Abstract
Human replication factor C (hRFC) is a five-subunit protein complex (p140, p40, p38, p37, and p36) that acts to catalytically load proliferating cell nuclear antigen onto DNA, where it recruits DNA polymerase δ or ɛ to the primer terminus at the expense of ATP, leading to processive DNA synthesis. We have previously shown that a subcomplex of hRFC consisting of three subunits (p40, p37, and p36) contained DNA-dependent ATPase activity. However, it is not clear which subunit(s) hydrolyzes ATP, as all five subunits include potential ATP binding sites. In this report, we introduced point mutations in the putative ATP-binding sequences of each hRFC subunit and examined the properties of the resulting mutant hRFC complex and the ATPase activity of the hRFC or the p40·p37·p36 complex. A mutation in any one of the ATP binding sites of the p36, p37, p40, or p140 subunits markedly reduced replication activity of the hRFC complex and the ATPase activity of the hRFC or the p40·p37·p36 complex. A mutation in the ATP binding site of the p38 subunit did not alter the replication activity of hRFC. These findings indicate that the replication activity of hRFC is dependent on efficient ATP hydrolysis contributed to by the action of four hRFC subunits.
Replication factor C (RFC) functions as an accessory factor for proliferating cell nuclear antigen (PCNA)-dependant processive DNA synthesis catalyzed by DNA polymerase δ or ɛ (pol δ or ɛ) (1–7). The assembly of the processive pol δ or ɛ holoenzyme initiates when RFC binds the 3′-OH terminus of a primer template and in the presence of ATP recruits PCNA forming a PCNA-RFC-DNA complex. After ATP hydrolysis, PCNA is assembled around DNA, pol δ or ɛ is recruited and RFC presumably dissociates from DNA.
Human RFC (hRFC) contains five different subunits that are 140, 40, 38, 37, and 36 kDa in apparent molecular masses. Reconstitution of hRFC from its five subunits has been achieved in baculovirus-infected insect cells (8–10) and in coupled in vitro transcription-translation reactions (11). By using these expression systems, we have shown that a functional hRFC complex could be assembled in vitro in two steps. (i) The p40, p37, and p36 subunits interact with each other to form a core p40·p37·p36 complex (11), which (ii) then binds the p38 subunit, which further recruits p140 to form the five-subunit RFC complex. This in vitro assembled complex was shown to be identical to native hRFC in supporting PCNA-dependent DNA synthesis catalyzed by pol δ (11, 12). Other possible assembly pathways have also been proposed based on the formation of various subcomplexes between hRFC subunits in the baculovirus expression system (9, 10).
Our understanding as to how individual hRFC subunits contribute to the catalytic loading of PCNA onto DNA has been facilitated by the overexpression and purification of individual hRFC subunits in Escherichia coli and baculovirus-infected insect cells. The p140 subunit mediates the binding of hRFC to the DNA primer terminus (13), but additional DNA binding activity associated with the p40·p37·p36 complex is required for stimulation of the DNA-dependent ATPase activity of the p40·p37·p36 complex (12). At least four hRFC subunits (p140, p40, p38, and p36) have been found to interact with PCNA (14–17; data not shown), but how these interactions contribute to the opening of the PCNA ring is unknown. The p40 subunit has been shown to interact with ATP (13, 18), but p40 purified from E. coli or baculovirus-infected cells failed to show significant ATPase activity (12, 18).
An unusual feature of RFC is the amino acid sequence similarities shared by all five subunits. These similarities are grouped in seven regions in each RFC subunit, referred to as RFC boxes (box II–VII; ref. 19). Two of these boxes include putative ATP-binding sequences (boxes III and V) known as Walker A and B motifs, respectively. Based on studies of known ATPases, the Walker A motif forms a phosphate-binding loop that interacts with the triphosphate tail of ATP, whereas the B motif interacts with Mg2+ required for ATP hydrolysis (20–25). Examination of these sequences reveals that the p38 subunit contains slight variations from the consensus sequences (GXXXXGKK as opposed to the consensus GXXXXGKT in box III and TEVD rather than the consensus DEAD in box V). The other four subunits contain perfect matches to the consensus sequences in both boxes.
We (12) and others (10, 26) have demonstrated that the p40·p37·p36 complex is a DNA-dependent ATPase, but the subunit(s) that catalyzes ATP hydrolysis in this complex remain unidentified as no subunit alone exhibits significant ATPase activity (12, 26). In addition, because the p40·p37·p36 complex is less effective than hRFC in hydrolyzing ATP, it is likely that the p140 and p38 subunits also contribute to the overall ATPase activity of hRFC.
To understand whether the ATPase activity of hRFC depends on the ability of all subunits to bind and hydrolyze ATP, we introduced point mutations into the conserved Walker A motif of each subunit and examined the ATPase activity of the resulting p40·p37·p36 complex and hRFC reconstituted with mutated subunits. In this report, we show that mutations in any single subunit of the p40·p37·p36 complex markedly reduced ATP hydrolysis catalyzed by this complex, as well as the replication activity of hRFC. A mutation in the p140 subunit also reduced the ATPase and replication activities of hRFC, whereas a mutation in p38 had no influence on these activities. These results suggest that optimal ATP hydrolysis in hRFC require the cooperative action of multiple subunits. Similar results have recently been reported by Podust et al. (26).
MATERIALS AND METHODS
Preparations of DNAs and Proteins.
Poly(dA)4500 annealed to oligo(dT)12–18 at a nucleotide ratio of 20:1, φX174 single-stranded circular (ssc) viral DNA, singly primed M13 ssc DNA, human single-stranded DNA binding protein, pol δ, PCNA, and hRFC were either obtained from commercial sources or purified as described previously (1, 3, 27).
ATPase Assay.
ATPase activity was assayed as described (12).
Molecular Cloning.
