GGG GCT GAG AGT GCT AAG 3′ |
Transformation of strain PCC 7942 and amt1 mutant CSF72 (49) with plasmid pCSP31 was performed as described previously (18). Transformants were selected in BG11 solid medium supplemented with 7 μg chloramphenicol ml−1, 2 μg spectinomycin ml−1, and 2 μg streptomycin ml−1 for CSF72 and with 2 μg spectinomycin ml−1 and 2 μg streptomycin ml−1 for strain PCC 7942. To test whether the resulting mutant strains were homozygous for the mutant chromosomes, a PCR analysis was performed for transformants using primers amtB-7942-1 and amtB-7942-2 (see above). Double-recombinant, homozygous clones were obtained for the two transformations and designated strains CSP11 (genotype, amtB::C.S3) and CSP12 (genotype, amt1::C.C2 amtB::C.S3).
DNA and RNA isolation, manipulation, and analysis.
Isolation of genomic DNA from cyanobacteria was carried out as described previously (8). Isolation of total RNA from strain PCC 7942 and from mutants CS37 (ntcA::C.C2), CSF72, CSP11, and CSP12 was done as described previously (17).
For Northern blotting, 20 μg of RNA was loaded in each lane and electrophoresed in denaturing 1% agarose formaldehyde gels. DNA probes for S. elongatus genes were generated by PCR using plasmids carrying the corresponding genes as templates and oligonucleotide primers that produced the following gene fragments: for amt1, from bp 13 to bp 345 with respect to the start of the gene; and for amtB, from bp −7 with respect to the start of the gene to the end of the gene. Hybridizations were performed at 65°C in 1 mM EDTA-0.3 M sodium phosphate buffer (pH 6.8) containing 7% sodium dodecyl sulfate (SDS), and the filters were washed at 65°C successively with 2× SSC-0.1% SDS, 1× SSC-0.1% SDS, and 0.5× SSC-0.1% SDS (1× SSC is 150 mM NaCl and 15 mM sodium citrate dihydrate). As a control for RNA loading and transfer efficiency, the filters were reprobed with a 0.57-kb XhoI-PstI fragment that contained the RNase P RNA gene (rnpB) from strain PCC 7942 (4). Probes were labeled with a DNA labeling kit (Ready to Go; Amersham Pharmacia Biotech) and [α-32P]dCTP. Radioactive areas in Northern blot hybridizations were visualized and quantified with a Cyclone storage phosphor system (Packard).
Comparative RNA hybridization and quantitative reverse transcription-PCR (RT-PCR).
The DNA probes used were those described above for amt1 and amtB and probes for glnA (from bp 37 to bp 392 with respect to the start of the gene) and glnB (from bp −2 to bp 339 with respect to the start of the gene). For hybridization of blots of these probes (2 pmol of PCR products resolved by electrophoresis in agarose gels) with total cyanobacterial RNA (16), 15 μg of RNA from strain PCC 7942 was partially hydrolyzed by incubation in 125 mM NaOH for 40 min at 0 to 4°C and labeled with T4 polynucleotide kinase (Roche) and [γ-32P]dCTP (3). Hybridization was performed at 65°C in a solution containing 50 mM Tris-HCl (pH 7.4), 0.2% bovine serum albumin, 0.2% Ficoll, 0.1% sodium pyrophosphate, 1% SDS, 1 M NaCl, and 100 μg ml−1 yeast tRNA (Roche) (11). Filters were washed twice at 65°C for 30 min with 1× SSC-1% SDS and once at room temperature for 15 min with 0.2× SSC (pH 7.0).
For quantitative RT-PCR, total RNA was isolated (17) from five independent cultures of wild-type strain PCC 7942 incubated without combined nitrogen for 2 h. Retrotranscription, catalyzed by SuperScript II reverse transcriptase (Invitrogen), was carried out with 10 μg RNA and oligonucleotide primers amt1-qPCR-2, amtB-qPCR-2, glnA-qPCR-2, glnB-qPCR-2, and rpoD1-qPCR-2 (Table 1), which were designed using Primer Select from DNAStar software. After treatment with RNase H, cDNA was amplified by PCR with oligonucleotide primer pairs amt1-qPCR-1/amt1-qPCR-2, amtB-qPCR-1/amtB-qPCR-2, glnA-qPCR-1/glnA-qPCR-2, glnB-qPCR-1/glnB-qPCR-2, and rpoD1-qPCR-1/rpoD1-qPCR-2, respectively (Table 1), using a Quantimix EASY SYG kit from Biotools. The thermal cycling program consisted of an initial preheating step of 3 min at 95°C, followed by 40 cycles of 30 s at 95°C, 30 s at 61.4°C, and 30 s at 72°C. The generation of products was monitored after each extension step by measuring the fluorescence intensity of the double-stranded DNA-binding SYBR green I dye using the multicolor real-time PCR detection system of Bio-Rad. The cycle threshold (CT) value was determined for each gene and RNA sample in triplicate, and the relative values of expression were derived from the corresponding 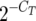 values using the rpoD1 gene as a reference.
values using the rpoD1 gene as a reference.
Methylammonium uptake assays, ammonium uptake, glutamine synthetase activity, and growth tests.
S. elongatus wild-type and mutant strains were grown in ammonium-containing medium bubbled with a mixture of CO2 (1%, vol/vol) and air, and the cells were harvested at room temperature, washed twice with BG110C, resuspended in BG110C, and incubated under culture conditions with CO2-enriched air. The cells were harvested at room temperature, washed with 0.5 mM (or 20 mM [see Fig. 7]) KH2PO4-10 mM NaHCO3-NaOH buffer (pH 7.1), and resuspended in the same buffer to obtain a cell density corresponding to 10 μg Chl ml−1. After preincubation at 30°C in the light (85 microeinsteins m−2 s−1) for 5 to 30 min, the assays were started by mixing the suspension of cells with a solution of 14CH3NH2·HCl (2.11 × 106 Bq μmol−1; Amersham) in phosphate-bicarbonate buffer. Alternatively (see Fig. 5), the cell suspension in BG110C was directly supplemented with a 14CH3NH2·HCl solution in water. After incubation for the time periods indicated below, 1-ml samples were filtered with 0.45-μm-pore-size Millipore HA filters. Without any further washing (22), the filters carrying the cells were immersed in scintillation cocktail, and the radioactivity was measured. The retention of radioactivity by boiled cells was used as a blank. For chase experiments, the cells were supplemented with 1 mM NH4Cl at the indicated time points before filtering.
