Abstract
3-Methyl adenine (3meA), a minor-groove DNA lesion, presents a strong block to synthesis by replicative DNA polymerases (Pols). To elucidate the means by which replication through this DNA lesion is mediated in eukaryotic cells, here we carry out genetic studies in the yeast Saccharomyces cerevisiae treated with the alkylating agent methyl methanesulfonate. From the studies presented here, we infer that replication through the 3meA lesion in yeast cells can be mediated by the action of three Rad6-Rad18-dependent pathways that include translesion synthesis (TLS) by Polη or -ζ and an Mms2-Ubc13-Rad5-dependent pathway which presumably operates via template switching. We also express human Pols ι and κ in yeast cells and show that they too can mediate replication through the 3meA lesion in yeast cells, indicating a high degree of evolutionary conservation of the mechanisms that control TLS in yeast and human cells. We discuss these results in the context of previous observations that have been made for the roles of Pols η, ι, and κ in promoting replication through the minor-groove N2-dG adducts.
Replicative DNA polymerase (Pols) are highly sensitive to geometric distortions in DNA; consequently, they are inhibited by the presence of DNA lesions in the template strand. A number of Pols that promote replication through DNA lesions exist in eukaryotes, and they are highly specialized for the roles they play in translesion synthesis (TLS) (40). For example, both yeast and human Polηs are highly adept at promoting error-free replication through UV-induced cyclobutane pyrimidine dimers (CPDs) (15, 20, 50, 52), and inactivation of Polη in humans causes the cancer-prone syndrome of the variant form of xeroderma pigmentosum (14, 30).
Although proficient replication through a DNA lesion such as a CPD can be accomplished by Polη alone, replication through many of the different lesions present in DNA requires the sequential action of two Pols, in which one polymerase inserts the nucleotide opposite the DNA lesion and another polymerase performs the subsequent extension reaction (40, 41). Yeast Polζ, comprised of the Rev3 catalytic and Rev7 accessory subunits (39), is highly specialized for performing the extension step of TLS (8, 19, 21, 35). Although humans also have the Rev3 and Rev7 proteins, there is no biochemical information available on the role of human Polζ in TLS.
In addition to Polη, humans have Pols ι and κ, which belong to the Y family of Pols (40). Polι differs strikingly from Pols η and κ and almost all other Pols in that it incorporates nucleotides opposite template purines with much higher efficiency and fidelity than opposite template pyrimidines (6, 19, 44, 49). Moreover, even opposite template purines, Polι exhibits higher catalytic efficiency and fidelity opposite template A than opposite template G. The ternary crystal structures of Polι bound to template A or G and the correct incoming deoxynucleoside triphosphate have shown that the purine template adopts a syn conformation in the Polι active site and forms a Hoogsteen base pair with the incoming nucleotide, which remains in the anti conformation (36-38). Hoogsteen base pairing explains the basis for the higher efficiency of correct nucleotide incorporation opposite template purines than opposite template pyrimidines, as only the purine bases have the Hoogsteen edge via which they can hydrogen bond with the correct incoming nucleotide.
The ability of the Polι active site to push the template purine into the syn conformation provides an elegant mechanism by which this polymerase can incorporate nucleotides opposite minor-groove DNA lesions. The minor-groove N2 group of guanine is highly reactive and can conjugate with a large variety of endogenously formed products. For example, the reaction of acrolein, an α,β-unsaturated aldehyde formed in cells as a product of lipid peroxidation, with the N2 group of guanine in DNA followed by ring closure at N-1, leads to the formation of the cyclic adduct γ-hydroxy-1,N2-propano-2′-deoxyguanosine (γ-HOPdG). Polι incorporates a C opposite this adduct with the same catalytic efficiency (kcat/Km) as it does opposite the nonadducted template G residue (51). Polι, however, is unable to carry out the subsequent extension reaction, and this step is modulated by Polκ (51). The sequential actions of Pols ι and κ can also promote replication through the structurally more complex aldehyde products of lipid peroxidation, such as trans-4-hydroxy-2-nonenal (HNE) (55).