The cDNAs coding for hRFC subunits were maintained in the pET series vectors (Promega) as described (11). The pET16a/p140N555 plasmid that expressed the p140 subunit with the N-terminal ligase domain removed was prepared as described (16). PET11a/p40·p37·p36 was constructed by subcloning DNA sequences coding for the p40, p37, and p36 genes from the above plasmids into the pET11a vector. In brief, the p36 gene was first inserted between the T7 promoter and terminator of the pET11a plasmid to generate pET11a/p36. The p40 gene was then inserted between the p36 gene and the T7 terminator to generate pET11a/p36·p40. The p37 gene was first subcloned into a separate pET11a vector between the T7 promoter and the terminator to generate pET11a/p37. A DNA fragment containing the p37 gene and the adjacent T7 promoter and terminator was then excised, blunt-ended, and inserted into the NruI site of the pET11a/p36·p40 plasmid to generate pET11a/p40·p37·p36. A 10-His coding sequence was added to the 5′-end of the p40 ORF.
Site-Directed Mutagenesis.
The conserved lysine residue in the Walker A motif of each hRFC subunit was changed to alanine by using a PCR-based overlap extension technique as previously described (28). Point mutations were introduced into the pET11a/p40·p37·p36 DNA for the creation of mutant p40, p37, and p36 subunits, pET19b/p38-His for the p38 subunit, and pET16a/p140 or pET16a/p140N555 for the p140 subunit.
UV Crosslinking.
Reaction mixtures (10 μl) containing 0.3 μg of the p40·p37·p36 complex, 50 mM Tris·HCl (pH 7.5), 7 mM MgCl2, 10% glycerol, 5% glucose, 5 mM DTT, 25 μg/ml BSA, and 2.5 μM [α-32P]ATP (400 Ci/mmol; 1 Ci = 37 GBq) were incubated at room temperature for 5 min, placed on ice, and then irradiated for 45 min under a germicidal UV lamp. The mixture was separated on a SDS/9% polyacrylamide gel followed by autoradiography.
Immunoprecipitation of hRFC Complex.
The hRFC complex, reconstituted after incubation of the p40·p37·p36 complex with in vitro translated p38 and p140 subunits was immunoprecipitated as described (11, 12, 16).
Replication Assays.
Replication assays using singly primed M13 DNA as the template were carried out as described (12). When in vitro translated products were used, the hRFC complex formed was adsorbed to protein A agarose beads (5 μl) bound to antibodies against the hRFC p37 subunit. The beads were then added to reaction mixtures (15 μl) containing 10 fmol of singly primed M13 DNA and other components required for the replication assay (12).
Purification of the p40·p37·p36 Complex.
The wt and mutant p40·p37·p36 complexes behaved identically in all chromatographic steps, though the yield and purity varied between preparations. Purification steps were monitored by analysis on SDS/9% polyacrylamide gels followed by Coomassie staining and Western blot analysis using antibodies specific for each subunit.
After isopropyl thiogalactoside induced overexpression of pET11a/p40·p37·p36 DNA, extracts (12 ml containing 130 mg of protein, prepared from 1L culture of E. coli cells after lysis, and resuspension in nickel column buffer (50 mM Tris⋅HCl, pH 8.0/20 mM sodium phosphate, pH 8.0/0.5 M NaCl/1 mM phenylmethylsulfonyl fluoride/0.2 μg/ml leupeptin/0.1 μg/ml antipain/0.2 μg/ml aprotinin) were incubated with 0.5 ml of nickel resin (Invitrogen) pre-equilibrated with the nickel column binding buffer for 1 hr. After washing twice with 10 ml of wash buffer (20 mM sodium phosphate, pH 8.0/0.5 M NaCl/50 mM imidazole), the nickel resin was packed into a 0.7 × 4 cm column and bound proteins were eluted with 20 mM sodium phosphate, pH 8.0/0.5 M NaCl/500 mM imidazole. The eluted material (6 mg, 2.5 ml) was then dialyzed against 2 liters of buffer A (25 mM Tris⋅HCl, pH 7.5/1 mM EDTA/0.01% Nonidet P-40/1 mM DTT/10% glycerol) plus 0.1 M NaCl for 3 hr, and chromatographed through a SP-Sepharose column (1.0 × 1.3 cm) equilibrated with the same buffer. After washing the column with 3 bed volumes of equilibration buffer, bound proteins were eluted by using a 10-ml gradient from 0.1 to 0.4 M NaCl in buffer A. Fractions containing the p40, p37, and p36 subunits eluted at 0.25 M NaCl and were pooled (2.5 mg of protein, 3 ml). After diluting the NaCl concentration to 0.05 M, the pooled fraction was loaded onto a Q-Sepharose column (1.0 × 1.3 cm) that was developed with a gradient (12 ml) of 0.05–0.4 M NaCl in buffer A. Fractions containing the p40·p37·p36 complex, which eluted at 0.3 M NaCl, were pooled (1.8 mg, 4.1 ml) and concentrated by using the Ultrafree-15 centrifuge filter membrane (10K, Millipore). An aliquot (66 μg, 150 μl) of the concentrated material was then sedimented through a 15–35% glycerol gradient (5 ml) in buffer A plus 250 mM NaCl at 250,000 × g for 24 hr. Pooled glycerol gradient fractions (50 mg, 0.66 ml) were stored at −80°C and showed no loss of DNA-dependent ATPase activity over 6 months with repeated cycles of freezing and thawing.
Preparation of hRFC Containing the Mutant p140 Subunit.
A DNA fragment containing the mutagenized p140 sequence in the pET16a/p140 plasmid was subcloned into pBlueBac2/p140-C-His to replace the wild-type (wt) p140 sequence. The resulting construct, pBlueBac2/p140K657A-C-His, was used to produce recombinant virus for expression of the mutant p140 subunit following the manufacturer’s procedure (Invitrogen). hRFC was reconstituted by coinfecting high five insect cells with baculoviruses that yielded the five RFC subunits as described (8), with the exception that mutant p140 virus was used in place of the wt p140 virus. The preparation of cell extracts and purification of mutant baculovirus RFC were as previously described (8).