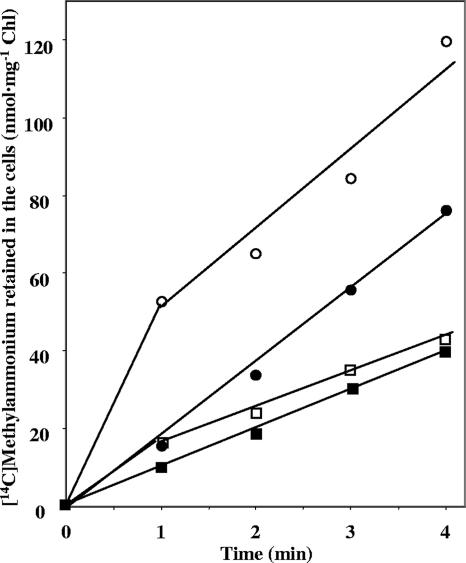
FIG. 7.
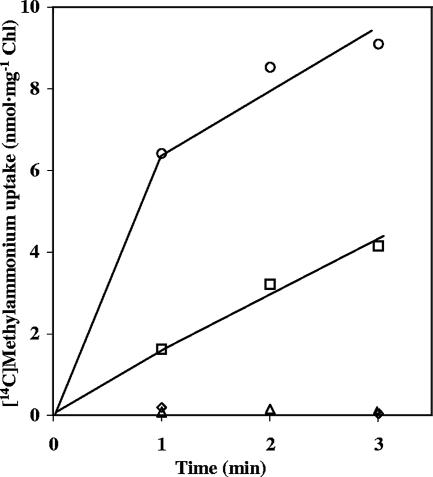
Uptake of [14C]methylammonium, supplied at a low concentration, by the S. elongatus wild type and mutants CSF72 (amt1), CSP11 (amtB), and CSP12 (amt1 amtB). Ammonium-grown cells incubated for 2 h in BG110C with CO2-enriched air were used in uptake assays in phosphate-bicarbonate buffer with 0.12 μM [14C]methylammonium. ○, wild-type S. elongatus; ▵, strain CSF72; □, strain CSP11; ⋄, strain CSP12.
FIG. 5.
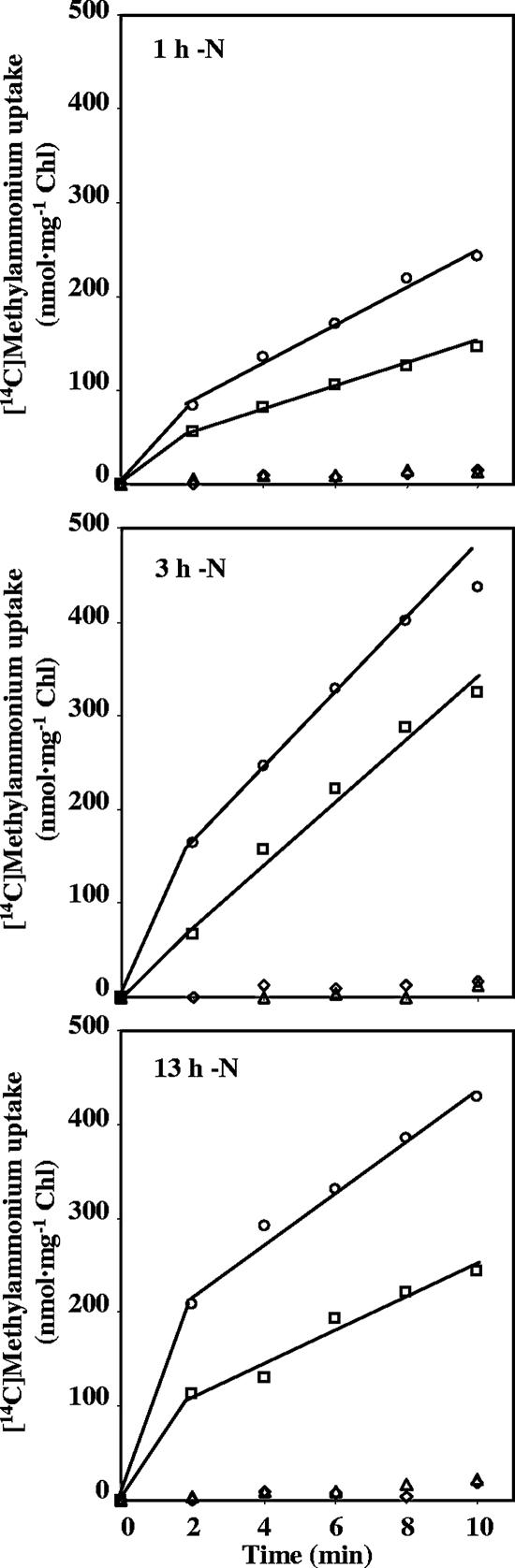
[14C]methylammonium uptake by the S. elongatus wild type and mutants CSF72 (amt1), CSP11 (amtB), and CSP12 (amt1 amtB). Cells from cultures grown with ammonium and CO2-enriched air (in the presence of antibiotics for the mutants) were incubated under the same conditions in BG110C for the indicated periods of time. Samples were then supplemented with 5 μM [14C]methylammonium, and retention of radioactivity by the cells was determined as specified in Materials and Methods. ○, wild-type S. elongatus; ▵, strain CSF72; □, strain CSP11; ⋄, strain CSP12.