γ-HOPdG adopts a ring-closed 1,N2-exo cyclic form when present as a templating residue, but upon pairing with a C, it changes from the closed cyclic form to a ring-opened conformation able to form a normal Watson-Crick base pair (3, 23, 24). Studies with the permanently ring-opened or ring-closed structural analogs of γ-HOPdG have yielded useful information about the relative abilities of Pols η, ι, and κ to replicate through such N2-dG adducts, and the combined results of these and other studies have indicated that these Pols vary in their response to different N2-dG adducts. For example, the ring-closed form of γ-HOPdG is a strong block for nucleotide incorporation by Pols η and κ, with only Polι being capable of inserting a C opposite from it (33, 54). And, even though Polκ can extend from a C opposite γ-HOPdG, which can adopt both the ring-opened and ring-closed forms (51), it is unable to extend from the permanently ring-closed analog of γ-HOPdG (54), suggesting that Polκ can extend only if γ-HOPdG is in the ring-opened conformation. The ability of Polι to push the ring-closed analog of γ-HOPdG into a syn conformation would allow for its Hoogsteen base pairing with dCTP, but since Polκ cannot accommodate such a structure in its active site (29), it will not carry out the subsequent extension reaction. Polκ can, however, proficiently extend from the C opposite γ-HOPdG, because the pairing with a C will trigger the change from the ring-closed to the ring-opened form of γ-HOPdG capable of forming a normal Watson-Crick base pair from which Polκ can extend. The proficient ability of Polκ to extend from a C opposite from the γ-HOPdG or HNE lesions could derive from the fact that the minor-groove edge of the templating residue at the template-primer junction is open to solvent and not obstructed by the Polκ active site (29).
In contrast to the inhibitory effects of the ring-closed analog of γ-HOPdG on DNA synthesis by the various TLS Pols, the permanently ring-opened form of γ-HOPdG does not present a significant block to synthesis by Pols η, ι, or κ, as they can each carry out both nucleotide incorporation and the extension reactions opposite from it (33, 54). Thus, we assume that an N2-dG minor-groove adduct, such as the ring-open form of γ-HOPdG, can be accommodated in the active site of these Pols at both the nucleotide insertion and subsequent extension steps.
To further investigate the ability of various polymerases to mediate TLS through minor-groove DNA adducts, we have carried out genetic experiments in the yeast Saccharomyces cerevisiae to examine the roles of yeast and human TLS Pols in promoting replication through an N-3 minor-groove adduct of adenine. Treatment of cells with methyl methanesulfonate (MMS) methylates the bases in DNA, particularly adenine at the N-3 position (3meA) and guanine at the N-7 position (7meG) (1, 26). In vitro studies have indicated that 3meA, which projects into the minor groove, is a strong inhibitor of DNA polymerases such as Escherichia coli Pol I or avian myeloblastosis virus reverse transcriptase, and DNA synthesis terminates one nucleotide before the lesion. 7meG projects into the major groove and does not block synthesis by these polymerases (25). Genetic studies in yeast have corroborated the blocking action of 3meA on synthesis by the replicative polymerases (see Results, below, for elaboration of this point). 3meA could be blocking replicative polymerases for a number of reasons. First, since the high-fidelity DNA polymerases use a minor-groove-sensing mechanism, in which certain residues from the polymerase form hydrogen bonds with the N-3 of a purine or the O2 of a pyrimidine to detect the correct Watson-Crick geometry of the postinsertion template-primer base pair (22, 53), 3meA would be inhibitory to these polymerases at the extension step. Second, the presence of a methyl group at N-3 could block synthesis by replicative polymerases at both the insertion and extension steps because of the steric constraints imposed in the active site adjacent to the minor-groove edge of the templating nucleotide as well as the template nucleotide at the template-primer junction.
Here we provide evidence for the role of yeast Pols η and ζ and of human Pols ι and κ in promoting replication through the 3meA adduct. We discuss the implications of these observations in the context of previous biochemical studies that have examined the role of different TLS Pols in promoting replication through the variety of minor-groove DNA lesions.
MATERIALS AND METHODS
Yeast strains.
All the deletion strains used in these studies were derived from the wild-type yeast strain EMY74.7, MATa his3-Δ1 leu2-3,112 trp1Δ ura3-52, by the one-step gene replacement method (42).
MMS survival and mutagenesis.
Cells grown overnight in yeast extract-peptone-dextrose (YPD) medium were washed with distilled water, sonicated to disperse clumps, and resuspended in 0.05 M KPO4 buffer, pH 7.0, at a density of 3 × 108 cells/ml. After treatment of cell suspensions with various concentrations of MMS for 20 min at 30°C with vigorous shaking, an equal volume of 10% sodium thiosulfate was added to inactivate the MMS. Appropriate dilutions were plated on YPD for viability determinations and on synthetic complete medium containing canavanine but lacking arginine for determinations of the frequency of canavanine-resistant colonies.
Expression of human Pols ι and κ in yeast cells.