RESULTS
Mutagenesis of the ATP Binding Sites in the hRFC p40·p37·p36 Complex.
The function of the Walker A motif has been extensively studied in various nucleoside triphosphate hydrolyzing enzymes by site-directed mutagenesis (29–32) and structural analyses (20–25). The lysine residue in the consensus GXXXGKT/S sequence of the phosphate-binding loop is involved in the electrostatic interactions with the triphosphate tail of ATP; a change of this residue to another amino acid (e.g., alanine) abolishes ATP binding and hydrolysis. To examine the role of the ATP binding sites in the p40·p37·p36 complex, we introduced a single amino acid change (from lysine to alanine) in each of the Walker A motifs of the p36, p37, and p40 subunits (Fig. 1A), and determined the properties of the p40·p37·p36 complex formed.
Figure 1.
(A) Mutagenesis in the Walker A motifs of the p36, p37, and p40 subunits. The amino acid sequences of the Walker A motifs in the p36, p37, and p40 subunits are aligned. The conserved lysine residue (boxed) in each subunit was changed to alanine by site-directed mutagenesis. (B) Purification of wt and mutant p40·p37·p36 complexes. The purified wt and mutant p40·p37·p36 complexes (described in Materials and Methods) were analyzed by SDS/9% polyacrylamide gels. Lane 1, 3 μg of wt p40·p37·p36 complex; lanes 2–4, 3 μg of the mutant p40·p37·p36 complex with a mutation in p36 (p36K66A), p37 (p37K84A), and p40 (p40K82A), respectively. (C) ATPase activity of wt and mutant p40·p37·p36 complexes. ATPase measurements were carried out as described in Materials and Methods. Reaction mixtures contained wt and mutant p40·p37·p36 complexes in amounts as indicated, in the presence of 12.5 μM (as nucleotides) φX174 ssc DNA.
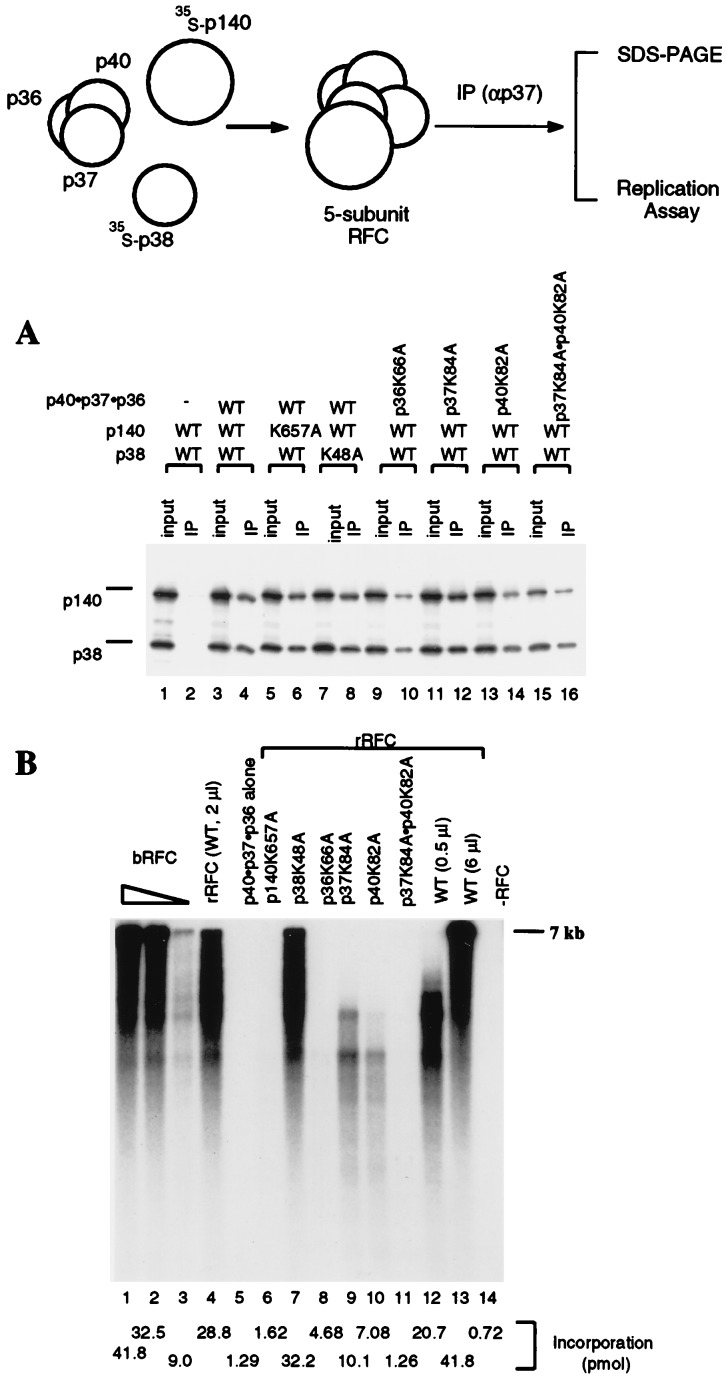
We expressed the p40·p37·p36 complex in E. coli from an expression vector containing all three genes. The properties of p40·p37·p36 complex purified from baculovirus-infected cells and from E. coli were identical. Both preparations contained stoichiometric amounts of the three subunits (Fig. 1B), hydrolyzed ATP (see Fig. 1C) and were converted to the five-subunit hRFC complex when incubated with the other two subunits (see Fig. 3A).
Figure 3.