Ammonium uptake assays were performed with cells grown in ammonium-containing medium bubbled with a mixture of CO2 (1%, vol/vol) and air. The cells were harvested at room temperature, washed twice with BG110C, resuspended in BG110C (pH 8) to obtain a cell density corresponding to 2.5 μg Chl ml−1, and incubated for 1.5 h under culture conditions with CO2-enriched air. The experiments were started by addition of NH4Cl (100 μM) to cell suspensions. Ammonium disappearance was determined by estimating the concentration of ammonium in the cell suspension after rapid removal of the cells by filtration of 2.5-ml samples (with 0.45-μm-pore-size Millipore HA filters) after incubation for different time periods up to 40 min. Ammonium contents were determined with glutamate dehydrogenase (6).
Glutamine synthetase transferase activity was determined in cell extracts as described previously (9). Cell extracts were prepared from an amount of cells containing about 7.5 μg of Chl. The cells were harvested by centrifugation (16,100 × g, 4°C, 5 min) and resuspended in 200 μl of a buffer containing 50 mM Tris-HCl (pH 7.4), 4 mM EDTA, 1 mM dithiothreitol, and 0.5 mM benzamidine, which was supplemented with about 150 mg of glass beads. After three cycles of 3 min of vortexing and incubation on ice, each suspension was centrifuged as described above to remove the glass beads and cell debris, and the resulting supernatant was used to assay the enzyme activity. One unit corresponded to the formation of 1 μmol of γ-glutamylhydroxamate per min.
To test growth of the mutants on solid medium, plates of BG110 that were not supplemented or were supplemented with 100 μM or 2 mM NH4Cl and buffered with 5 mM TES at pH 7.5 were prepared. Because of its ferric ammonium citrate content, BG110 contains an unspecified concentration of ammonium that may be as high as about 20 μM. Drops (10 μl) of cell suspensions of strains PCC 7942, CSF72, CSP11, and CSP12 at a concentration corresponding to 1 μg Chl ml−1 were spotted on the medium, and the plates were incubated under culture conditions for 2 weeks.
Calculations.
To apply the Nernst equation, the intracellular concentration of methylammonium was calculated by using an intracellular cell volume of 125 μl mg Chl−1 (21, 39). Passive influx of NH3 was calculated by applying Fick's diffusion equation, J = P·Δc (where J is the flux and Δc is the concentration gradient), using a conservative permeability coefficient for ammonia (P) of 10−5 m s−1 (2, 24) (for ammonia permeability coefficients in S. elongatus, see reference 43). To compare the deduced diffusion value to observed ammonium uptake rates, it was assumed that an S. elongatus cell has a surface area of about 10−11 m2 and that 1 μg of Chl corresponds to about 5 × 107 Synechococcus cells (E. Flores, unpublished).
Phylogenetic analysis.
Predicted polypeptide sequences were aligned with the program ClustalX 1.8 (47). Phylogenetic trees were visualized with the NJplot program (38).
RESULTS
Identification of amtB.
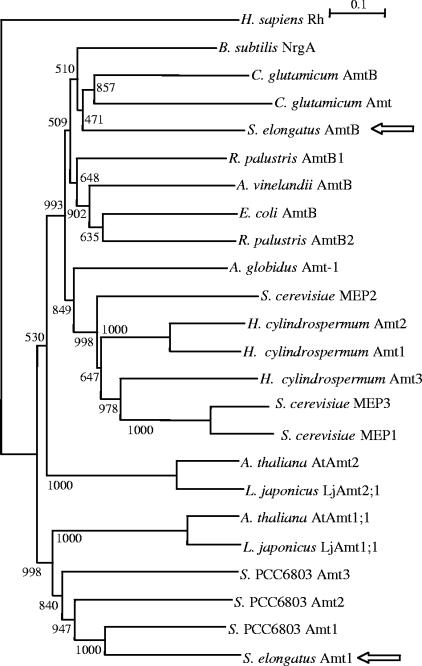
Inspection of the genomic sequence of S. elongatus strain PCC 7942 (http://img.jgi.doe.gov/cgi-bin/pub/main.cgi) showed the presence of two putative amt genes. Open reading frame Syn_pcc79420442 is the previously characterized amt1 gene (49). Open reading frame Syn_pcc79422279 encodes a protein that exhibits 50.9% identity with Amt1. However, a phylogenetic analysis of Amt proteins from a number of different biological sources placed the Syn_pcc79422279 product in the branch where the AmtB proteins from E. coli and other organisms are also located rather than in the branch containing S. elongatus Amt1 or Synechocystis Amt1, Amt2, or Amt3 (Fig. 1). Therefore, the Syn_pcc79422279 protein appears to be more closely related to some bacterial Amt proteins, of which Escherichia AmtB is the best-known example, than to other characterized cyanobacterial Amt proteins. We refer to Syn_pcc79422279 here as the S. elongatus amtB gene.
FIG. 1.
Phylogenetic relationships of some Amt/MEP proteins. The analysis was restricted to some proteins that have been experimentally characterized, and a distantly related human Rh protein was used as the outgroup. Bootstrap values based on 1,000 trials are indicated at the nodes. The arrows indicate the S. elongatus Amt proteins.
Expression of amtB.
