Abstract
Resistance genotyping provides an important resource for the clinical management of patients infected with human immunodeficiency virus type 1 (HIV-1). However, resistance to protease (PR) inhibitors (PIs) is a complex phenotype shaped by interactions among nearly half of the residues in HIV-1 PR. Previous studies of the genetic basis of PI resistance focused on fixed substitutions among populations of HIV-1, i.e., host-specific adaptations. Consequently, they are susceptible to a high false discovery rate due to founder effects. Here, we employ sequencing “mixtures” (i.e., ambiguous base calls) as a site-specific marker of genetic variation within patients that is independent of the phylogeny. We demonstrate that the transient response to selection by PIs is manifested as an excess of nonsynonymous mixtures. Using a sample of 5,651 PR sequences isolated from both PI-naive and -treated patients, we analyze the joint distribution of mixtures and eight PIs as a Bayesian network, which distinguishes residue-residue interactions from direct associations with PIs. We find that selection for resistance is associated with the emergence of nonsynonymous mixtures in two distinct groups of codon sites clustered along the substrate cleft and distal regions of PR, respectively. Within-patient evolution at several positions is independent of PIs, including those formerly postulated to be involved in resistance. These positions are under strong positive selection in the PI-naive patient population, implying that other factors can produce spurious associations with resistance, e.g., mutational escape from the immune response.
The human immunodeficiency virus type 1 (HIV-1) protease (PR) cleaves itself and other viral proteins from the gag-pol polypeptide precursor (37). Ever since its essential role in the formation of mature viral particles was experimentally confirmed (41), HIV-1 PR has been productively exploited as a target for the development of antiretroviral agents. There are currently nine PR inhibitors (PI) approved for clinical use, with several more in development (23). PIs have become an important component of highly active antiretroviral therapy, which can successfully reduce viral load and extend the life expectancy of HIV-infected patients (55). However, an HIV-1 population that is exposed to PIs will rapidly acquire mutations that render its PR resistant to one or more PIs (14). PI monotherapy, for instance, is characterized by a transient suppression of viral load that is inevitably followed by the emergence of resistant virus.
Resistance to PIs is a complex phenotype of HIV-1 that tends to require the fixation of multiple mutations within the PR-encoding region of pol (14, 49). Nevertheless, the genetic sequence of HIV-1 PR can be used to predict its resistance to PIs and anticipate the evolutionary response of the virus population, e.g., genotypic resistance testing (80). Genotypic testing provides results within a shorter period of time and is generally less expensive than directly assaying the resistance phenotype of a variant. Retrospective and prospective studies demonstrated that resistance genotyping contributes beneficially to the clinical management of patients undergoing highly active antiretroviral therapy regimens (2, 21, 75). Investigators at both the Los Alamos National Laboratory (57) and Stanford University (60) maintain public databases of PR and reverse transcriptase (RT) genotypes associated with drug treatment, resistance phenotype, or clinical outcome. In addition, a volunteer panel of experts in the field has regularly updated a list of PR and RT mutations associated with resistance, which was compiled from a consensus review of the empirical literature (35).
Nevertheless, it is difficult to formulate accurate clinical guidelines for interpreting results from HIV-1 resistance genotyping. Roughly half of the 99 codon sites in the HIV-1 PR sequence have been implicated in the evolution of PI resistance. For example, Wu et al. (78) previously reported statistically significant associations with PI treatment at 45 different sites from an analysis of 2,248 subtype B HIV-1 PR sequences isolated from both PI-naive and PI-active patients. A more conservative list of resistance mutations compiled by an expert panel refers to 36 different sites in PR (35), and the Stanford HIV Drug Resistance Database currently assigns resistance scores to amino acid substitutions affecting 32 different sites (60). The evolution of resistance to PIs in HIV-1 typically requires several mutations to attain substantial levels of resistance (13, 49). HIV-1 populations tend to acquire such mutations in a particular order, so that primary mutations, which are the first to emerge, are followed by secondary mutations that often compensate for the fitness costs of the primary mutations (30, 54, 74, 78). Finally, combination therapy has become more prevalent than monotherapy, confounding the individual effects of PIs on genetic variation.
A comprehensive array of statistical methods, many of which are computationally intensive, has been used to map from the multidimensional space of PR sequences to the resistance phenotype, including pairwise correlation tests (34, 78), linear regression (5, 77), support vector machines (6), decision trees (68), neural networks (20, 76), hierarchical clustering (71), and Bayesian networks (1, 18, 19). However, these methods uniformly neglect important features of the evolution of HIV-1. First of all, the data are mostly population-based (i.e., bulk) sequences obtained from the direct sequencing of each patient isolate, i.e., “standard” genotyping. As a result, subsequent analyses can detect only trends in HIV-1 sequence variation among patients, i.e., fixed substitutions in divergent virus populations. The analysis of population-based sequences is sufficient to find associations between PI resistance and the strongly selected mutations that rapidly become incorporated into the dominant genotype of each HIV-1 population. On the other hand, the evolution of HIV-1 within patients is characterized by extensive genetic variation that is commensurate with the level of divergence in the patient population. Minority variants can play an important role in the evolution of PI resistance by enabling the population to explore alternative evolutionary pathways to resistant genotypes, which may eventually replace the dominant variant over the course of an infection (11, 22, 65). Although the clinical significance of persistent minority variants remains unresolved, they are clearly a common feature of failed drug regimens (22, 48). Therefore, there is a clear mandate to determine what associations may exist between minority variants and clinical outcomes. Secondly, comparative studies of PI resistance in HIV-1 have neglected the common ancestry of sequences (24). Bhattacharya et al. (8) recently demonstrated the impact of common ancestry in their reanalysis of a landmark comparative study associating the diversity of human leukocyte antigens (HLAs) with the evolution of HLA-restricted epitopes in HIV-1 (50). They found that the majority of statistically significant associations described in that study were false-positive results caused by founder effects in which epitope variants and HLA alleles became linked within a clade through identity by descent and epidemiological mixing, respectively. In other words, a virus is often likely to possess a specific variant of an epitope by inheriting it from an ancestor rather than evolving it de novo in response to selection. Although founder effects are often caused by divergence among HIV-1 subtypes, their influence is also evident in variation within subtypes (10). Similar mechanisms must also influence statistical associations between genetic variation in HIV-1 PR and the use of PIs. As a result, a set of HIV-1 PR sequences cannot be treated as a random sample, which previous work has customarily done.
We propose to address both of these limitations by using sequencing mixtures to quantify the evolution of resistance within patients. A sequencing mixture occurs when multiple peaks occur at the same point in a sequencing electropherogram such that the “correct” nucleotide is ambiguous. The application of mixtures to study evolutionary processes has several advantages. First, mixtures can provide a measure of the transient response to selection within patients. Mixtures have been used successfully to screen HIV-1 populations for minority variants above a frequency threshold that ranges from 10% to 25% (32, 43, 46, 67). Population genetic models predict that a new mutation that reaches this intermediate range of frequencies in the population is more likely to be driven by selection (17). Although many within-host polymorphisms go undetected due to the inherent sampling variability of population-based sequencing, the range of polymorphisms most likely to become sampled as mixtures is enriched for variants under selection. Mixtures are frequently used to diagnose the likelihood that an HIV-1 population will become resistant to a new drug regimen (67). As noted above, they have yet to be applied to the mapping of genetic determinants of resistance. Secondly, the genetic variation of an HIV-1 population within a patient is independent of founder effects. Thirdly, mixtures are easy to count (i.e., twofold ambiguous base calls are encoded by the characters “W,” “R,” “K,” “Y,” “S,” and “M”), which allows us to process very large samples (n > 1,000) of population-based sequences. For a large sample of patients, clonal sequencing remains a prohibitively expensive and time-consuming technique for assaying within-patient genetic variation.
To evaluate the influence of PI therapy on the site-specific rates of evolution in HIV-1 PR, we analyzed the distribution of mixtures in 5,651 subtype B sequences that were isolated either from patients undergoing a regimen of one or more PIs (“PI treated”) or from patients who have not used any PIs (“PI naive”). We validate the interpretation of mixtures as a site-specific proxy for the evolution of resistance within patients by contrasting the distribution of mixtures against the level of diversifying selection among patients. By convention, the latter quantity is measured by the statistic dN − dS or dN/dS, where dN is the observed number of nonsynonymous substitutions, scaled by the expected number of nonsynonymous substitutions at the codon site, and dS is the equivalent for synonymous substitutions (38). In a previous study, we proposed the use of an analogous statistic, mN − mS, to quantify selection within patients as a function of the nonsynonymous and synonymous mixture frequencies (59). By applying this statistic to HIV-1 PR sequences from PI-naive patients, we found that the relative excess of nonsynonymous mixtures at a codon site predicted the relative excess of nonsynonymous substitutions from diversifying selection (59). Here, we investigate associations between the site-specific frequency of nonsynonymous mixtures and specific PIs by associating each sequence in the current data set with the PI regimen at time of isolation. These observations were analyzed as a joint probability distribution of discrete-valued random variables encoded as a Bayesian network model. A Bayesian network is a compact graphical representation of the joint distribution of random variables, where an edge between nodes in the graph corresponds to a conditional dependency between the corresponding variables (58). We employ our model to identify the genetic determinants of resistance that emerge within patients in the context of the entire PR sequence.