Coupled in vitro transcription-translation reactions with plasmid DNA containing the coding sequence for the hRFC p140 and p38 subunits were carried out in the presence or absence of the p40·p37·p36 complex followed by immunoprecipitation using a polyclonal antibody against the hRFC p37 subunit. Because the C-terminal half of the p140 subunit (amino acids 555-1147) forms an hRFC complex that is 5–10 fold more active than that formed with the full-length p140 subunit, p140N555 was used in the experiments described here. The immunoprecipitated products were analyzed on SDS/9% polyacrylamide gels followed by autoradiography to visualize the 35S-labeled p140 and p38, or assayed for their replication activities. (A) Reconstitution of hRFC from its five subunits. Reactions containing 20% of the input material (lanes 1, 3, 5, 7. 9, 11, 13, and 15) and the immunoprecipitates (lanes 2, 4, 6, 8, 10, 12, 14, and 16) were separated by SDS/9% polyacrylamide gels. The reactions contained wt or mutant hRFC subunits or subcomplexes as indicated. (B) Replication activities of mutant hRFCs. Mutant hRFCs, immunoprecipitated on protein A beads, were examined for their ability to elongate singly primed M13 DNA as described in Materials and Methods. Reactions shown in lanes 1–3 were carried out in the presence 70, 14, and 1.4 fmol of baculovirus RFC, respectively. Reactions shown in lanes 4–11 were carried out after immunoprecipitation of 2 μl of the reconstitution mixture as follows: lane 4, RFC reconstituted with wt p140, p38, and wt p40·p37·p36 complex; lane 5, the wt p40·p37·p36 complex only; lane 6, mutant p140 (p140K657A), wt p38, and wt p40·p37·p36 complex; lane 7, wt p140, mutant p38 (p38K48A), and wt p40·p37·p36 complex; lane 8, wt p140 and p38, mutant p40·p37·p36 complex (p36K66a); lane 9, wt p140 and p38, mutant p40·p37·p36 complex (p37K84A); lane 10, wt p140 and p38, mutant p40·p37·p36 complex (p40K82A); lane 11, wt p140 and p38, mutant p40·p37·p36 complex (p37K84Ap40K82A). Reactions shown in lanes 12 and 13 were carried out after immunoprecipitation of two levels of the reconstituted mixture (0.5 and 6 μl, respectively) containing wt p140, p38, and wt p40·p37·p36 complex. The reaction shown in lane 14 was carried out in the absence of RFC. Total nucleotide incorporation (pmol) obtained in the above reactions, detected after acid precipitation and liquid scintillation counting, are shown at the bottom of the figure.
Optimal ATP Hydrolysis by the p40·p37·p36 Complex Requires All Three Subunits.
DNA-dependent ATPase activities of purified wt and mutant p40·p37·p36 complexes were examined (Fig. 1C). The wt p40·p37·p36 complex contained DNA-dependent ATPase activity comparable to that purified from baculovirus-infected insect cells (12). However, complexes reconstituted with any single mutated subunit were only ≈10–20% as active as the wt complex, indicating that functional ATP binding sites in all three subunits are required for maximal ATP hydrolysis and suggesting that the three subunits contribute to the hydrolysis of ATP in a cooperative manner.
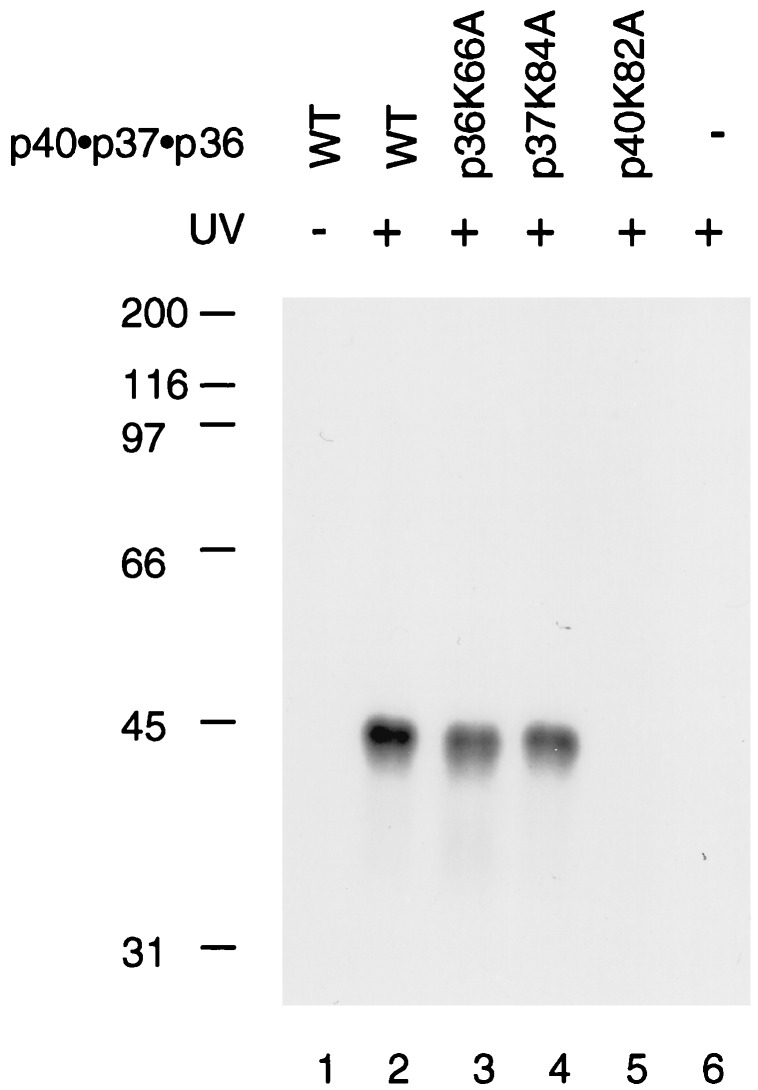
A Functional Walker A Motif Is Required for ATP Binding to the p40 Subunit.