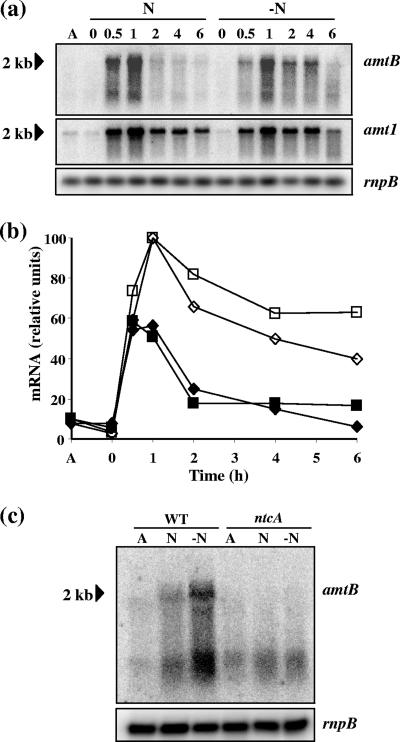
The expression of amtB was studied by Northern analysis. The level of expression was low in ammonium-grown cells of S. elongatus (Fig. 2a), and induction was observed after transfer of the cells to media with nitrate or no combined nitrogen. Expression was detected as soon as 30 min after withdrawal of ammonium but declined after 1 h of incubation, especially in nitrate-supplemented cultures. For comparison, the filter was rehybridized with an amt1 probe. The expression of amtB and the expression of amt1 were parallel in both nitrate-containing medium and medium lacking any source of combined nitrogen (Fig. 2a and b), although the levels of expression of amt1 were higher than those of amtB (see below). The amtB gene was expressed as an approximately 2-kb transcript. Since amtB is 1,410 bp long and inspection of the S. elongatus genome showed no putative accompanying gene, amtB appears to be transcribed monocistronically.
FIG. 2.
Expression of the amtB gene in S. elongatus. (a) Northern analysis performed with RNA isolated from cells of wild-type S. elongatus grown with ammonium (lane A) or grown with ammonium and incubated for the numbers of hours indicated above the lanes in the presence of nitrate (N) or with no combined nitrogen (−N). The filter was successively hybridized with the amtB probe (upper panel), the amt1 probe (middle panel), and an rnpB probe that was used as a loading and transfer control (lower panel). (b) Comparison of the time course patterns of expression of amtB and amt1. The amtB and amt1 signals in panel a were quantified and normalized with the rnpB signals. For each probe (amtB or amt1), the signal obtained after 1 h of incubation without combined nitrogen was defined as 100. Diamonds, amtB; squares, amt1; open symbols, without N; filled symbols, with nitrate. Note that the relative levels of expression of the genes with respect to each other cannot be deduced from these data because visualization of amtB required a longer exposure than visualization of amt1. (c) Northern analysis performed with RNA isolated from cells of the wild type (WT) or the S. elongatus ntcA mutant grown with ammonium (lanes A) or grown with ammonium and incubated for 2 h in the presence of nitrate (lanes N) or with no combined nitrogen (lanes −N), using the amtB probe (upper panel) or an rnpB probe (lower panel).
As is the case for amt1 (49), induction of amtB was not observed in an S. elongatus ntcA mutant in medium containing nitrate or in the absence of combined nitrogen (Fig. 2c). This shows that like amt1, amtB is subject to NtcA-mediated nitrogen control. A putative NtcA binding site, GTAN8TAC, is located 84 to 72 bp upstream from the translation start of the gene, which could be involved in NtcA-dependent regulation. Unfortunately, however, we were unable to determine the transcription start point of amtB, perhaps because of its low level of expression.
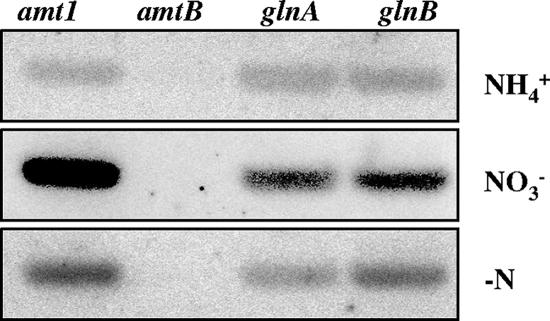
To compare the level of expression of amtB to that of other nitrogen assimilation genes, DNA probes of amt1, amtB, glnA, and glnB were fixed in a membrane filter and hybridized with 32P-labeled S. elongatus total RNA. RNA isolated from cultures incubated with ammonium, nitrate, or no combined nitrogen was used. The hybridization signal of the amtB probe was much lower than that of any of the other probes for the three RNA preparations (Fig. 3). These results indicate that amtB is expressed at lower levels than the other nitrogen assimilation genes tested under any of the nitrogen regimens used in our analysis. The large difference between expression of amtB and expression of amt1 is remarkable.
FIG. 3.
Expression of amtB compared to expression of other nitrogen assimilation genes (amt1, glnA, and glnB). DNA fragments corresponding to the indicated genes of S. elongatus were bound to a membrane filter and hybridized with 32P-labeled RNA extracted from cells grown with ammonium or grown with ammonium and incubated with nitrate or without combined nitrogen (−N) for 2 h. The presence of the amtB probe in the filters was corroborated by Southern analysis (not shown). Note that hybridizations with the different labeled RNA preparations (isolated from cells subjected to the different nitrogen regimens) cannot be quantitatively compared to each other because they required different exposure times.
To quantify the differences in expression of the amtB, amt1, glnA, and glnB genes, quantitative RT-PCR was performed, as described in Materials and Methods, using RNA samples isolated from cultures of S. elongatus incubated for 2 h in the absence of combined nitrogen. The relative levels of expression determined with five independent RNA samples, using the RNA polymerase rpoD1 gene as a reference, were 0.10 ± 0.02 for amtB, 1 for rpoD1, 3.47 ± 2.27 for glnA, 5.82 ± 1.40 for glnB, and 10.32 ± 2.59 for amt1. Therefore, a difference of over 100-fold was found between the expression of amtB and the expression of amt1 after 2 h of nitrogen deprivation.
Inactivation of amtB.