MATERIALS AND METHODS
Data.
We retrieved 5,651 full-length HIV-1 subtype B PR sequences from the Stanford HIV Drug Resistance Database (60), where each sequence represented a unique patient. Within this sample, 2,648 sequences had been isolated from PI-treated patients, i.e., undergoing a drug regimen including at least one PI. The regimens were comprised of eight different PIs: amprenavir (APV) (n = 233), atazanavir (ATV) (n = 24), indinavir (IDV) (n = 1,407), lopinavir (n = 128), nelfinavir (NFV) (n = 1,335), ritonavir (n = 925), saquinavir (SQV) (n = 1,007), and tipranavir (n = 92). Roughly half of these regimens (n = 1,316) included two or more PIs. The remaining sequences (n = 3,003) in our sample had been isolated from PI-naive patients, omitting sequences corresponding to patients who were already represented in the “PI-treated” subset. We further screened the PI-naive set for resistant sequences according to the Stanford algorithm (60). The algorithm identified 21 sequences from the PI-naive set (0.7%) with resistance scores exceeding a cutoff of 30. Removal of these sequences had no discernible effect on our results. The sequence alignment was adjusted manually using the alignment editor Se-Al (Andrew Rambaut [http://evolve.zoo.ox.ac.uk/software.html]) (the alignment is available upon request). Only 64 out of 5,651 sequences (1.1%) contained gaps, which occurred exclusively at the 3′ ends of the sequences, suggesting that these gaps were caused by the truncation of low-quality sequence regions rather than deletions.
The sequences were screened for twofold mixtures (i.e., an ambiguous base call that can be resolved as two different nucleotides at the same position) using a custom Python script. Out of the 10,039 mixtures found, we omitted 77 threefold mixtures (0.8%) from further analysis. Each mixture was classified as a nonsynonymous or synonymous polymorphism in the context of the codon in which it occurred. For example, the codon “ATR” contains a nonsynonymous mixture in the third position, indicating that a fraction of the sequences in the population encode an isoleucine (“ATA”) at that position, while others encode methionine (“ATG”). Conversely, the codon “AAR” contains a synonymous mixture because both “AAA” and “AAG” encode the amino acid lysine. When more than one mixture occurred in the same codon, the mixtures were omitted from the analysis unless every possible resolution was synonymous, e.g., serine, leucine, or arginine. Following this procedure, we converted our alignment into two 5,651-by-99 binary-valued matrices for the presence or absence of nonsynonymous or synonymous mixtures, respectively. For instance, a codon without any mixtures was encoded as a “0” in either case. For each codon site, we tallied the frequency of nonsynonymous or synonymous mixtures and normalized each quantity by the mean number of nonsynonymous or synonymous sites in the codon (59), herein indicated as mN and mS, respectively.
Selection analysis.
To estimate the rates of nonsynonymous (dN) and synonymous (dS) substitutions (as a site-specific marker of diversifying selection among hosts), we employed the method of single-likelihood ancestor counting (SLAC) as implemented in HyPhy (38, 40). First, we estimated a neighbor-joining tree in HyPhy using Tamura-Nei nucleotide distances (73) with rate variation across sites, parameterized by a gamma distribution with the shape parameter α = 0.5 (representative of HIV-1-derived values) and the scale parameter β = α. For analyzing selection in sequences isolated from PI-active patients, we excluded codon sites that have been associated with primary PI resistance mutations (35) to minimize the influence of convergent evolution on the reconstruction of the phylogeny (45).
We fit a Muse-Gaut codon substitution model (52), crossed with a general time-reversible model of nucleotide substitution (42), to the nucleotide alignment and tree by maximum likelihood, thereby reconstructing the ancestral sequences at the internal nodes of the tree. On the basis of this ancestral reconstruction, the SLAC analysis inferred the expected number of nonsynonymous or synonymous substitutions at each codon site, which were scaled by the expected number of nonsynonymous or synonymous sites in the codon to yield estimates of the quantities dN and dS, respectively. Ambiguous codons containing sequencing mixtures were resolved to the most frequent codon at that site. This procedure for resolving mixtures yields estimates of substitution rates that are independent of mixture frequencies but may underestimate the numbers of nonsynonymous and synonymous substitutions per site as a result. To evaluate the extent of this bias, we ran an additional SLAC analysis with resolution of mixtures by averaging over the relative frequencies of codons at that site (38). We found that resolution to the most frequent codon indeed underestimated the number of substitutions but only by a fractional amount (<1% on average).
Bayesian network inference.
A Bayesian network is a graph encoding a set of conditional independence assertions over a joint probability distribution of random variables (58). Each random variable is represented by a node in the network. A directed edge originating from node A and terminating at node B (A→B) indicates that the outcome of B is conditionally dependent on A, i.e., P(B|A) ≠ P(B). In other words, a directed edge can be interpreted as the hypothesis that A “causes” B (58). Conversely, the lack of an edge between nodes indicates that the nodes are conditionally independent. Conditional independence is an important concept, particularly when dealing with complex systems. For example, if there are two nodes, B and C, that are both dependent on A (B←A→C), then B can appear to be directly influenced by C when failing to account for A. Hence, B is conditionally independent of C, expressed formally by the equation P(B∩C|A) = P(B|A)P(C|A). Pairwise association tests are particularly susceptible to false-positive results in this example, because the outcome of A is masked from the test. In contrast, such conditional dependence relations have an explicit representation in a Bayesian network, providing a more accurate reproduction of biological causation. The set of directed edges encoding dependence relationships is referred to as the “structure” of a Bayesian network. Bayesian networks have previously been applied to detect associations between PIs and genetic variation in PR at the level of the patient population (1, 18, 19) but have not been able to account for the lack of phylogenetic independence among sequences.
In our analysis, each PR sequence was encoded as a binary vector indicating the presence or absence of a nonsynonymous mixture at every codon site. We omitted 16 out of 99 codon sites (alignment consensus residues P1, G27, A28, D29, G40, W42, P44, G52, V56, Q59, G78, T80, P81, G94, T96, and F99) at which nonsynonymous mixtures occurred in fewer than three sequences. In addition, each mixture vector was concatenated with a binary vector encoding the presence or absence of the eight PIs upon isolation of the corresponding sequence. Hence, all sequences isolated from PI-naive patients were associated with zero vectors. The resulting vectors were combined as rows to form a binary matrix comprised of 5,651 rows and 91 columns. We carried out a Bayesian network analysis of this matrix in order to detect associations between the presence or absence of specific PIs (encoded by “drug” nodes) and the presence or absence of a nonsynonymous mixture at a codon site (encoded by “mixture” nodes). This model also accounted for associations between codon nodes, such as those that would result from a compensatory interaction between residues in PR (54). We assumed that sequences associated with PIs in the Stanford HIV database had been isolated after the onset of the drug regimen. This assumption was expressed by a ban on all networks containing directed edges that originated at codon nodes and terminated in drug nodes, which would imply that the emergence of a nonsynonymous mixture influenced the composition of the associated drug regimen (i.e., a reversal of chronological order). We also carried out an analysis of a Bayesian network without any banned edges to evaluate the sensitivity of our results to this assumption.
We implemented a Monte Carlo Markov chain (MCMC) procedure for the inference of Bayesian network structures (26) as a component of the software package HyPhy (40). In practice, it is unlikely that the available data will favor a single structure because the number of possible structures is a combinatorial function of the number of nodes (64). For example, there are approximately 10276 possible structures for a network comprised of 40 nodes only. Consequently, Friedman and Koller proposed an MCMC-based procedure to identify the most robust interactions among nodes through Bayesian model averaging over subsets of structures (26). We employed this procedure to estimate the marginal posterior probability for each potential edge of the network. A vague prior probability of 0.5 was assigned to every edge. To calculate the posterior probabilities of structures, we employed the K2 scoring metric (15), which tends to favor structures comprised of fewer edges, thereby generating a more parsimonious and interpretable network.
We ran a single Markov chain for 3 × 105 iterations. The first 5 × 104 iterations were discarded as a burn-in period, and the marginal posterior probabilities of edges were sampled at every 2,500 iterations of the remainder. Our burn-in period and sampling frequency were derived from MCMC settings described previously by Friedman and Koller (26). We found that this sampling frequency sufficiently reduced autocorrelation in our sample of the chain. In addition, we ran replicate chains initialized with randomized states to evaluate convergence behavior (Gelman-Rubin convergence diagnostic of 1.03; 97.5% quantile, 1.17) (29). A consensus network structure was assembled from all edges, with marginal posterior probabilities exceeding a threshold value of 0.9. To quantify our level of confidence in edges of the consensus network, we employed a nonparametric bootstrap method by resampling the data with replacement to generate 100 samples comprised of 5,651 sequences each. We repeated the MCMC-based analysis of Bayesian networks for each bootstrap sample and recorded the frequency across samples that edges occurred with a posterior probability exceeding a threshold of 0.5.