The hRFC p40 subunit has been shown to interact with ATP (13, 18). To examine whether other subunits in the p40·p37·p36 complex also interact with ATP, the wt or mutant p40·p37·p36 complexes were incubated with [α-32P]ATP, followed by UV-irradiation as described in Materials and Methods. Although all three subunits contain putative ATP binding sites, only the p40 subunit was efficiently crosslinked to ATP (Fig. 2, lanes 2–4. This interaction was dependent on a functional Walker A motif because mutation in this region abolished ATP binding (lane 5).
Figure 2.
ATP binds to the p40 subunit in the p40·p37·p36 complex. Reactions containing [α-32P]ATP and the wt or various mutant p40·p37·p36 complexes (0.3 μg) were UV-irradiated and subjected to SDS-PAGE as described in the Materials and Methods. Lane 1, wt p40·p37·p36 complex not UV-irradiated; lane 2, wt p40·p37·p36; lane 3, p36K66A; lane 4, p37K84A; lane 5, p40K82A; lane 6, no protein added.
Mutations in p40, p37, and p36 Subunits Markedly Reduce the Replication Activity of hRFC.
To understand whether the mutations in p40, p37, and p36 also affected the replication activity of hRFC, we assembled the five-subunit hRFC complex by mixing the wt or mutant p40·p37·p36 complex with 35S-labeled p140 and p38 subunits that were synthesized in coupled in vitro transcription-translation reactions (Fig. 3). Formation of the five-subunit complex and the replication activity of the reconstituted hRFC were then examined after immunoprecipitation by using polyclonal antibodies specific for the p37 subunit. As shown in Fig. 3A, p140 and p38 subunits coprecipitated with the wt (lanes 3 and 4) as well as mutant p40·p37·p36 complexes (lanes 9 and 10, p36K66A; lanes 11 and 12, p37K84A; lanes 13 and 14, p40K82A; lanes 15 and 16, double mutant p37K84A·p40K82A).
The reconstituted five-subunit hRFC complexes were examined for their ability to support PCNA-dependent DNA elongation of singly primed M13 DNA catalyzed by pol δ (Fig. 3B). Consistent with our previous findings, hRFC reconstituted with the wt p40·p37·p36 complex supported M13 DNA synthesis (lanes 4, 12, and 13). However, mutations in any single subunit of the p40·p37·p36 complex abolished synthesis of full-length DNA, and reduced DNA synthesis by 70–90% (lanes 8–10), indicating that the replication activity of hRFC correlates with the ability of the p40·p37·p36 complex to hydrolyze ATP. The finding that a mutation in any single subunit reduced the replication activity by more than one third suggests a cooperative mechanism between the p40, p37, and p36 subunits. In support of this, RFC reconstituted with subunits containing mutations in both p40 and p37 supported DNA synthesis much less efficiently than RFC containing one of the two mutations (lane 11). The replication activities of the above mutant hRFC complexes were also confirmed in replication assays by using poly(dA)·oligo(dT) as the template (data not shown).
We examined the ability of these mutant hRFC complexes to load PCNA onto singly-nicked plasmid DNA as described (8). As observed in the replication assay, a mutation in any one subunit of the p40·p37·p36 complex reduced the PCNA loading activity of the reconstituted hRFC by 70–80% (data not shown). Thus, these mutations affected the replication activity of hRFC by altering its ability to load PCNA onto DNA.
The p140 Subunit Walker A Motif Is Required for Replication and the ATPase Activities of hRFC.
We examined whether the p140 and p38 subunits also participate in ATP hydrolysis in hRFC by introducing similar point mutations (lysine to alanine) in the Walker A motifs of these subunits. hRFC was reconstituted by incubating the p40·p37·p36 complex and 35S-labeled p140 and p38 subunits as described in Fig. 3. The formation of the five-subunit hRFC complex and the replication activity of reconstituted hRFC were then determined after immunoprecipitation. As shown in Fig. 3A, mutations in p140 and p38 did not affect their ability to form the five-subunit hRFC complex (lanes 5–8), however, a point mutation in the Walker A motif of the p140 subunit abolished synthesis of full-length M13 DNA and reduced total DNA synthesis by ≈95% (Fig. 3B, lane 6). In contrast to mutations in other hRFC subunits, the mutation in p38 did not alter the replication activity of the reconstituted hRFC complex (Fig. 3B, lane 7).
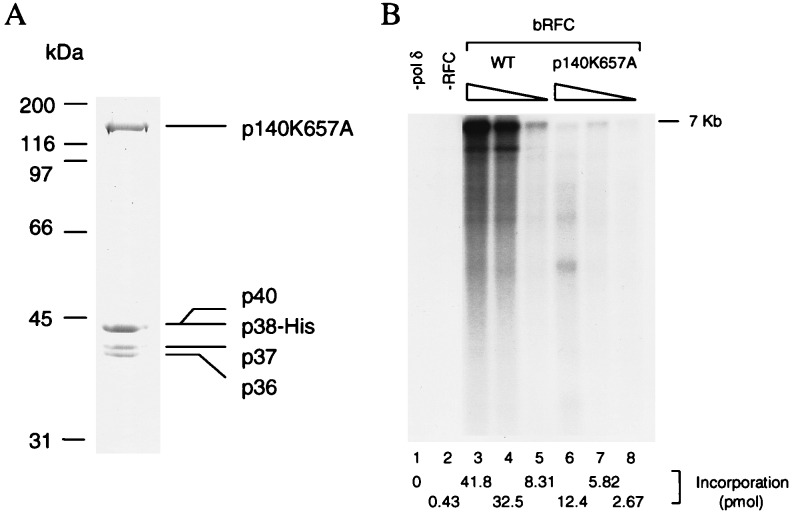
The contribution of the p140 subunit to the ATPase activity of hRFC was examined. For this purpose, a recombinant baculovirus expressing the mutant p140 was constructed and the properties of hRFC reconstituted with this mutant subunit determined. The purified mutant hRFC contained a stoichiometric amount of each of the five subunits (Fig. 4A) and behaved identically to its wt counterpart during purification and in its ability to preferentially bind to primed DNA in a filter binding assay (data not shown). Consistent with the observations described in Fig. 3B (lane 6), the purified mutant hRFC was only ≈5% as active as the wt hRFC in the M13 DNA elongation assay (Fig. 4B). This low activity was not due to contamination of the preparation with RFC from baculovirus-infected insect cells as omission of the baculovirus encoding the large subunit during infection of insect cells yielded preparations devoid of RFC replication activity (data not shown). The weak DNA synthetic activity observed with RFC containing the mutated p140 subunit has been largely attributed to its reduced ability to load PCNA onto DNA in the PCNA loading assay (10–15% as active as wt, data not shown).