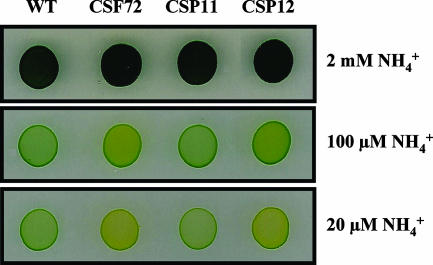
An interrupted amtB::C.S3 gene was transferred by transformation to wild-type strain PCC 7942 and mutant strain CSF72 (amt1::C.C2), generating mutant strains CSP11 (amtB::C.S3) and CSP12 (amt1::C.C2 amtB::C.S3), respectively. Growth tests in solid medium supplemented with different concentrations of ammonium as the sole nitrogen source at pH 7.5 were performed with four strains, PCC 7942, CSF72, CSP11, and CSP12. No differences in growth were observed when the ammonium concentration was 2 mM (Fig. 4). Impairment of ammonium-dependent growth of the amtB mutant (strain CSP11), indicated by yellow-green coloration, was barely observed only with the lowest ammonium concentration tested (ca. 20 μM). In contrast, the ammonium-dependent growth of the amt1 mutant (strain CSF72) was impaired with ammonium concentrations of 100 and 20 μM (49). The results for the amt1 amtB double mutant (strain CSP12) were similar to those for the amt1 mutant.
FIG. 4.
Growth of the S. elongatus wild type (WT) and mutants CSF72 (amt1), CSP11 (amtB), and CSP12 (amt1 amtB) on solid medium with ammonium as the nitrogen source. Cells grown with ammonium (in the presence of antibiotics for the mutants) were washed with BG110 and spotted on BG110 (buffered at pH 7.5) with the indicated concentrations of ammonium. Each spot was inoculated with an amount of cells containing 0.01 μg of Chl.
Uptake of methylammonium.
The uptake of [14C]methylammonium was determined for the wild-type strain and the single and double amt mutants in time course induction experiments. The results of a representative experiment in which ammonium-grown cells that had been incubated in the absence of nitrogen for 1, 3, or 13 h were used in uptake assays with 5 μM [14C]methylammonium are shown in Fig. 5. As previously described (49), the amt1 mutant showed a very low rate of methylammonium uptake. In contrast, the amtB mutant, although its activity was impaired, still showed a substantial level of uptake. The results of eight different experiments using 5 μM [14C]methylammonium as the substrate showed that in the amtB mutant the uptake activity was about 55% ± 9% of the activity in the wild type. Like the amt1 mutant, the amt1 amtB double mutant showed about 5% of the activity of the wild type. Some of the data in Fig. 5 also reflect the two phases that have been distinguished for methylammonium uptake in S. elongatus: quick uptake essentially reflecting transport, followed by a steady phase corresponding to methylamine incorporation into methylglutamine (7).
Because the steady phase of methylammonium uptake is dependent on glutamine synthetase, the activity of this enzyme was determined in the wild-type and mutant strains. The transferase activity of glutamine synthetase was measured in cell extracts from ammonium-grown cells that had been incubated for 2 h in the absence of combined nitrogen. The activities obtained were 63.1 ± 3.1 U mg of Chl−1for strain PCC 7942, 64.4 ± 6.9 U mg of Chl−1 for strain CSP11, 63.2 ± 3.5 U mg of Chl−1 for strain CSP12, and 67.2 ± 1.2 U mg of Chl−1 for strain CSF72 (means ± standard deviations for the results of three independent assays). Therefore, no significant differences in glutamine synthetase activity were evident.
In cyanobacteria, methylammonium uptake is a membrane potential-dependent process that permits accumulation of the substrate within the cell (7, 35, 40). The effect of inactivation of amtB on the ability of S. elongatus to retain [14C]methylammonium was investigated in chase experiments with ammonium. As shown in Fig. 6, a fraction of the methylammonium taken up was displaced by ammonium. This fraction must correspond to the methylammonium accumulated within the cells, and the remaining radioactivity must correspond to [14C]methylglutamine. A calculation based on the Nernst equation indicated that the observed concentration gradient of [14C]methylammonium in the wild type corresponded to a free energy change of 115 mV, roughly corresponding to the accumulation permitted by the membrane potential of actively photosynthesizing Synechococcus cells, which is about −110 to −130 mV (42). A similar chase experiment performed with the amtB mutant showed that the amount of methylammonium that this mutant could accumulate was only about 10% of the amount observed in the wild type (Fig. 6).
FIG. 6.
[14C]methylammonium uptake and chase with ammonium in the S. elongatus wild type and mutant CSP11 (amtB). Ammonium-grown cells incubated for 2 h in BG110C with CO2-enriched air were used in [14C]methylammonium uptake assays in phosphate-bicarbonate buffer and supplemented (filled symbols) or not supplemented (open symbols) with 1 mM ammonium before filtration. Circles, wild type S. elongatus; squares, strain CSP11.
Using a different experimental approach, the uptake of very low concentrations of [14C]methylammonium was investigated. Figure 7 shows the results of a representative experiment in which the uptake of 0.12 μM [14C]methylammonium was tested. Whereas the two phases of [14C]methylammonium uptake were observed for the wild type, only the lineal phase was observed for the amtB mutant. The amt1 mutant and the amt1 amtB double mutant showed negligible uptake activities. Therefore, AmtB appears to be needed for accumulation of [14C]methylammonium within the cells at this very low methylammonium concentration.
Uptake of ammonium.
The amt1 amtB double mutant (strain CSP12) can grow using ammonium as a nitrogen source. In direct ammonium uptake assays (100 μM ammonium, CO2-supplemented cells, pH 8), uptake rates of 1 μmol min−1 mg Chl−1 were observed for the four strains analyzed in this work: the wild-type S. elongatus strain, CSF72, CSP11, and CSP12. This uptake rate corresponds to an influx into the cells of about 3 × 10−8 mol m−2 s−1, which in the amt1 amtB double mutant could take place by diffusion of ammonia. A calculation based on Fick's diffusion equation showed that the value for ammonia diffusion would be on the order of 10−8 mol m−2 s−1 for a difference in the NH3 concentration of 1 μM. Thus, to account for the observed uptake rates, a difference in the NH3 concentration of 3 μM (intracellular concentration lower than the extracellular concentration) would be necessary. Assimilation of ammonia by glutamine synthetase, which in cyanobacteria exhibits Km values for ammonia as low as 20 μM (13), could maintain this ammonia gradient, permitting diffusion to take place.