RESULTS
Frequency distribution of mixtures.
We counted 9,962 nucleotide mixtures in our sample of 5,651 HIV-1 PR sequences. There were 287 codons containing multiple mixtures that were omitted from subsequent analyses as a result. Mixtures were slightly more abundant overall in sequences isolated from PI-active patients (averaging 1.9 mixtures per sequence) than in sequences from PI-naive patients (1.6 mixtures); this difference was statistically significant (W = 3.58 × 106 by Wilcoxon rank sum test; P < 2 × 10−7). A significantly greater proportion of mixtures were nonsynonymous in PI-active sequences (χ2 = 242.9 [logistic regression, likelihood ratio]; P ≪ 0.001). We also observed greater variation among codon sites in the number of nonsynonymous than synonymous mixtures (coefficients of variation of 1.32 and 0.89, respectively).
We calculated mN − mS for each codon site from mixture frequencies. We found that this quantity was negative when averaged across codon sites for both PI-naive and PI-active patients, indicating that the net effect of selection within patients was to suppress nonsynonymous mixtures in the population. Nevertheless, we observed a considerable excess of nonsynonymous mixtures (mN − mS > 0.01) in PI-active sequences at positions 13, 71, 77, and 93. All four of these positions were previously associated with minor resistance to PIs (35). The P value of this outcome occurring by chance was 0.001.
Mixtures indicate selection for resistance.
We detected significant positive (i.e., diversifying) selection (dN − dS > 0) among patients on active PI regimens at 16 codon sites in PR (positions 12, 13, 19, 33, 35, 37, 54, 62, 71, 73, 74, 77, 82, 84, 90, and 93) after applying a conservative Bonferroni correction for multiple comparisons (α = 5.0 × 10−4). These sites were consistent with those found in previous analyses of selection in HIV-1 PR (12, 39), and the majority of sites (13 out of 16) were previously associated with PI resistance (35). The largest excess of nonsynonymous substitutions occurred at sites 37, 82, and 90 (dN − dS > 3.0). Primary or secondary resistance-associated mutations have previously been described for all eight PIs at sites 82 and 90. However, codon site 37 has no known association with PI resistance. A similar analysis of selection on sequences isolated from PI-naive patients detected significant positive selection at nine codon sites in PR (positions 12, 13, 19, 35, 37, 63, 64, 77, and 93) after correcting for multiple comparisons. Again, we found strong positive selection at codon site 37 (dN − dS = 5.3), suggesting that variation at this site was shaped by divergent immune selection in the patient population irrespective of drug therapy. None of other values for dN − dS at sites under significant positive selection exceeded 3.0 in the PI-naive sample (median dN − dS = 1.7).
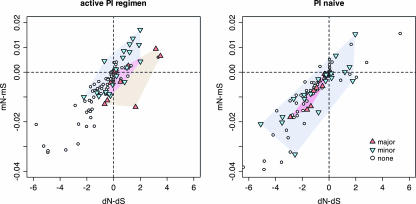
We found strong positive correlations across codon sites between dN − dS and mN − mS in both PI-naive and PI-active patient samples (ρactive = 0.87 and ρnaive = 0.83 [Spearman's rank correlation]; P ≪ 0.001) (Fig. 1) such that codon sites under stronger host-specific selection also had a greater excess of nonsynonymous mixtures. Sites with documented primary or secondary resistance mutations were clustered in the upper-right limit of the joint distribution for PI-active sequences (Fig. 1), implying that resistance to PIs was a major influence on selection at both levels of the population. Also, the cluster of primary resistance sites appeared to be displaced towards the lower-right quadrant of the PI-active plot, indicating that nonsynonymous mixtures were less abundant than expected for this class of codon site. This observation was consistent with stronger selection for primary resistance mutations, suppressing nonsynonymous mixtures by rapidly driving the fixation of favorable variants within patients (59). In contrast, within the joint distribution of dN − dS and mN − mS from PI-naive sequences, sites associated with primary or secondary resistance mutations were indistinguishable from other sites (Fig. 1).
FIG. 1.
Correlations for variation in HIV-1 protease within and among patients. Each point corresponds to a unique codon site in the HIV-1 PR sequence. The red triangles indicate codon sites that have been associated with major effects on resistance to PIs (35). Inverted blue triangles indicate positions with minor effects on resistance to PIs. These clusters are highlighted by colored polygons (determined by the convex hulls). The x axis corresponds to diversifying selection among patients quantified by the difference in nonsynonymous and synonymous rates of substitution (dN − dS). Similarly, the y axis corresponds to directional selection within patients quantified by the difference in the frequencies of nonsynonymous and synonymous mixtures (mN − mS). The left plot displays estimates of substitution rates and mixture frequencies for patients undergoing PI regimens, whereas the right plot displays estimates for patients whom were PI naive upon isolation of HIV-1 sequences. Both plots were clipped to the same range along each axis to better resolve the central distribution of points, i.e., omitting position Q2 (coordinates −7.3, and −0.04) from the PI-active plots and positions Q2, E65, and I93 (−9.7 and −0.04, −8.1 and −0.05, and −8.0 and −0.03, respectively) from the PI-naive plots.
Anatomy of a drug mixture network.
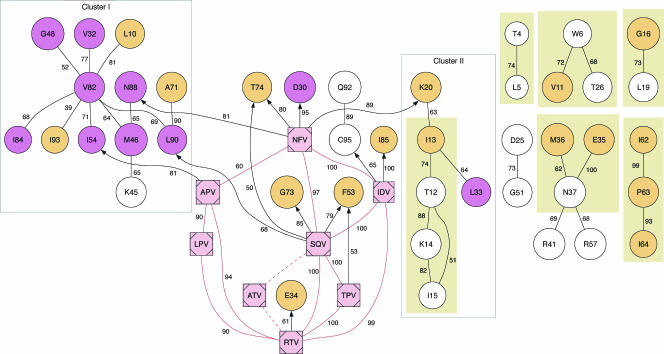
We analyzed the joint distribution of nonsynonymous mixtures and the composition of drug regimens using an MCMC-based Bayesian network model (26). The resulting distribution of marginal posterior probabilities for all edges in the network was distinctly U shaped, indicating that our sample size was sufficient to distinguish real associations from background variation. A consensus network structure, assembled from a total of 56 edges with marginal posterior probabilities exceeding 0.9, is shown in Fig. 2. Nearly all edges in the consensus network represented “positive” interactions (odds ratio [OR] of >1), such that nonsynonymous mixtures at one position tended to be either accompanied by mixtures at other positions or associated with a specific PI. Negative interactions were restricted to edges between drug nodes. For example, there were no patients on regimens combining saquinavir with tipranavir in our sample (OR of 0). The lack of edges between mixture nodes that represented negative interactions (OR of <1) was most likely caused by relatively low frequencies of nonsynonymous mixtures per codon site. Hence, our power to detect mutually exclusive mixtures was limited by our sample size.
FIG. 2.
Consensus Bayesian network for the joint distribution of mixtures and PIs. A consensus Bayesian network was assembled from edges with a marginal posterior probability exceeding 0.9. Each circular “mixture” node encodes the presence or absence of a nonsynonymous mixture at the codon site identified by the node label. Mixture nodes are color coded violet if major resistance mutations to one or more PIs have been described at that position and are color coded orange for minor mutations, according to an expert panel (35). Each square “drug” node encodes the presence or absence of a protease inhibitor upon isolation of the sequence, where the node label is the abbreviation of the inhibitor name. Undirected edges indicate the occurrence of either directed edge in the MCMC sample. Directed edges from mixture nodes to drug nodes were banned from the network. Each edge is labeled with the support value from a nonparametric bootstrap analysis of 100 samples. Edges connecting the drug node ATV are dashed to indicate that these edges were not observed in bootstrap networks. Filled rectangles enclose edges that were also recovered in a Bayesian network of mixtures from only PI-naive sequences. LPV, lopinavir; TPV, tipranavir; RTV, ritonavir.
The consensus network was comprised of seven components, including a large component that contained all eight drug nodes, which we henceforth refer to as the “resistance” component of the network. Drug nodes were highly interconnected in a distinct cluster within the “resistance” component, reflecting the predominant use of multiple PIs in combination therapy, e.g., the use of ritonavir as a pharmacologic booster of other PIs (79). Edges between drug nodes tended to have high levels of bootstrap support, with the exception of edges connecting ATV (Fig. 2). Only 23 sequences in our sample corresponded to patients on drug regimens that included ATV; as a result, very few networks inferred from bootstrap samples contained edges between ATV and other drug nodes.