The genes encoding human Polι (19) and Polκ (previously designated Polθ [16]) were cloned into the expression vector pMA91 (32), in which expression is under the control of the efficiently expressed yeast PGK1 promoter. The plasmids generated, pBJ1129 for expression of Polι and pPOL15 for expression of Polκ, were introduced into various yeast strains, such as mag1Δ and other mutant strains, and tested for their ability to restore resistance to MMS by the serial dilution spot test. For these studies, cells were grown to mid-logarithmic phase in synthetic complete medium lacking leucine to maintain selection of the plasmid, washed, and resuspended in water to a density of 2 × 108/ml. Aliquots (200 μl) of serial 10-fold dilutions were pipetted into a 96-well microtiter dish, followed by transfer to YPD plates containing different concentrations of MMS, and plates were incubated at 30°C for 2 days before photographing.
RESULTS
Involvement of yeast DNA Pols η and ζ in promoting replication through the 3meA lesion.
Genetic studies in yeast have indicated that replication through UV-induced DNA lesions can be handled via at least three different Rad6-Rad18-dependent pathways (45) that include TLS by Pols η and ζ and a postreplicational repair pathway, which presumably involves template switching and a copy choice type of DNA synthesis and requires the Mms2-Ubc13 ubiquitin-conjugating enzyme (12), together with the Rad5 protein, which harbors a DNA-dependent ATPase and a ubiquitin ligase activity (17, 48). To determine the contribution that these three pathways make to 3meA bypass during replication, we examined the MMS sensitivity of yeast strains deleted for the genes that function in the Rad6-Rad18-dependent lesion bypass process.
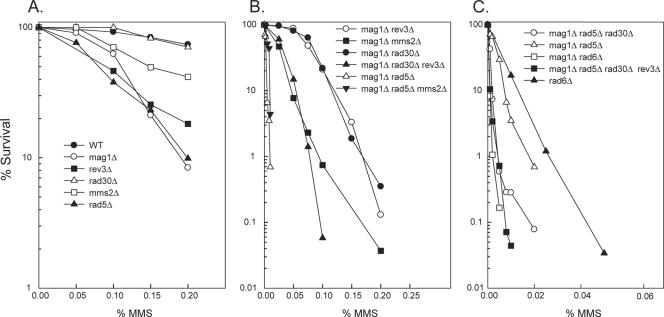
We found that deletion of the REV3 gene, which encodes the catalytic subunit of Polζ, confers a higher level of sensitivity to MMS than does deletion of RAD30, encoding Polη, whereas the mms2Δ mutation confers an intermediate level of MMS sensitivity between that resulting from the rad30Δ and rev3Δ mutations, and the rad5Δ mutation elicits a somewhat higher level of MMS sensitivity than the rev3Δ mutation (Fig. 1A).
FIG. 1.
MMS sensitivity of yeast strains carrying deletions of genes belonging to the RAD6 epistasis group. Cells were treated with the indicated concentrations of MMS (percent [vol/vol]) at 30°C for 20 min, followed by inactivation of MMS with sodium thiosulfate. Appropriate dilutions of cells were plated on YPD for viability determinations. The survival curves represent an average of at least three experiments for each strain. WT, wild type. (A) MMS sensitivity of strains deleted for the TLS polymerases η and ζ and for the MMS2 and RAD5 genes, involved in postreplicational repair. (B) MMS sensitivity of the mag1Δ strain in combination with deletions of genes encoding the TLS polymerase η or ζ or genes that function in PRR. (C) MMS sensitivity of the mag1Δ strain in combination with deletions of genes, so that all three pathways of Rad6-Rad18-dependent lesion bypass have been inactivated.
The methylated bases 3meA and 7meG, formed in DNA upon treatment with MMS, are removed by an N-methyl purine DNA glycosylase encoded in yeast by the MAG1 gene (2), and deletion of MAG1 confers a high level of MMS sensitivity upon yeast cells (Fig. 1A). Since 3meA would be a block to synthesis by the replicative polymerases, the enhanced MMS sensitivity of the mag1Δ strain must result from the blocking effects of 3meA on replication. The experiments indicating that the treatment of yeast or human cells with Me-lex, a compound which selectively generates 3meA, confers a high level of cytotoxicity and that this cytotoxicity is further enhanced in the mag1Δ yeast strain have provided corroborative evidence that if not repaired, 3meA is inhibitory to normal DNA synthesis during replication (4, 34). The introduction of the rad30Δ or the rev3Δ mutation into the mag1Δ strain led to a synergistic enhancement of MMS sensitivity in the mag1Δ rad30Δ or mag1Δ rev3Δ double mutants compared to that in the respective single mutants (compare Fig. 1A and B); furthermore, a synergistic enhancement of MMS sensitivity occurred in the mag1Δ rad30Δ rev3Δ triple mutant strain compared to the MMS sensitivity of the mag1Δ rad30Δ or mag1Δ rev3Δ strains (Fig. 1B). We infer from these observations that Pols η and ζ provide alternate means by which replication through the 3meA lesion can be accomplished.