Figure 4.
(A) Purification of hRFCp140K657A. Mutant hRFC was purified from baculovirus-infected high five insect cells cells as described in the Materials and Methods. The purified product (2 μg) was separated by SDS/9% polyacrylamide gels followed by Coomasie staining. (B) Replication activity of mutant hRFC. The wt and mutant hRFCs were examined for their ability to support DNA synthesis from singly primed M13 DNA as described in Materials and Methods. Lanes 1 and 2 represent reactions carried out in the absence of pol δ or RFC, respectively. Wild-type baculovirus RFC was added to the reactions in amount as follows: lane 3, 70 fmol; lane 4, 14 fmol; lane 5, 1.4 fmol. Mutant baculovirus RFC was added to the reactions in amounts as follows: lane 6, 333 fmol; lane 7, 67 fmol; lane 8, 23 fmol. Total nucleotide incorporation (pmol), detected after acid precipitation and liquid scintillation counting, is shown at the bottom of the figure.
The DNA-dependent ATPase activity of the mutant hRFC was determined and compared with those of the wt hRFC and the p40·p37·p36 complex (Table 1). A mutation in the p140 subunit reduced ATP hydrolysis by ≈75% (≈5 molecules of ATP hydrolyzed per molecule of protein per min) in the presence of φX174 ssc DNA and by ≈85% (0.34 molecule of ATP hydrolyzed per molecule of protein per min) in the absence of DNA, efficiencies similar to that of the wt p40·p37·p36 complex (4.5 and 0.15 molecules of ATP hydrolyzed per molecule of protein per min in the presence and absence of φX174 ssc DNA, respectively). This indicates that p140 subunit also contributes significantly to optimal ATP hydrolysis by hRFC.
Table 1.
Comparison of ATPase activity of the p140K656A mutant hRFC, wt hRFC, and wt p40·p37·p36 complex
| Protein | ATP hydrolyzed per molecule of protein per
min
|
|
|---|---|---|
| −DNA | +DNA | |
| Mutant hRFC | 0.34 | 5 |
| wt hRFC178 | 2.4 | 19.5 |
| wt p40·p37·p36* | 0.15 | 4.5 |
DISCUSSION
We have examined the role of each hRFC subunit in ATP hydrolysis after site-directed mutagenesis of the conserved Walker A motifs. Mutations in any one subunit of the p40·p37·p36 complex or the p140 subunit markedly reduced both the ATPase activity of the complex and the replication activity of hRFC. These findings are consistent with results reported recently by Podust et al. (26) that similar mutations in any of the four hRFC subunits impaired the replication activities of the mutant hRFC complexes. In this report, we provide evidence that the reduced replication efficiency of each of the four mutant hRFC complexes was a result of the reduced ATPase activities in these mutants; similar observations were reported for only one of the hRFC subunits (p37) by Podust et al. (26).
We also showed that mutations in the ATP binding sites of hRFC subunits affected only the ATPase and the replication activities of hRFC, but not other properties including assembly of the hRFC complex (Fig. 3A), the ability to bind DNA or PCNA (data not shown). Specifically, we observed that expression of hRFC containing the mutant p140 subunit was nearly identical to that of the wt hRFC in baculovirus-infected insect cells, and the yield of the mutant hRFC was comparable to that of the wt hRFC after purification. Podust et al. (26) reported that mutations in the ATP binding sites of the p140, p40 and p36 subunits interfered with assembly of the hRFC complex (5–10% complex formed compared with wt hRFC). It should be noted that whereas the conserved lysine residue in the Walker A motif was changed to an alanine in our study, a more dramatic substitution of lysine for glutamate was carried out by Podust et al. (26). Whether this accounts for the different observations, however, is not known.
Whereas the p140, p40, p37, and p36 subunits all contain perfect matches to the consensus Walker A motif, the p38 subunit does not (GXXXXGKK instead of GXXXXGKT). It should be noted, that the sequence of the p38 Walker A motif contains GXXXXGKK followed by a threonine (T). This raised the question of whether the second lysine residue could substitute for the first lysine residue when mutated. We therefore addressed this possibility by making a mutant form of p38 in which both lysine residues were changed to alanines. However, the five-subunit hRFC complex did not form efficiently in the presence of this double mutant p38 (data not shown), suggesting that a resulting structural change may have impaired hRFC complex assembly.
Like RFC of human and yeast cells, the clamp loaders of E. coli and bacteriophage T4 also contain multiple subunits: the γ, δ, δ′, χ, and ϕ subunits for the E. coli γ complex, and four copies of gene product 44 (gp44) and one copy of gp62 for the T4 gp44/gp62 complex (33). Though it appears that only one subunit in the E. coli or T4 clamp loaders possesses ATPase activity [γ subunit and gp44, respectively (34–36)], both γ and gp44 are present as multiple copies in the corresponding clamp loaders (33, 36). Thus the clamp loaders in all three species use multiple copies of subunits to carry out ATP hydrolysis.