The resistance component also contained 13 directed edges originating from a drug node and terminating at a mixture node (Fig. 2). Such edges implied that the use of a specific PI directly favored the emergence of nonsynonymous mixtures at the corresponding codon site, i.e., primary resistance mutations. For instance, the edges NFV→D30 and SQV→L90 corresponded to well-characterized associations between PIs and primary mutations. Other directed edges with robust bootstrap support (e.g., NFV→K20 and IDV→I85) represented novel associations between PIs and codon sites. Among the drug nodes, SQV and NFV were assigned the largest number of directed edges to mixture nodes (four), whereas ATV and lopinavir were assigned none. To evaluate the robustness of these edges to relaxing our assumption that drug nodes could not be conditionally dependent on codon nodes, we carried out a replicate analysis without this constraint. The unconstrained Bayesian network consensus structure recovered 10 out of the 13 directed edges from drug nodes to codon nodes (IDV→positions 85 and 95, NFV→positions 20 and 74, SQV→positions 73, 74, and 90, ritonavir→position 34, tipranavir→position 53, and APV→position 54) with high marginal posterior probabilities (>0.9) (data not shown). Only one edge between drug and codon nodes in the unconstrained network was oriented in the opposite direction (position 45→ATV). This reversed edge was likely a spurious association due to the low frequency of ATV (n = 24) among patient drug regimens. All other features of the original network structure were also recovered intact, including all clusters of codon nodes. Consequently, we will continue to refer to the original consensus network structure in this section.
The majority of mixture nodes in the resistance component fell into one of two clusters separated by drug nodes. The larger cluster was comprised predominantly of a hub centered around the mixture node V82, which was connected by eight different edges to the mixture nodes L10, V32, M46, G48, I54, I84, L90, and I93. Eight out of 12 mixture nodes in the V82 cluster corresponded to positions where primary resistance mutations have been documented (35), including all three of the mixture nodes connected to drug nodes (NFV→N88, SQV→L90, and APV→I54). In addition, many of the mixture nodes in this cluster have previously been implicated in conferring cross-resistance to multiple PIs. However, several these mixture nodes (e.g., L10 and I84) were blocked from drug nodes by one or more intervening mixture nodes, suggesting that many ostensible associations between mutations and cross-resistance could be explained by residue-residue interactions with broadly compensating mutations at other sites, e.g., V82.
A smaller cluster within the resistance component was comprised of six mixture nodes (T12, I13, K14, I15, K20, and L33) connected to the hub of drug nodes by a single edge (NFV→K20). Several edges in this cluster formed a causal chain across the mixture nodes T12, I13, K14, and I15. These edges were also recovered in another Bayesian network that was trained exclusively on mixtures from PI-naive sequences, indicating that the edges represented residue-residue interactions within the native structure of PR irrespective of PIs.
The remaining network components were comprised of residue-residue interactions that were not influenced by the presence of PIs; 8 out of 11 edges in these components were also recovered by the PI-naive network (Fig. 2). Furthermore, mixture nodes in the “PI-independent” components were disproportionately represented in the set of codon sites under statistically significant diversifying selection among PI-naive patients (i.e., dN − dS > 0) (see above). This result implied that diversification at these sites was being driven by factors other than selection for resistance, such as the immune response mediated by cytotoxic T lymphocytes (CTLs) (51).
Structural context of PI resistance.
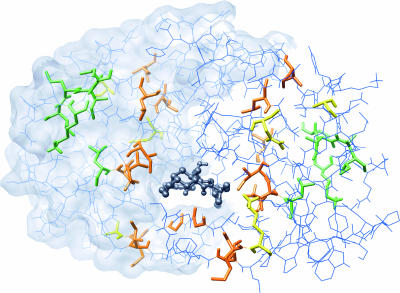
We mapped the resistance component of the Bayesian network to a structural model of an HIV-1 subtype B protease molecular dimer complexed with the inhibitor nelfinavir (Protein Data Bank accession number 1OHR) (36). From the resistance component, mixture nodes forming a hub with V82 at its center corresponded to residues that were located within a distinct layer spanning the flap and the substrate cleft (Fig. 3). This cluster included several residues near the active site (i.e., V82 and I84) that can potentially form direct contacts with either protease inhibitors or substrate molecules. However, residues that were identified as being sites of primary resistance mutations by directed edges in the network (I54, N88, and L90) were generally located further away from the substrate cleft. A second cluster of mixture nodes from the resistance component mapped to residues that were all localized in the “hinge” region of the PR molecule, located distally from the inhibitor binding site (Fig. 3). Residues within this cluster were either adjacent in the primary structure or separated by no more than 4 Å (i.e., minimum distance between atoms in the respective side chains). In sum, the two clusters of mixture nodes from the resistance component of our network mapped to distinct regions in the structural model of PR, suggesting that the genetic basis of resistance to PIs could be partitioned into at least two distinct functional modules.
FIG. 3.
Visualization of a structural model of HIV-1 protease. This image depicts a structural model of the HIV-1 protease dimer complexed with a molecule of the inhibitor nelfinavir (Protein Data Bank accession number 1OHR). The model is oriented such that the binding cavity containing the inhibitor molecule (emphasized as a dark gray ball-and-stick model) is visible. Residues corresponding to the V82-containing cluster of mixture nodes in the “resistance” component of the Bayesian network are orange or yellow, where the latter indicates residues under the direct influence of PIs. Residues corresponding to the K20-containing cluster of mixture nodes are green.
DISCUSSION
We have taken a new approach to unraveling PI resistance in HIV-1 using nonsynonymous mixtures in PR to quantify the evolutionary response within patients to the onset of drug therapy. First, we screened population-based sequences of HIV-1 protease for mixtures, which can represent nucleotide polymorphisms in the virus population. Second, we determined the validity of using mixtures as a signature of viral evolution within hosts by comparing variation in mixture frequencies to nonsynonymous and synonymous substitution rates at each site in the protease gene. The substitution rates were estimated by reconstructing the substitution history of the sequences according to their relationships within a hypothetical phylogeny. We found that site-specific selection for different variants across the patient population was recapitulated by an excess of nonsynonymous mixtures within hosts at those sites. This outcome was unique to patients under PI therapy, which implies that the distribution of nonsynonymous mixtures is being shaped by selection for PI resistance. Third, we analyzed this entire distribution at once using a Bayesian network to evaluate the effect of different PIs on the evolution of HIV-1 within hosts.
Using sequence and drug regimen data obtained from the Stanford HIV Drug Resistance Database, we were able to recover well-documented features of this response, such as the cluster of residues associated with cross-resistance (33) and the specific association between nelfinavir and mutations at site 30, and uncover several novel features as well. However, our use of mixtures to dissect the genetic basis of PI resistance in HIV-1 is subject to some unique caveats. First, the frequency of nonsynonymous mixtures at a given codon site is sensitive to the strength of directional selection (59). Although nonsynonymous mixtures are enriched by weak selection, they can also be removed by the rapid fixation of favorable variants in the population. Because we omitted codon sites at which nonsynonymous mixtures occurred in fewer than 3 out of 5,651 sequences, we may have inadvertently excluded sites under strong selection. However, none of the sites affected by this criterion (P1, G27, A28, D29, G40, W42, P44, G52, V56, Q59, G78, T80, P81, G94, T96, and F99) has previously been implicated in resistance to PIs (35). Furthermore, we found a strong correlation between the quantities mN − mS and dN − dS, indicating that sites deficient in nonsynonymous mixtures were conserved, i.e., under strong purifying selection.
Second, we encoded the presence of any nonsynonymous mixture within a codon site as a single observation without making a distinction between mixtures that could be resolved into different amino acids. The mixture-containing codons GYA and GSA can both be resolved to alanine, for example, but the alternate resolution of GYA is valine, whereas GSA resolves to glycine. This omission may confound residue-specific associations between codon sites or between a codon site and a specific PI. For instance, residue-residue interactions between codon sites 46, 54, and 88 in HIV-1 PR are conditional based on the actual residues involved (61). Although it is possible to encode different types of nonsynonymous mixtures for each codon site, doing so greatly inflates the complexity of the Bayesian network. It is also impossible to distinguish the ancestral and derived nucleotides for a given mixture. A mixture does not inform us about the direction of evolution, and the occurrence of multiple mixtures in a sequence does not reveal the linkage relationship between the mutations, i.e., whether the mutations reside on the same nucleic acid. Because the rate of recombination exceeds the mutation rate in HIV-1 (62), the linkage disequilibrium between mutations can be rapidly broken down. However, recombination may be suppressed by the bottleneck in genetic variation caused by selection for resistance to PIs (53).