Role for the Mms2-Ubc13-Rad5-dependent postreplication repair pathway in 3meA bypass.
The Mms2-Ubc13-Rad5-dependent pathway promotes the repair of discontinuities that form in the newly synthesized DNA from UV-damaged templates (45). This postreplicational repair (PRR) pathway presumably occurs by template switching and involves a copy choice type of synthesis in which replication through the DNA lesion is mediated by using the newly synthesized strand of the undamaged sister duplex as the template (10). The operation of this pathway requires Mms2-Ubc13-Rad5-dependent lysine 63-linked polyubiquitylation of PCNA, in which the ubiquitin ligase function of Rad5 promotes the polyubiquitylation of PCNA by Mms2-Ubc13 (7, 11, 43). In addition to its role as a ubiquitin ligase, Rad5 is likely to also function in postreplication repair in a more direct manner wherein its ATPase activity would modulate the template switching process (5).
We found that a synergistic enhancement in MMS sensitivity occurs in the mag1Δ mms2Δ and mag1Δ rad5Δ double mutant strains compared to that in the respective single mutant strains (compare Fig. 1A and B). The mag1Δ rad5Δ strain, however, displays a much higher level of MMS sensitivity than the mag1Δ mms2Δ strain, whereas the MMS sensitivity of the mag1Δ rad5Δ mms2Δ strain remained the same as that of the mag1Δ rad5Δ strain, which is in keeping with the epistasis of the rad5Δ mutation over the mms2Δ mutation (Fig. 1B). For UV damage also, although Rad5 functions in Mms2-Ubc13-dependent PRR, the rad5Δ mutation causes a much higher level of UV sensitivity than the mms2Δ and ubc13Δ mutations, and we have previously ascribed the increased UV sensitivity of the rad5Δ mutation to the additional role of Rad5 in affecting the efficiency of TLS mediated by Pols η and ζ (5). However, how Rad5 contributes to TLS by these Pols is not understood.
To verify that the TLS mediated by Pols η and ζ and that lesion bypass mediated by the Rad5-dependent pathway provide three different means for promoting replication through the 3meA lesion, we compared the MMS sensitivity of the triple mutant strain, in which all three pathways have been inactivated, with that of the double mutant strains, in which only two of the pathways have been inactivated. As expected, the MMS sensitivity of the mag1Δ rad5Δ strain was greatly enhanced upon the introduction of the rad30Δ mutation, and introduction of the rev3Δ mutation into the mag1Δ rad5Δ rad30Δ strain led to a further increase in MMS sensitivity (Fig. 1C). Since the MMS sensitivity of the mag1Δ rad5Δ rad30Δ rev3Δ strain was nearly the same as that of the mag1Δ rad6Δ strain (Fig. 1C), Polη- and -ζ-dependent TLS and Rad5-dependent lesion bypass provide three different means of Rad6-Rad18-dependent bypass of 3meA during replication.
TLS mediated by Pols η and ζ through the 3meA lesion is predominantly error free.
In yeast cells, inactivation of Polη confers a large enhancement in the incidence of mutagenesis induced by UV light (31, 57), consistent with the role of Polη in the error-free bypass of CPDs, and inactivation of Polζ confers a large decrease in the frequency of UV-induced mutations (27, 28), which accords with its role in promoting the mutagenic bypass of UV lesions.
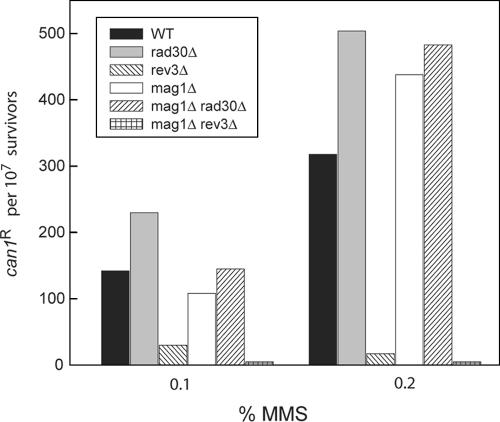
To determine if replication through the 3meA lesion involves a mutagenic process, we first examined the frequency of MMS-induced mutations in the mag1Δ strain. Our observation that MMS induced can1r mutations occur at about the same rate in the mag1Δ strain as in the wild-type strain (Fig. 2), however, suggested that replication through the 3meA lesion by Pols η and ζ was mediated in a predominantly error-free way.