DISCUSSION
The S. elongatus genome carries two amt genes, the previously described amt1 gene (49) and the amtB gene characterized in this work. These two genes are subjected to N control mediated by the NtcA transcriptional regulator, so that they are expressed in response to withdrawal of ammonium. For both genes, expression is higher in the absence of combined nitrogen than in the presence of nitrate, and the latter conditions lead to a significant decrease in expression soon after induction (Fig. 2a and b). This decrease in expression has also been observed in S. elongatus for other nitrogen assimilation genes (37), and it might be related to a feedback effect of the ammonium resulting from nitrate assimilation. Expression of NtcA-activated genes in nitrogen-starved S. elongatus cells has been shown, on the other hand, to be dependent on the signal transduction protein PII (the glnB gene product [37]).
The results presented in this work show that whereas Amt1 is essential for [14C]methylammonium uptake, AmtB appears to be needed only for maximum uptake levels. It is possible that the optimal Amt complex in the S. elongatus cytoplasmic membrane consists of both Amt1 and AmtB subunits, but whereas an Amt1 homooligomer is functional, a complex consisting of only AmtB subunits is not. This would be consistent with the differences observed in the expression levels of amt genes that support a scenario where Amt1 subunits would be more represented than AmtB subunits in Amt trimers. Nonetheless, given the much lower expression of amtB, the contribution of AmtB to activity seems to be substantial. Additionally, AmtB is important for providing S. elongatus cells with the ability to concentrate methylammonium and, hence, probably also ammonium. This can be especially important when the cells encounter very low external concentrations of ammonium, conditions under which accumulation of ammonium within the cells (to the level permitted by the membrane potential) can favor the functioning of glutamine synthetase. NtcA-dependent expression of the amt genes under nitrogen deficiency conditions is consistent with a role for the Amt complex(es) in scavenging nitrogen from low ammonium concentrations by S. elongatus cells. Although we have not been able to study growth dependent on very low concentrations of ammonium, AmtB might be needed for optimal growth with ammonium concentrations below 20 μM (Fig. 4).
The physiological studies described in this work do not permit derivation of mechanistic conclusions regarding the functioning of the Amt proteins. They raise, however, novel questions. It is noteworthy that AmtB, which allows greater efficiency in methylammonium uptake and permits accumulation of methylammonium within the cells, cannot support this activity alone (Fig. 6 and 7). Whether this protein has a structural role in Amt trimers or has an activity complementary to that of the Amt1 proteins is unknown. Parallel expression of the amtB and amt1 genes (Fig. 2) is consistent with possible joint action of their corresponding protein products. Whatever the mechanism, the possibility that AmtB is necessary for the Amt complex to transport the methylammonium ion (especially when methylammonium is present at low concentrations in the outer medium), whereas Amt1 is sufficient to make a complex that can mediate translocation of methylamine, is intriguing. In this scenario, Amt1-dependent uptake of methylamine pulled by glutamine synthetase would take place, as shown for the amtB mutant in Fig. 6 and 7.
A phylogenetic analysis of cyanobacterial Amt proteins recently performed by Luque and Forchhammer (28) showed that Amt1 is the Amt protein that is widespread in the cyanobacteria, consistent with its major role in methylammonium uptake (35, 49). In contrast, proteins that group with S. elongatus AmtB are found only in the heterocyst-forming cyanobacteria (28), which are not closely related to S. elongatus (46). The amtB gene might have been imported into S. elongatus and the heterocyst formers from noncyanobacterial sources by lateral gene transfer, which is a recognized mechanism in cyanobacterial genome evolution (15). One possible scenario, consistent with our experimental results, is that amtB is a recently acquired secondary gene in the S. elongatus genome whose protein product has been adapted to provide a function complementary to the function of Amt1.
We have also considered the possibility that AmtB has a regulatory role, specifically mediating some regulatory effects of ammonium on gene transcription and the activity of the nitrate permease. The repressor effect of ammonium was checked, using the glnA gene as a probe, in ammonium-grown cells that had been incubated for 60 or 90 min in the absence of combined nitrogen. Addition of 50 to 500 μM ammonium inhibited glnA expression, and the effects were similar in the wild type and the amtB mutant, as well as in the amt1 mutant and the amt1 amtB double mutant (results not shown). Inhibition by ammonium of nitrate uptake (12) was also tested, using 50 to 150 μM nitrate as a substrate and adding 100 μM ammonium, but again no differences were found between the wild type and the mutants (results not shown). These negative results do not support any possible regulatory role of the Amt proteins in S. elongatus.
With an ammonium concentration of 100 μM, a relatively high rate of ammonium uptake was observed in this work for the S. elongatus amt1 amtB double mutant, similar to that observed for the wild type and the amt single mutants. As mentioned above, diffusion of ammonia pulled by glutamine synthetase can account for such an uptake rate. Therefore, although the existence of ammonium uptake systems other than Amt cannot be ruled out, growth of S. elongatus on high ammonium concentrations can be explained based on ammonia diffusion. For similar concentrations of ammonium (around 100 μM), diffusion of ammonia also appears to be sufficient to mediate regulatory effects such as the repression of N-regulated genes and the inhibition of nitrate uptake. The Amt complex might be needed, however, for regulation promoted by more limiting concentrations of ammonium.
Acknowledgments
We thank Vicente Mariscal for help with the quantitative RT-PCR and Ignacio Luque for critical reading of the manuscript.
This work was supported by grant BFU2005-07672 from the Ministerio de Educación y Ciencia (Spain).
Footnotes
Published ahead of print on 17 August 2007.