Third, sequencing mixtures are a considerably error-prone measure of within-host polymorphism. This error affects the interpretation of absolute numbers of mixtures, such as mean differences between treated and untreated patient groups, but not the detection of covariation among sites or associations with drug regimens. Nevertheless, it is important to diagnose this source of uncertainty. The probability of observing a mixture is dependent on the frequency of the minority variant in the population (32, 43, 46, 67, 70). It is also sensitive to differences in experimental conditions and protocols (e.g., location of primers), sequence quality, and base-calling criteria among laboratories. Shafer et al. (69) previously showed that most discordances between two laboratories were due to inherent sampling variation in sequencing a heterogeneous population rather than exogenous differences in the sequencing process. However, Sayer et al. (66) found that nine laboratories processing the same set of HIV-1 protease and RT samples varied consistently in their rates of reporting mixtures. They were unable to resolve whether differences in reporting mixtures were due to experimental procedures or subsequent sequence editing. Their assessment also found that a laboratory's rate of reporting mixtures correlated with its success at reporting substitutions associated with resistance, highlighting the need for a standardization of laboratory protocols. Establishing standards will require that we understand which aspects of the sequencing protocol contribute the most variability in detecting mixtures. For instance, Galli et al. (28) previously noted that most discordances were due to the extraction and reverse transcription steps of processing patient samples.
Moreover, within-host polymorphisms do not necessarily represent the effect of selection on emerging advantageous mutations. For instance, many patients become superinfected or coinfected with multiple variants of HIV-1 (31, 72), which may be manifested by an excess of mixtures in population-based sequences. However, the overall incidence of superinfection or coinfection appears to remain low in patient populations, with estimates ranging from 0.5 to 5% (16, 72). Infection by multiple variants of HIV-1 may also occur by transmission of a multiply infected cell (63), but these variants immediately undergo a severe selective bottleneck (44) and are unlikely to contribute to the frequency of mixtures. Drug-naive patients may also become infected by resistant variants of HIV-1 transmitted from individuals undergoing drug therapy (47) such that mixtures may reflect the reversion of drug resistance mutations. On the other hand, we note several lines of evidence showing that this process does not significantly influence the evolution of HIV-1 within hosts. First, the reversion of drug-resistant variants would be manifested as a surplus of nonsynonymous mixtures at resistance-associated sites in sequences from PI-naive patients, but we find no such trend in our data (Fig. 1). Second, the reversion of resistant variants transmitted to a drug-naive host is relatively slow and unlikely to be observed as mixtures (4, 9, 56), and the removal of the 21 putatively PI-resistant sequences from the PI-naive data set had no effect on the outcome of our analysis. Third, patients that have received successful drug treatments should not be infectious. Although several processes can influence the frequency of mixtures, we were nevertheless able to recover a strong signature of within-host evolution shaped by selection for resistance.
Despite these caveats, our analysis of mixtures provides a number of significant improvements on previous attempts to map the genetic basis of PI resistance. First, mixtures are phylogenetically independent observations. Previous studies (7, 18, 78) invariably focused on the variation in amino acid sequences at the level of the patient population, i.e., fixed substitutions in divergent HIV-1 populations. However, substitutions found within sequences in the presence of PI therapy may have occurred before the onset of PI therapy or may have been transmitted from a previous host, thereby producing spurious associations with resistance in a comparative study. In contrast, sequence mixtures directly manifest the evolution of HIV-1 within each patient. Second, our application of Bayesian networks to analyze the joint distribution of mixtures and PIs enables us to distinguish between the direct influence of a PI from an indirect association mediated by residue-residue interactions in PR (18). For instance, a mutation may appear to be associated with a specific PI because it compensates for a resistance mutation. Hence, our methods can also be used to detect covariation among sites. Third, all eight PIs were encoded by nodes in the Bayesian network explicitly recognizing the prevalence of combination therapy. Thus, associations between specific PIs and codon sites in PR are evaluated in the context of other PIs. Previous studies have either analyzed PIs on a case-by-case basis (7, 18) or grouped patients by the number of PIs in their regimens (78). As a result, associations between mutations and specific PIs were confounded by the effects of other PIs (e.g., the use of ritonavir as a pharmacologic booster). Fourth, our use of mixture data reveals trends in the evolution of resistance within patients and is therefore less susceptible to becoming confounded by trends in the evolution of HIV-1 at the level of the patient population. For example, the immune response mediated by CTLs can influence the frequency of mutations within CTL epitopes (50) that often coincide with sites where resistance mutations occur (51).
The structure of the Bayesian network inferred in this study indicates that residue-residue interactions are more likely to be responsible for the statistical associations of these sites with a cross-resistant phenotype. For instance, codon site V82 appears to be a site for “global” compensatory mutations; through its interactions with other sites, V82 becomes indirectly associated with resistance to multiple PIs. Although this distinction between correlation and causation does not necessarily lessen the predictive value of mutations at V82, it provides insight into the mechanistic basis of cross-resistance. The V82 side chain can directly contact the PI molecule, but its mutation can also cause considerable structural rearrangements in PR (3). The lack of edges between drug nodes and V82 in the network suggests that the emergence of a mutation at V82 within a patient is not itself sufficient to confer resistance, which implies that the conformational effect of the mutant residue is more important than its contact with the inhibitor. Additional mixture nodes that correspond to sites customarily associated with primary resistance (e.g., V32, G48, and I84) (35) are similarly “blocked” from drug nodes in the network and also occur in the substrate cleft of the PR structure, suggesting a common mechanism of resistance based on mutational effects on the structural conformation of protease.
We also found several codon sites at which variation within patients is independent of PI therapy. This result appears to contradict previous work because seven of the codon sites represented by nodes in the PI-independent network components were previously associated with minor resistance to PIs (35). In addition, these sites tend to be under strong diversifying selection in the patient population (i.e., dN − dS > 0), and they overlap remarkably with those reported to be associated with HLA variation in the patient population (10, 51). For example, Brumme et al. (10) recently identified significant associations between HLA types and sites L10, T12, K14, I15, E35, N37, P63, I64, and I93. Although E35, P63, and I64 have been characterized as being sites of minor resistance mutations, our network indicates that they are PI independent. Consequently, we propose that these associations with resistance are false positives caused by founder effects driven by population level selection for CTL escape variants (39). We also find extensive diversifying selection at sites N37 and I93 (12, 38); N37 is independent of PIs, and the edge joining I93 to V82 in the resistance network is supported in only a minority of bootstrap samples. Similarly, we found that site V77 was under significant diversifying selection (dN − dS > 0) in both patient groups but was not associated with any PIs in our network. Mutations at this site have been associated with minor resistance to PIs (35), but it is also a potential anchor position within an HLA B57-restricted epitope (25). Contributions to resistance and CTL escape are not necessarily mutually exclusive effects of site-specific variation (51). However, failure to account for the phylogeny exposes population-level resistance association studies to misinterpreting the genetic divergence driven by the patient-specific immune response.
Sequencing mixtures are a poorly understood phenomenon. Although they are an inherently noisy sort of observation, they can nevertheless retain useful information about the evolution of HIV-1 within hosts. In previous work, we demonstrated that mixtures in population-based HIV-1 and hepatitis C virus sequences recapitulate the adaptation of the circulating virus population to immune variation in the host population owing to the similar time scales of transmission and selection in these viruses (59). The accurate interpretation of mixtures will ultimately require a comprehensive model that addresses the relative contributions of both population genetic (59) and experimental (28) processes. This model may be further complicated by the infrequent occurrence of coinfection or superinfection. Such a model remains outside the scope of this paper. Nevertheless, we show here that the evolution of resistance to PI in HIV-1 can be gainfully investigated using mixtures. Our analysis identifies many potential genetic interactions within HIV-1 protease from covariation in the site-specific frequencies of nonsynonymous mixtures. Put simply, the coincidence of nonsynonymous mixtures at pairs of sites across sequences implies that the corresponding residues are structurally or functionally related. We find that M46 and V82 participate in interactions with several other sites, for instance, and may play an important role in conditioning which genetic pathway the virus population will traverse to evolve resistance to PIs. For example, codon node I93 is conditionally dependent on both V82 and IDV (indinavir), so a preceding mutation at V82 may determine whether the population will use I93 or an alternative pathway such as I85 to acquire resistance (data not shown). This finding corroborates similar conclusions derived from structural studies of HIV-1 protease (3). Similarly, clinical studies of HIV-1 protease reported that multiple resistance mutations may need be accumulated in a specific order (49). Using our current model and data set, however, it is difficult to identify mutational orders from the distribution of mixtures. This important objective will require extensive longitudinal sequence data as well as an extension of these methods into dynamic Bayesian networks (27), which would be better suited for detecting trends in the distribution of mixtures or substitutions over time.
Acknowledgments
We thank Andrew J. Leigh Brown and Fraser I. Lewis for insightful discussions and providing the source code for a Monte Carlo Markov chain analysis of Bayesian networks, from which our implementation in HyPhy was derived, and two anonymous reviewers for their insightful comments on a previous version of the manuscript.