Although ATP plays an essential role in loading PCNA onto DNA, it may not be required for the ring opening step as indicated by the finding that the hRFC-p40 subunit alone (albeit inefficiently) can unload PCNA from DNA in the absence of ATP (12; data not shown), a process that involves opening of the PCNA clamp. Thus, the PCNA/p40 interaction may be sufficient to open the PCNA ring, but ATP may be required to induce a conformational change in RFC that renders the p40 subunit accessible for interaction with PCNA. In E. coli, the δ subunit alone interacts with β and can unload PCNA efficiently from DNA. The δ subunit, however, is sequestered within the γ complex in the absence of ATP and is exposed for interaction with β only when ATP is present (M.O’D., unpublished data).
RFC can interact with PCNA in the absence of ATP (12). However, ATP is also known to enhance this interaction substantially (ref. 37; data not shown). Thus it is possible that the PCNA-interacting subunits in hRFC (p140, p40, p38, and p36) fall into two classes: class A subunits whose interaction with PCNA is ATP independent and may recruit PCNA to the primer terminus and class B subunits (e.g., p40) that by analogy to the δ subunit of E. coli, requires ATP to interact with PCNA and bring about ring opening. Although PCNA and DNA do not interact stably in solution, these two molecules are held together by RFC during the loading process, making any possible interaction between them intramolecular. It is known that the inner region of the PCNA ring is heavily populated with positively charged amino acids that can interact with the negatively charged DNA once DNA is juxtaposed with these amino acids. Therefore clamp loading may result from the trapping of DNA that moves randomly from outside to inside of PCNA after reclosure of the ring. This hypothesis predicts that movement of DNA from outside to the inside of the PCNA ring is a rate limiting step in the loading process. Indeed, an analysis of the E. coli clamp loading process demonstrated that the rate limiting step involves an intramolecular reaction (38). It is also possible that the movement of DNA to the inner region of the PCNA ring is facilitated by RFC.
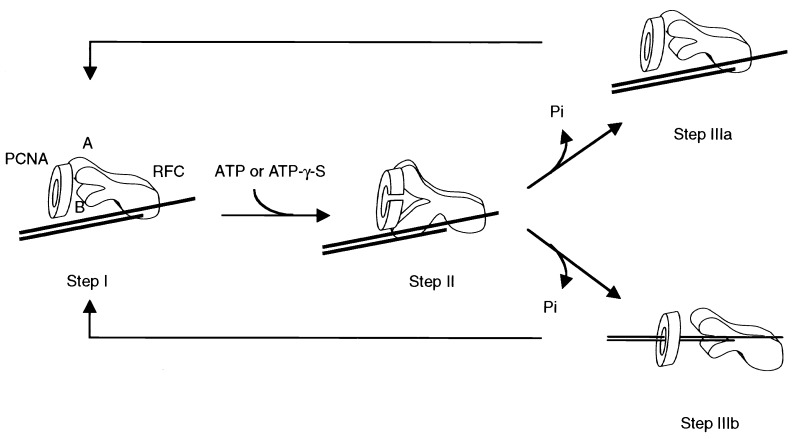
Based on these assumptions, we proposed the following model in which loading of PCNA onto DNA occurs in three steps (Fig. 5). First, RFC locates the primer terminus and recruits PCNA by using the class A subunits (step I). ATP binding to RFC induces conformational changes that permit class B subunits to interact with PCNA, resulting in PCNA ring opening (step II). Next, hydrolysis of ATP restores class B subunits to their original state, and PCNA ring closes as a result of the dissociation of the class B subunits. If DNA is not enclosed within PCNA after ring closure, RFC reverts to its original state in step I, repeating the cycle as new molecules of ATP are bound and hydrolyzed (step IIIa). However, if DNA moves into the PCNA ring before ring closure, the loading process is complete (step IIIb). RFC is then released from PCNA and returns to its original state in step I to catalyze a new cycle of clamp loading. If the movement of DNA into the opened PCNA ring were inefficient, multiple cycles of ATP binding and hydrolysis (steps I–II–IIIa) could be required before PCNA is successfully loaded onto DNA (step I–II–IIIb).
Figure 5.
A model for the loading of PCNA onto DNA catalyzed by RFC. The RFC subunits that interact with PCNA are indicated in the figure (A and B). Class A subunits can interact with PCNA in the absence of ATP. ATP binding induces a conformational change in RFC that permit the interaction of class B subunits with PCNA. The PCNA ring opens and recloses in response to interactions with the class B subunits governed by ATP binding and hydrolysis. Multiple rounds of ATP binding and hydrolysis may be required before PCNA is finally loaded onto DNA.
This model is consistent with our findings presented here, as well as the similarities between human RFC and E. coli γ complex. However, a more detailed understanding of RFC function awaits the elucidation of its crystal structure.
Acknowledgments
The authors wish to thank Frank Uhlmann for helpful discussions and Yixin Lin for technical support. These studies were supported by the National Institutes of Health Grants GM 38559 (to J.H.) and GM 38839 to (M.O’D.).
ABBREVIATIONS
- pol
DNA polymerase
- RFC
replication factor C
- hRFC
human RFC
- PCNA
proliferating cell nuclear antigen
- ssc DNA
single-stranded circular DNA
- gp
gene product
- wt
wild-type.