REFERENCES
- 1.Andrade, S. L., A. Dickmanns, R. Ficner, and O. Einsle. 2005. Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. Proc. Natl. Acad. Sci. USA 102: 14994-14999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antonenko, Y. N., P. Pohl, and G. A. Denisov. 1997. Permeation of ammonia across bilayer lipid membranes studied by ammonium ion selective microelectrodes. Biophys. J. 72: 2187-2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apte, S. K., and R. Haselkorn. 1990. Cloning of salinity stress-induced genes from the salt-tolerant nitrogen-fixing cyanobacterium Anabaena torulosa. Plant Mol. Biol. 15: 723-733. [DOI] [PubMed] [Google Scholar]
- 4.Banta, A. B., E. S. Haas, J. W. Brown, and N. R. Pace. 1992. Sequence of the ribonuclease P RNA gene from the cyanobacterium Anacystis nidulans. Nucleic Acids Res. 20: 911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnes, E. M., Jr., P. Zimniak, and A. Jayakumar. 1983. Role of glutamine synthetase in the uptake and metabolism of methylammonium by Azotobacter vinelandii. J. Bacteriol. 156: 752-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergmeyer, H. U. 1974. Methoden der enzymatischen Analyse, 2nd ed. Verlag Chemie, Weinheim, Germany.
- 7.Boussiba, S., W. Dilling, and J. Gibson. 1984. Methylammonium transport in Anacystis nidulans R-2. J. Bacteriol. 160: 204-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai, Y. P., and C. P. Wolk. 1990. Use of a conditionally lethal gene in Anabaena sp. strain PCC 7120 to select for double recombinants and to entrap insertion sequences. J. Bacteriol. 172: 3138-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dobrogosz, W. J. 1981. Enzymatic activity, p. 365-392. In P. Gerhardt, R. G. E. Murray, R. N. Costilow, E. W. Nester, W. A. Wood, N. R. Krieg, and G. B. Phillips (ed.), Manual of methods for general bacteriology. American Society for Microbiology, Washington, DC.
- 10.Elhai, J., and C. P. Wolk. 1988. A versatile class of positive-selection vectors based on the nonviability of palindrome-containing plasmids that allows cloning into long polylinkers. Gene 68: 119-138. [DOI] [PubMed] [Google Scholar]
- 11.Fellay, R., X. Perret, V. Viprey, W. J. Broughton, and S. Brenner. 1995. Organization of host-inducible transcripts on the symbiotic plasmid of Rhizobium sp. NGR234. Mol. Microbiol. 16: 657-667. [DOI] [PubMed] [Google Scholar]
- 12.Flores, E., M. G. Guerrero, and M. Losada. 1980. Short-term ammonium inhibition of nitrate utilization by Anacystis nidulans and other cyanobacteria. Arch. Microbiol. 128: 137-144. [Google Scholar]
- 13.Flores, E., and A. Herrero. 1994. Assimilatory nitrogen metabolism and its regulation, p. 487-517. In D. A. Bryant (ed.), The molecular biology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 14.Flores, E., and A. Herrero. 2005. Nitrogen assimilation and nitrogen control in cyanobacteria. Biochem. Soc. Trans. 33: 164-167. [DOI] [PubMed] [Google Scholar]
- 15.Flores, E., A. M. Muro-Pastor, and J. C. Meeks. Gene transfer to cyanobacteria in the laboratory and in nature, p. 45-57. In A. Herrero and E. Flores (ed.), The cyanobacteria: molecular biology, genomics and evolution, in press. Caister Academic Press, Norfolk, United Kingdom.
- 16.Frías, J. E., E. Flores, and A. Herrero. 1997. Nitrate assimilation gene cluster from the heterocyst-forming cyanobacterium Anabaena sp. strain PCC 7120. J. Bacteriol. 179: 477-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.García-Domínguez, M., and F. J. Florencio. 1997. Nitrogen availability and electron transport control the expression of glnB gene (encoding PII protein) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol. Biol. 35: 723-734. [DOI] [PubMed] [Google Scholar]
- 18.Golden, S. S., and L. A. Sherman. 1984. Optimal conditions for genetic transformation of the cyanobacterium Anacystis nidulans R2. J. Bacteriol. 158: 36-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hackette, S. L., G. E. Skye, C. Burton, and I. H. Segel. 1970. Characterization of an ammonium transport system in filamentous fungi with methylammonium-14C as the substrate. J. Biol. Chem. 245: 4241-4250. [PubMed] [Google Scholar]
- 20.Herrero, A., A. M. Muro-Pastor, and E. Flores. 2001. Nitrogen control in cyanobacteria. J. Bacteriol. 183: 411-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ihlenfeldt, M. J., and J. Gibson. 1975. CO2 fixation and its regulation in Anacystis nidulans (Synechococcus). Arch. Microbiol. 102: 13-21. [DOI] [PubMed] [Google Scholar]
- 22.Javelle, A., G. Thomas, A. M. Marini, R. Kramer, and M. Merrick. 2005. In vivo functional characterization of the Escherichia coli ammonium channel AmtB: evidence for metabolic coupling of AmtB to glutamine synthetase. Biochem. J. 390: 215-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khademi, S., J. O'Connell III, J. Remis, Y. Robles-Colmenares, L. J. Miercke, and R. M. Stroud. 2004. Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 Å. Science 305: 1587-1594. [DOI] [PubMed] [Google Scholar]
- 24.Kleiner, D. 2000. Ammonium uptake and ammonia excretion by bacteria—a review. Rec. Res. Dev. Microbiol. 4: 467-479. [Google Scholar]
- 25.Ludewig, U. 2006. Ion transport versus gas conduction: function of AMT/Rh-type proteins. Transfus. Clin. Biol. 13: 111-116. [DOI] [PubMed] [Google Scholar]
- 26.Ludewig, U., S. Wilken, B. Wu, W. Jost, P. Obrdlik, M. El Bakkoury, A. M. Marini, B. Andre, T. Hamacher, E. Boles, N. von Wiren, and W. B. Frommer. 2003. Homo- and hetero-oligomerization of ammonium transporter-1 NH4+ uniporters. J. Biol. Chem. 278: 45603-45610. [DOI] [PubMed] [Google Scholar]
- 27.Luque, I., E. Flores, and A. Herrero. 1994. Molecular mechanism for the operation of nitrogen control in cyanobacteria. EMBO J. 13: 2862-2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luque, I., and K. Forchhammer. Nitrogen assimilation and C/N balance sensing, p. 335-382. In A. Herrero and E. Flores (ed.), The cyanobacteria: molecular biology, genomics and evolution, in press. Caister Academic Press, Norfolk, United Kingdom.