This work was supported by grants AI43638, AI47745, AI57167, and AI57167 from the National Institutes of Health, developmental awards AI36214 to S.D.W.F. and S.L.K.P. from the University of California San Diego Centers for AIDS Research and the National Institute of Allergy and Infectious Disease, and the Veterans Affairs San Diego Healthcare System.
Footnotes
Published ahead of print on 3 October 2007.
REFERENCES
- 1.Abecasis, A. B., K. Deforche, J. Snoeck, L. T. Bacheler, P. McKenna, A. P. Carvalho, G. Perpetua, R. J. Camacho, and A.-M. Vandamme. 2005. Protease mutation M89I/V is linked to therapy failure in patients infected with the HIV-1 non-B subtypes C, F, or G. AIDS 19:1799-1806. [DOI] [PubMed] [Google Scholar]
- 2.Badri, S. M., O. M. Adeyemi, B. E. Max, B. M. Zagorski, and D. E. Barker. 2003. How does expert advice impact genotypic resistance testing in clinical practice? Clin. Infect. Dis. 37:708-713. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin, E. T., T. N. Bhat, B. Liu, N. Pattabiraman, and J. W. Erickson. 1995. Structural basis of drug resistance for the V82A mutant of HIV-1 protease. Nat. Struct. Biol. 2:244-249. [DOI] [PubMed] [Google Scholar]
- 4.Barbour, J. D., F. M. Hecht, T. Wrin, T. J. Liegler, C. A. Ramstead, M. P. Busch, M. R. Segal, C. J. Petropoulos, and R. M. Grant. 2004. Persistence of primary drug resistance among recently HIV-1 infected adults. AIDS 18:1683-1689. [DOI] [PubMed] [Google Scholar]
- 5.Baxter, J. D., J. M. Schapiro, C. A. Boucher, V. M. Kohlbrenner, D. B. Hall, J. R. Scherer, and D. L. Mayers. 2006. Genotypic changes in human immunodeficiency virus type 1 protease associated with reduced susceptibility and virologic response to the protease inhibitor tipranavir. J. Virol. 80:10794-10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beerenwinkel, N., T. Lengauer, J. Selbig, B. Schmidt, H. Walter, K. Korn, R. Kaiser, and D. Hoffmann. 2001. Geno2pheno: interpreting genotypic HIV drug resistance tests. IEEE Intell. Syst. 16:35-41. [Google Scholar]
- 7.Beerenwinkel, N., B. Schmidt, H. Walter, R. Kaiser, T. Lengauer, D. Hoffmann, K. Korn, and J. Selbig. 2002. Diversity and complexity of HIV-1 drug resistance: a bioinformatics approach to predicting phenotype from genotype. Proc. Natl. Acad. Sci. USA 99:8271-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhattacharya, T., M. Daniels, D. Heckerman, B. Foley, N. Frahm, C. Kadie, J. Carlson, K. Yusim, B. McMahon, B. Gaschen, S. Mallal, J. I. Mullins, D. C. Nickle, J. Herbeck, C. Rousseau, G. H. Learn, T. Miura, C. Brander, B. Walker, and B. Korber. 2007. Founder effects in the assessment of HIV polymorphisms and HLA allele associations. Science 315:1583-1586. [DOI] [PubMed] [Google Scholar]
- 9.Brenner, B. G., J.-P. Routy, M. Petrella, D. Moisi, M. Oliveira, M. Detorio, B. Spira, V. Essabag, B. Conway, R. Lalonde, R.-P. Sekaly, and M. A. Wainberg. 2002. Persistence and fitness of multidrug-resistant human immunodeficiency virus type 1 acquired in primary infection. J. Virol. 76:1753-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brumme, Z. L., C. J. Brumme, D. Heckerman, B. T. Korber, M. Daniels, J. Carlson, C. Kadie, T. Bhattacharya, C. Chui, J. Szinger, T. Mo, R. S. Hogg, J. S. G. Montaner, N. Frahm, C. Brander, B. D. Walker, and P. R. Harrigan. 2007. Evidence of differential HLA class I-mediated viral evolution in functional and accessory/regulatory genes of HIV-1. PLoS Pathog. 3:e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charpentier, C., D. E. Dwyer, F. Mammano, D. Lecossier, F. Clavel, and A. J. Hance. 2004. Role of minority populations of human immunodeficiency virus type 1 in the evolution of viral resistance to protease inhibitors. J. Virol. 78:4234-4247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, L., A. Perlina, and C. J. Lee. 2004. Positive selection detection in 40,000 human immunodeficiency virus (HIV) type 1 sequences automatically identifies drug resistance and positive fitness mutations in HIV protease and reverse transcriptase. J. Virol. 78:3722-3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Condra, J. H., D. J. Holder, W. A. Schleif, O. M. Blahy, R. M. Danovich, L. J. Gabryelski, D. J. Graham, D. Laird, J. C. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, T. Yang, J. A. Chodakewitz, P. J. Deutsch, R. Y. Leavitt, F. E. Massari, J. W. Mellors, K. E. Squires, R. T. Steigbigel, H. Teppler, and E. A. Emini. 1996. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J. Virol. 70:8270-8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Condra, J. H., W. A. Schleif, O. M. Blahy, L. J. Gabryelski, D. J. Graham, J. Quintero, A. Rhodes, H. L. Robbins, E. Roth, M. Shivaprakash, D. Titus, T. Yang, H. Tepplert, K. E. Squires, P. J. Deutsch, and E. A. Emini. 1995. In vivo emergence of HIV-1 variants resistant to multiple protease inhibitors. Nature 374:569-571. [DOI] [PubMed] [Google Scholar]
- 15.Cooper, G., and E. Herskovits. 1992. A Bayesian method for the induction of probabilistic networks from data. Mach. Learn. 9:309-347. [Google Scholar]
- 16.Courgnaud, V., R. Seng, P. Becquart, A. Boulahtouf, C. Rouzioux, F. Boufassa, C. Deveau, P. Van De Perre, L. Meyer, and V. Foulongne. 2007. HIV-1 co-infection prevalence in two cohorts of early HIV-1 seroconverters in France. AIDS 21:1055-1056. [DOI] [PubMed] [Google Scholar]
- 17.Crow, J. F., and M. Kimura. 1970. An introduction to population genetics theory. Harper & Row, New York, NY.
- 18.Deforche, K., R. Camacho, Z. Grossman, T. Silander, M. A. Soares, Y. Moreau, R. W. Shafer, K. Van Laethem, A. P. Carvalho, B. Wynhoven, P. Cane, J. Snoeck, J. Clarke, S. Sirivichayakul, K. Ariyoshi, A. Holguin, H. Rudich, R. Rodrigues, M. B. Bouzas, P. Cahn, L. F. Brigido, V. Soriano, W. Sugiura, P. Phanuphak, L. Morris, J. Weber, D. Pillay, A. Tanuri, P. R. Harrigan, J. M. Shapiro, D. A. Katzenstein, R. Kantor, and A.-M. Vandamme. 2007. Bayesian network analysis of resistance pathways against HIV-1 protease inhibitors. Infect. Genet. Evol. 7:382-390. [DOI] [PubMed] [Google Scholar]
- 19.Deforche, K., T. Silander, R. Camacho, Z. Grossman, M. A. Soares, K. Van Laethem, R. Kantor, Y. Moreau, A.-M. Vandamme, et al. 2006. Analysis of HIV-1 pol sequences using Bayesian networks: implications for drug resistance. Bioinformatics 22:2975-2979. [DOI] [PubMed] [Google Scholar]
- 20.Drãghici, S., and R. B. Potter. 2003. Predicting HIV drug resistance with neural networks. Bioinformatics 19:98-107. [DOI] [PubMed] [Google Scholar]
- 21.Durant, J., P. Cleverbergh, P. Haifon, P. Delgiudice, S. Porsin, P. Simonet, N. Montagne, C. A. B. Boucher, J. M. Schapiro, and P. Dellamonica. 1999. Drug-resistance genotyping in HIV-1 therapy: the VIRADAPT randomised controlled trial. Lancet 353:2195-2199. [DOI] [PubMed] [Google Scholar]
- 22.Dykes, C., J. Najjar, R. J. Bosch, M. Wantman, M. Furtado, S. Hart, S. M. Hammer, and L. M. Demeter. 2004. Detection of drug-resistant minority variants of HIV-1 during virologic failure of indinavir, lamivudine, and zidovudine. J. Infect. Dis. 189:1091-1096. [DOI] [PubMed] [Google Scholar]
- 23.Eder, J., U. Hommel, F. Cumin, B. Martoglio, and B. Gerhartz. 2007. Aspartic proteases in drug discovery. Curr. Pharm. Des. 13:271-285. [DOI] [PubMed] [Google Scholar]
- 24.Felsenstein, J. 1985. Phylogenies and the comparative method. Am. Nat. 125:1-15. [Google Scholar]
- 25.Frahm, N., C. Linde, and C. Brander. 2007. Identification of HIV-derived, HLA class I restricted CTL epitopes: insights into TCR repertoire, CTL escape and viral fitness, p. 3-28. In B. Korber, C. Brander, B. F. Haynes, R. Koup, J. P. Moore, B. D. Walker, and D. I. Watkins (ed.), HIV molecular immunology 2006, LA-UR 07-4752. Los Alamos National Laboratory, Los Alamos, NM.