References
- 1. Lee S-H, Eki T, Hurwitz J. Proc Natl Acad Sci USA. 1989;86:7361–7365. doi: 10.1073/pnas.86.19.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsurimoto T, Stillman B. Mol Cell Biol. 1989;9:609–619. doi: 10.1128/mcb.9.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee S-H, Kwong A D, Pan Z-Q, Hurwitz J. J Biol Chem. 1991;266:584–602. [PubMed] [Google Scholar]
- 4.Tsurimoto T, Stillman B. Proc Natl Acad Sci USA. 1990;87:1023–1027. doi: 10.1073/pnas.87.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoder B L, Burgers P M. J Biol Chem. 1991;266:22689–22697. [PubMed] [Google Scholar]
- 6.Fien K, Stillman B. Mol Cell Biol. 1992;12:155–163. doi: 10.1128/mcb.12.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Podust V N, Georgaki A, Strack B, Hübscher U. Nucleic Acids Res. 1992;20:4159–4165. doi: 10.1093/nar/20.16.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai J, Uhlmann F, Gibbs E, Flores-Rozas H, Lee C-G, Phillips B, Finkelstein J, Yao N, O’Donnell M, Hurwitz J. Proc Natl Acad Sci USA. 1996;93:12896–12901. doi: 10.1073/pnas.93.23.12896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Podust V N, Fanning E. J Biol Chem. 1997;272:6303–6310. doi: 10.1074/jbc.272.10.6303. [DOI] [PubMed] [Google Scholar]
- 10.Ellison V, Stillman B. J Biol Chem. 1998;273:5979–5987. doi: 10.1074/jbc.273.10.5979. [DOI] [PubMed] [Google Scholar]
- 11.Uhlmann F, Cai J, Flores-Rozas H, Dean F B, Finkelstein J, O’Donnell M, Hurwitz J. Proc Natl Acad Sci USA. 1996;93:6521–6526. doi: 10.1073/pnas.93.13.6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai J, Gibbs E, Uhlmann F, Phillips B, Yao N, O’Donnell M, Hurwitz J. J Biol Chem. 1997;272:18974–18981. doi: 10.1074/jbc.272.30.18974. [DOI] [PubMed] [Google Scholar]
- 13.Tsurimoto T, Stillman B. J Biol Chem. 1991;266:1950–1960. [PubMed] [Google Scholar]
- 14.Pan Z-Q, Chen M, Hurwitz J. Proc Natl Acad Sci USA. 1993;90:6–10. doi: 10.1073/pnas.90.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fotedar R, Mossi R, Fitzgerald P, Rousselle T, Maga G, Brichner H, Messier H, Kasibhatla S, Hübscher U, Fotedar A. EMBO J. 1996;15:4423–4433. [PMC free article] [PubMed] [Google Scholar]
- 16.Uhlmann F, Cai J, Gibbs E, O’Donnell M, Hurwitz J. J Biol Chem. 1997;272:10058–10064. doi: 10.1074/jbc.272.15.10058. [DOI] [PubMed] [Google Scholar]
- 17.Mossi R, Jonsson Z O, Allen B L, Hardin S H, Hübscher U. J Biol Chem. 1997;272:1769–1776. doi: 10.1074/jbc.272.3.1769. [DOI] [PubMed] [Google Scholar]
- 18.Chen M, Pan Z-Q, Hurwitz J. Proc Natl Acad Sci USA. 1992;89:2516–2520. doi: 10.1073/pnas.89.7.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cullmann G, Fien K, Kobayashi R, Stillman B. Mol Cell Biol. 1995;15:4661–4671. doi: 10.1128/mcb.15.9.4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker J E, Saraste M, Runswich M J, Gay N J. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schulz G E. Curr Opin Struct Biol. 1992;2:61–67. [Google Scholar]
- 22.Story R M, Steitz T A. Nature (London) 1992;355:374–376. doi: 10.1038/355374a0. [DOI] [PubMed] [Google Scholar]
- 23.Cronet P, Bellsolell L, Sander C, Coll M, Serrano L. J Mol Biol. 1995;249:654–664. doi: 10.1006/jmbi.1995.0326. [DOI] [PubMed] [Google Scholar]
- 24.Abrahams J P, Leslie A G W, Lutter R, Walker J E. Nature (London) 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 25.Subramanya H S, Bird L E, Brannigan J A, Wigley D B. Nature (London) 1996;384:379–383. doi: 10.1038/384379a0. [DOI] [PubMed] [Google Scholar]
- 26.Podust V N, Tiwari N, Ott R, Fanning E. J Biol Chem. 1998;273:12935–12942. doi: 10.1074/jbc.273.21.12935. [DOI] [PubMed] [Google Scholar]
- 27.Kenny M K, Lee S-H, Hurwitz J. Proc Natl Acad Sci USA. 1989;86:9757–9761. doi: 10.1073/pnas.86.24.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho S N, Hunt H P, Horton R M, Pullen J K, Pease L R. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 29.Pause A, Sonenberg N. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bourne H R, Sanders D A, McCormick F. Nature (London) 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 31.Shen H, Yao B, Mueller D M. J Biol Chem. 1994;269:9424–9428. [PubMed] [Google Scholar]
- 32.Seeley T W, Grossman L. Proc Natl Acad Sci USA. 1989;86:6577–6581. doi: 10.1073/pnas.86.17.6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelman Z, O’Donnell M. Curr Opin Genet Dev. 1994;4:185–195. doi: 10.1016/s0959-437x(05)80044-9. [DOI] [PubMed] [Google Scholar]
- 34.Xiao H, Naktinis V, O’Donnell M. J Biol Chem. 1995;270:13378–13383. doi: 10.1074/jbc.270.22.13378. [DOI] [PubMed] [Google Scholar]
- 35.Rush J, Lin T-C, Quinones M, Spicer E K, Douglas I, Williams K R, Konigsberg W H. J Biol Chem. 1989;264:10943–10953. [PubMed] [Google Scholar]
- 36.Onrust R, Finkelstein J, Natkinis V, Turner J, Fang L, O’Donnell M. J Biol Chem. 1995;270:13348–13357. doi: 10.1074/jbc.270.22.13348. [DOI] [PubMed] [Google Scholar]
- 37.Gerik K J, Gary S L, Burgers P M. J Biol Chem. 1997;272:1256–1262. doi: 10.1074/jbc.272.2.1256. [DOI] [PubMed] [Google Scholar]
- 38.Bloom L B, Turner J, Kelman Z, Beechem J M, O’Donnell M, Goodman M F. J Biol Chem. 1996;271:30699–30708. doi: 10.1074/jbc.271.48.30699. [DOI] [PubMed] [Google Scholar]