- 29.Mackinney, G. 1941. Absorption of light by chlorophyll solutions. J. Biol. Chem. 140: 109-112. [Google Scholar]
- 30.Marini, A. M., S. Soussi-Boudekou, S. Vissers, and B. André. 1997. A family of ammonium transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 17: 4282-4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marini, A. M., J.-Y. Springael, W. B. Frommer, and B. André. 2000. Cross-talk between ammonium transporters in yeast and interference by the soybean SAT1 protein. Mol. Microbiol. 35: 378-385. [DOI] [PubMed] [Google Scholar]
- 32.Markwell, M. A., S. M. Haas, L. L. Bieber, and N. E. Tolbert. 1978. A modification of the Lowry procedure to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 87: 206-210. [DOI] [PubMed] [Google Scholar]
- 33.Mayer, M., G. Schaaf, I. Mouro, C. López, Y. Colin, P. Neumann, J. P. Cartron, and U. Ludewig. 2006. Different transport mechanisms in plant and human AMT/Rh-type ammonium transporters. J. Gen. Physiol. 127: 133-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meier-Wagner, J., L. Nolden, M. Jakoby, R. Siewe, R. Kramer, and A. Burkovski. 2001. Multiplicity of ammonium uptake systems in Corynebacterium glutamicum: role of Amt and AmtB. Microbiology 147: 135-143. [DOI] [PubMed] [Google Scholar]
- 35.Montesinos, M. L., A. M. Muro-Pastor, A. Herrero, and E. Flores. 1998. Ammonium/methylammonium permeases of a cyanobacterium. Identification and analysis of three nitrogen-regulated amt genes in Synechocystis sp. PCC 6803. J. Biol. Chem. 273: 31463-31470. [DOI] [PubMed] [Google Scholar]
- 36.Ninnemann, O., J. C. Jauniaux, and W. B. Frommer. 1994. Identification of a high affinity NH4+ transporter from plants. EMBO J. 13: 3464-3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paz-Yepes, J., E. Flores, and A. Herrero. 2003. Transcriptional effects of the signal transduction protein PII (glnB gene product) on NtcA-dependent genes in Synechococcus sp. PCC 7942. FEBS Lett. 543: 42-46. [DOI] [PubMed] [Google Scholar]
- 38.Perrière, G., and M. Gouy. 1996. WWW-query: an on-line retrieval system for biological sequence banks. Biochimie 78: 364-369. [DOI] [PubMed] [Google Scholar]
- 39.Raboy, B., and E. Padan. 1978. Active transport of glucose and alpha-methylglucoside in the cyanobacterium Plectonema boryanum. J. Biol. Chem. 253: 3287-3291. [PubMed] [Google Scholar]
- 40.Rai, A. N., P. Rowell, and W. D. P. Stewart. 1984. Evidence for an ammonium transport system in free-living and symbiotic cyanobacteria. Arch. Microbiol. 137: 241-246. [Google Scholar]
- 41.Rippka, R., J. Deruelles, J. B. Waterbury, M. Herdman, and R. Y. Stanier. 1979. Generic assignments, strain histories and properties of pure cultures of cyanobacteria. J. Gen. Microbiol. 111: 1-61. [Google Scholar]
- 42.Ritchie, R. J. 1991. Membrane potential and pH control in the cyanobacterium Synechococcus R-2 (Anacystis nidulans) PCC 7942. J. Plant Physiol. 137: 409-418. [Google Scholar]
- 43.Ritchie, R. J., and J. Gibson. 1987. Permeability of ammonia, methylamine and ethylamine in the cyanobacterium, Synechococcus R-2 (Anacystis nidulans). J. Membr. Biol. 95: 131-142. [Google Scholar]
- 44.Sohlenkamp, C., M. Shelden, S. Howitt, and M. Udvardi. 2000. Characterization of Arabidopsis AtAMT2, a novel ammonium transporter in plants. FEBS Lett. 467: 273-278. [DOI] [PubMed] [Google Scholar]
- 45.Soupene, E., L. He, D. Yan, and S. Kustu. 1998. Ammonia acquisition in enteric bacteria: physiological role of the ammonium/methylammonium transport B (AmtB) protein. Proc. Natl. Acad. Sci. USA 95: 7030-7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swingley, W. D., R. E. Blankenship, and J. Raymond. Insights into cyanobacterial evolution from comparative genomics, p. 21-43. In A. Herrero and E. Flores (ed.), The cyanobacteria: molecular biology, genomics and evolution, in press. Caister Academic Press, Norfolk, United Kingdom.
- 47.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vázquez-Bermúdez, M. F., A. Herrero, and E. Flores. 2002. 2-Oxoglutarate increases the binding affinity of the NtcA (nitrogen control) transcription factor for the Synechococcus glnA promoter. FEBS Lett. 512: 71-74. [DOI] [PubMed] [Google Scholar]
- 49.Vázquez-Bermúdez, M. F., J. Paz-Yepes, A. Herrero, and E. Flores. 2002. The NtcA-activated amt1 gene encodes a permease required for uptake of low concentrations of ammonium in the cyanobacterium Synechococcus sp. PCC 7942. Microbiology. 148: 861-869. [DOI] [PubMed] [Google Scholar]
- 50.Zheng, L., D. Kostrewa, S. Berneche, F. K. Winkler, and X. D. Li. 2004. The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc. Natl. Acad. Sci. USA 101: 17090-17095. [DOI] [PMC free article] [PubMed] [Google Scholar]