- 26.Friedman, N., and D. Koller. 2003. Being Bayesian about network structure. A Bayesian approach to structure discovery in Bayesian networks. Mach. Learn. 50:95-125. [Google Scholar]
- 27.Friedman, N., K. Murphy, and S. Russell. 1998. Learning the structure of dynamic probabilistic networks, p. 139-147. In G. F. Cooper and S. Moral (ed.), Proceedings of the Fourteenth Conference on Uncertainty in Artificial Intelligence. Morgan Kaufmann, San Francisco, CA.
- 28.Galli, R. A., B. Sattha, B. Wynhoven, M. V. O'Shaughnessy, and P. R. Harrigan. 2003. Sources and magnitude of intralaboratory variability in a sequence-based genotypic assay for human immunodeficiency virus type 1 drug resistance. J. Clin. Microbiol. 41:2900-2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gelman, A., and D. B. Rubin. 1992. Inference from iterative simulation using multiple sequences. Stat. Sci. 7:457-511. [Google Scholar]
- 30.Gonzales, M. J., T. D. Wu, J. Taylor, I. Belitskaya, R. Kantor, D. Israelski, S. Chou, A. R. Zolopa, W. J. Fessel, and R. W. Shafer. 2003. Extended spectrum of HIV-1 reverse transcriptase mutations in patients receiving multiple nucleoside analog inhibitors. AIDS 17:791-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gottlieb, G. S., D. C. Nickle, M. A. Jensen, K. G. Wong, J. Grobler, F. Li, S.-L. Liu, C. Rademeyer, G. H. Learn, S. S. A. Karim, C. Williamson, L. Corey, J. B. Margolick, and J. I. Mullins. 2004. Dual HIV-1 infection associated with rapid disease progression. Lancet 363:619-622. [DOI] [PubMed] [Google Scholar]
- 32.Günthard, H. F., J. K. Wong, C. C. Ignacio, D. V. Havlir, and D. D. Richman. 1998. Comparative performance of high-density oligonucleotide sequencing and dideoxynucleotide sequencing of HIV type 1 pol from clinical samples. AIDS Res. Hum. Retrovir. 14:869-876. [DOI] [PubMed] [Google Scholar]
- 33.Hertogs, K., S. Bloor, S. D. Kemp, C. Van den Eynde, T. M. Alcorn, R. Pauwels, M. Van Houtte, S. Staszewski, V. Miller, and B. A. Larder. 2000. Phenotypic and genotypic analysis of clinical HIV-1 isolates reveals extensive protease inhibitor cross-resistance: a survey of over 6000 samples. AIDS 14:1203-1210. [DOI] [PubMed] [Google Scholar]
- 34.Hoffman, N. G., C. A. Schiffer, and R. Swanstrom. 2003. Covariation of amino acid positions in HIV-1 protease. Virology 314:536-548. [DOI] [PubMed] [Google Scholar]
- 35.Johnson, V. A., F. Brun-Vezinet, B. Clotet, D. R. Kuritzkes, D. Pillay, J. M. Schapiro, and D. D. Richman. 2006. Update of the drug resistance mutations in HIV-1: Fall 2006. Top. HIV Med. 14:125-130. [PubMed] [Google Scholar]
- 36.Kaldor, S. W., V. J. Kalish, J. F. Davies, B. V. Shetty, J. E. Fritz, K. Appelt, J. A. Burgess, K. M. Campanale, N. Y. Chirgadze, D. K. Clawson, B. A. Dressman, S. D. Hatch, D. A. Khalil, M. B. Kosa, P. P. Lubbehusen, M. A. Muesing, A. K. Patick, S. H. Reich, K. S. Su, and J. H. Tatlock. 1997. Viracept (nelfinavir mesylate, AG1343): a potent, orally bioavailable inhibitor of HIV-1 protease. J. Med. Chem. 40:3979-3985. [DOI] [PubMed] [Google Scholar]
- 37.Kohl, N. E., E. A. Emini, W. A. Schleif, L. J. Davis, J. C. Heimbach, R. A. F. Dixon, E. M. Scolnick, and I. S. Sigal. 1988. Active human immunodeficiency virus protease is required for viral infectivity. Proc. Natl. Acad. Sci. USA 85:4686-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kosakovsky Pond, S. L., and S. D. W. Frost. 2005. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22:1208-1222. [DOI] [PubMed] [Google Scholar]
- 39.Kosakovsky Pond, S. L., S. D. W. Frost, Z. Grossman, M. B. Gravenor, D. D. Richman, and A. J. L. Brown. 2006. Adaptation to different human populations by HIV-1 revealed by codon-based analyses. PLoS Comput. Biol. 2:e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosakovsky Pond, S. L., S. D. W. Frost, and S. V. Muse. 2005. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21:676-679. [DOI] [PubMed] [Google Scholar]
- 41.Kramer, R. A., M. D. Schaber, A. M. Skalka, K. Ganguly, F. Wong-Staal, and E. P. Reddy. 1986. HTLV-III Gag protein is processed in yeast cells by the virus Pol-protease. Science 231:1580-1584. [DOI] [PubMed] [Google Scholar]
- 42.Lanave, C., G. Preparata, C. Saccone, and G. Serio. 1984. A new method for calculating evolutionary substitution rates. J. Mol. Evol. 20:86-93. [DOI] [PubMed] [Google Scholar]
- 43.Larder, B. A., A. Kohli, P. Kellam, S. D. Kemp, M. Kronick, and R. D. Henfrey. 1993. Quantitative detection of HIV-1 drug resistance mutations by automated DNA sequencing. Nature 365:671-673. [DOI] [PubMed] [Google Scholar]
- 44.Learn, G. H., D. Muthui, S. J. Brodie, T. Zhu, K. Diem, J. I. Mullins, and L. Corey. 2002. Virus population homogenization following acute human immunodeficiency virus type 1 infection. J. Virol. 76:11953-11959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leitner, T., D. Escanilla, C. Franzén, M. Uhlén, and J. Albert. 1996. Accurate reconstruction of a known HIV-1 transmission history by phylogenetic tree analysis. Proc. Natl. Acad. Sci. USA 93:10864-10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leitner, T., E. Halapi, G. Scarlatti, P. Rossi, J. Albert, E. M. Fenyö, and M. Uhlén. 1993. Analysis of heterogeneous viral populations by direct DNA-sequencing. Biotechniques 15:120-127. [PubMed] [Google Scholar]
- 47.Little, S. J., E. S. Daar, R. T. D'Aquila, P. H. Keiser, E. Connick, J. M. Whitcomb, N. S. Hellmann, C. J. Petropoulos, L. Sutton, J. A. Pitt, E. S. Rosenberg, R. A. Koup, B. D. Walker, and D. D. Richman. 1999. Reduced antiretroviral drug susceptibility among patients with primary HIV infection. JAMA 282:1142-1149. [DOI] [PubMed] [Google Scholar]
- 48.Mellors, J., S. Palmer, D. Nissley, M. Kearney, E. Halvas, C. Bixby, L. Demeter, S. Eshleman, K. Bennett, S. Hart, F. Vaida, M. Wantman, J. Coffin, S. Hammer, et al. 2004. Low-frequency NNRTI-resistant variants contribute to failure of efavirenz-containing regimens, abstr. 134. Abstr. 11th Conf. Retrovir. Opportun. Infect.
- 49.Molla, A., M. Korneyeva, Q. Gao, S. Vasavanonda, P. J. Schipper, H. M. Mo, M. Markowitz, T. Chernyavskiy, P. Niu, N. Lyons, A. Hsu, G. R. Granneman, D. D. Ho, C. A. Boucher, J. M. Leonard, D. W. Norbeck, and D. J. Kempf. 1996. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat. Med. 2:760-766. [DOI] [PubMed] [Google Scholar]
- 50.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439-1443. [DOI] [PubMed] [Google Scholar]
- 51.Mueller, S. M., B. Schaetz, K. Eismann, S. Bergmann, M. Bauerle, M. Schmitt-Haendle, H. Walter, B. Schmidt, K. Korn, H. Sticht, B. Spriewald, E. G. Harrer, and T. Harrer. 2007. Dual selection pressure by drugs and HLA class I-restricted immune responses on human immunodeficiency virus type 1 protease. J. Virol. 81:2887-2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muse, S. V., and B. S. Gaut. 1994. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol. Biol. Evol. 11:715-724. [DOI] [PubMed] [Google Scholar]
- 53.Nijhuis, M., C. A. Boucher, P. Schipper, T. Leitner, R. Schuurman, and J. Albert. 1998. Stochastic processes strongly influence HIV-1 evolution during suboptimal protease-inhibitor therapy. Proc. Natl. Acad. Sci. USA 95:14441-14446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nijhuis, M., R. Schuurman, D. de Jong, J. Erickson, E. Gustchina, J. Albert, P. Schipper, S. Gulnik, and C. A. B. Boucher. 1999. Increased fitness of drug resistant HIV-1 protease as a result of acquisition of compensatory mutations during suboptimal therapy. AIDS 13:2349-2359. [DOI] [PubMed] [Google Scholar]
- 55.Palella, F. J., Jr., K. M. Delaney, A. C. Moorman, M. O. Loveless, J. Fuhrer, G. A. Satten, D. J. Aschman, S. D. Holmberg, et al. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N. Engl. J. Med. 338:853-860. [DOI] [PubMed] [Google Scholar]
- 56.Pao, D., U. Andrady, J. Clarke, G. Dean, S. Drake, M. Fisher, T. Green, S. Kumar, M. Murphy, A. Tang, S. Taylor, D. White, G. Underhill, D. Pillay, and P. Cane. 2004. Long-term persistence of primary genotypic resistance after HIV-1 seroconversion. J. Acquir. Immune Defic. Syndr. 37:1570-1573. [DOI] [PubMed] [Google Scholar]
- 57.Parikh, U., C. Calef, B. A. Larder, R. Schinazi, and J. W. Mellors. 2002. Mutations in retroviral genes associated with drug resistance, p. 95-183. In C. Kuiken, B. Foley, E. Freed, B. Hahn, B. Korber, P. Marx, F. McCutchan, and J. W. Mellors (ed.), HIV sequence compendium 2002. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, NM.
- 58.Pearl, J. 1988. Probabilistic reasoning in intelligent systems: networks of plausible inference. Morgan Kaufmann, San Mateo, CA.
- 59.Poon, A. F. Y., S. L. Kosakovsky Pond, P. Bennett, D. D. Richman, A. J. L. Brown, and S. D. W. Frost. 2007. Adaptation to human populations is revealed by within-host polymorphisms in HIV-1 and hepatitis C virus. PLoS Pathog. 3:e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rhee, S.-Y., M. J. Gonzales, R. Kantor, B. J. Betts, J. Ravela, and R. W. Shafer. 2003. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 31:298-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rhee, S.-Y., T. F. Liu, S. P. Holmes, and R. W. Shafer. 2007. HIV-1 subtype B protease and reverse transcriptase amino acid covariation. PLoS Comput. Biol. 3:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rhodes, T., H. Wargo, and W.-S. Hu. 2003. High rates of human immunodeficiency virus type 1 recombination: near-random segregation of markers one kilobase apart in one round of viral replication. J. Virol. 77:11193-11200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ritola, K., C. D. Pilcher, S. A. Fiscus, N. G. Hoffman, J. A. E. Nelson, K. M. Kitrinos, C. B. Hicks, J. J. Eron, Jr., and R. Swanstrom. 2004. Multiple V1/V2 env variants are frequently present during primary infection with human immunodeficiency virus type 1. J. Virol. 78:11208-11218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robinson, R. 1971. Counting labeled acyclic digraphs, p. 239-273. In F. Harary (ed.) New directions in the theory of graphs. Proceedings of the Third Ann Arbor Conference on Graph Theory. Academic Press, New York, NY.
- 65.Roquebert, B., I. Malet, M. Wirden, R. Tubiana, M.-A. Valantin, A. Simon, C. Katlama, G. Peytavin, V. Calvez, and A.-G. Marcelin. 2006. Role of HIV-1 minority populations on resistance mutational pattern evolution and susceptibility to protease inhibitors. AIDS 20:287-289. [DOI] [PubMed] [Google Scholar]
- 66.Sayer, D. C., S. Land, L. Gizzarelli, M. French, G. Hales, S. Emery, F. T. Christiansen, and E. M. Dax. 2003. Quality assessment program for genotypic antiretroviral testing improves detection of drug resistance mutations. J. Clin. Microbiol. 41:227-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schuurman, R., L. Demeter, P. Reichelderfer, J. Tijnagel, T. de Groot, and C. Boucher. 1999. Worldwide evaluation of DNA sequencing approaches for identification of drug resistance mutations in the human immunodeficiency virus type 1 reverse transcriptase. J. Clin. Microbiol. 37:2291-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sevin, A. D., V. Degruttola, M. Nijhuis, J. M. Schapiro, A. S. Foulkes, M. F. Para, and C. A. Boucher. 2000. Methods for investigation of the relationship between drug-susceptibility phenotype and human immunodeficiency virus type 1 genotype with applications to AIDS clinical trials group 333. J. Infect. Dis. 182:59-67. [DOI] [PubMed] [Google Scholar]
- 69.Shafer, R. W., K. Hertogs, A. R. Zolopa, A. Warford, S. Bloor, B. J. Betts, T. C. Merigan, R. Harrigan, and B. A. Larder. 2001. High degree of interlaboratory reproducibility of human immunodeficiency virus type 1 protease and reverse transcriptase sequencing of plasma samples from heavily treated patients. J. Clin. Microbiol. 39:1522-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shafer, R. W., A. Warford, M. A. Winters, and M. J. Gonzales. 2000. Reproducibility of human immunodeficiency virus type 1 (HIV-1) protease and reverse transcriptase sequencing of plasma samples from heavily treated HIV-1-infected individuals. J. Virol. Methods 86:143-153. [DOI] [PubMed] [Google Scholar]
- 71.Sing, T., V. Svicher, N. Beerenwinkel, F. Ceccherini-Silberstein, M. Däumer, R. Kaiser, H. Walter, K. Korn, D. Hoffmann, M. Oette, J. K. Rockstroh, G. Fätkenheuer, C.-F. Perno, and T. Lengauer. 2005. Characterization of novel HIV drug resistance mutations using clustering, multidimensional scaling and SVM-based feature ranking, p. 285-296. In A. Jorge, L. Torgo, P. Brazil, R. Camacho, and J. Gama (ed.), Knowledge discovery in databases: PKDD 2005. Springer, New York, NY.
- 72.Smith, D. M., J. K. Wong, G. K. Hightower, C. C. Ignacio, K. K. Koelsch, E. S. Daar, D. D. Richman, and S. J. Little. 2004. Incidence of HIV superinfection following primary infection. JAMA 292:1177-1178. [DOI] [PubMed] [Google Scholar]
- 73.Tamura, K., and M. Nei. 1993. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10:512-526. [DOI] [PubMed] [Google Scholar]
- 74.Tisdale, M., S. D. Kemp, N. R. Parry, and B. A. Larder. 1993. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc. Natl. Acad. Sci. USA 90:5653-5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tural, C., L. Ruiz, C. Holtzer, J. Schapiro, P. Viciana, J. Gonzales, P. Domingo, C. Boucher, C. Rey-Joly, B. Clotet, and the Havana Study Group. 2002. Clinical utility of HIV-1 genotyping and expert advice: the Havana trial. AIDS 16:209-218. [DOI] [PubMed] [Google Scholar]
- 76.Wang, D., and B. Larder. 2003. Enhanced prediction of lopinavir resistance from genotype by use of artificial neural networks. J. Infect. Dis. 188:653-660. [DOI] [PubMed] [Google Scholar]
- 77.Wang, K., E. Jenwitheesuk, R. Samudrala, and J. E. Mittler. 2004. Simple linear model provides highly accurate genotypic predictions of HIV-1 drug resistance. Antivir. Ther. 9:343-352. [PubMed] [Google Scholar]
- 78.Wu, T. D., C. A. Schiffer, M. J. Gonzales, J. Taylor, R. Kantor, S. Chou, D. Israelski, A. R. Zolopa, W. J. Fessel, and R. W. Shafer. 2003. Mutation patterns and structural correlates in human immunodeficiency virus type 1 protease following different protease inhibitor treatments. J. Virol. 77:4836-4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zeldkin, R. K., and R. A. Petruschke. 2004. Pharmacological and therapeutic properties of ritonavir-boosted protease inhibitor therapy in HIV-1 infected patients. J. Antimicrob. Chemother. 53:4-9. [DOI] [PubMed] [Google Scholar]
- 80.Zolopa, A. R., R. W. Shafer, A. Warford, J. G. Montoya, P. Hsu, D. Katzenstein, T. C. Merigan, and B. Efron. 1999. HIV-1 genotypic resistance patterns predict response to saquinavir-ritonavir therapy in patients in whom previous protease inhibitor therapy had failed. Ann. Intern. Med. 131:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]