FIG. 2.
MMS-induced can1r mutations in various mutant strains. Cells were treated for 20 min at 30°C with the indicated MMS concentrations. Following inactivation of MMS, cells were spread onto the surface of YPD plates for viability determinations and onto synthetic complete medium plates containing canavanine but lacking arginine for determining the frequency of can1r mutations. Each histogram represents the average of two to three experiments. WT, wild type.
Although 3meA is the blocking lesion for synthesis by the replicative polymerases, 7meG is the predominant lesion formed in DNA upon MMS treatment (1, 26). Because spontaneous depurination of 7meG would lead to the formation of abasic sites, we presume that the increase in the can1r mutation frequency obtained in MMS-treated wild-type cells results from the mutagenic TLS that occurs opposite the abasic sites that happened to have escaped repair by the action of AP endonucleases or by nucleotide excision repair (46). In fact, the frequency of MMS-induced can1r mutations rises greatly in the apn1Δ apn2Δ strain, which lacks both the AP endonucleases, and it rises even further in the apn1Δ apn2Δ rad14Δ strain, which additionally lacks the nucleotide excision repair pathway (46). Also, since the elevated incidence of MMS-induced can1r mutations in the apn1Δ apn2Δ strain is not affected by the rad30Δ mutation, Polη does not contribute to TLS through the abasic sites in any significant way (8). By contrast, the rev3Δ mutation confers a large reduction in the frequency of MMS-induced can1r mutations in the apn1Δ apn2Δ strain, consistent with the role of Polζ in extending from the nucleotide inserted opposite the abasic site by another polymerase (8, 18). Since we observed that the frequency of can1r mutations is not significantly affected by the rad30Δ mutation in MMS-treated wild-type or mag1Δ cells, whereas the rev3Δ mutation confers a much-reduced level of mutagenesis in both these genetic backgrounds (Fig. 2), we infer that MMS-induced mutations in the wild-type or mag1Δ yeast cells emanate from the TLS mediated by Polζ opposite the abasic sites and, as expected from biochemical experiments indicating a highly inefficient bypass of abasic lesions by Polη (9), this Pol has no significant impact on the TLS through this lesion.
Human Pols ι and κ promote TLS through 3meA in yeast cells.
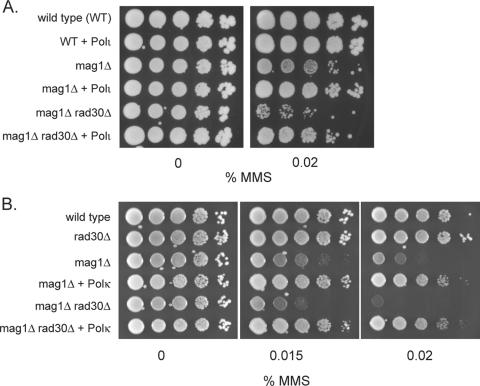
The genetic studies with the yeast rad30Δ and rev3Δ mutations reported here indicated that both Pols η and ζ can promote TLS through the 3meA lesion. In addition to these Pols, humans contain Pols ι and κ, able to promote replication through the DNA lesions. To examine if human Pols ι and κ can also support replication through the 3meA lesion, we expressed these Pols in yeast cells and tested whether they could restore MMS resistance to mag1Δ and mag1Δ rad30Δ yeast strains. As shown in Fig. 3, the expression of Pol ι or Pol κ in mag1Δ or mag1Δ rad30Δ yeast cells led to a large increase in the MMS resistance of these cells; thus, both these human Pols are able to function in yeast cells in promoting TLS through the 3meA lesions.
FIG. 3.
Enhanced MMS resistance conferred upon mag1Δ and mag1Δ rad30Δ strains by human Pols ι and κ. For each strain, a sample of cells containing a given 10-fold serial dilution was pipetted into a well of a 96-well microtiter dish as described in Materials and Methods. Cells were transferred to YPD plates containing different MMS concentrations and photographed after 2 days of incubation at 30°C.
DISCUSSION
Although MMS treatment causes the formation of 3meA and 7meG in DNA, 3meA presents a strong block to synthesis by the replicative polymerases; hence, the cytotoxicity of MMS results from 3meA and not from the 7meG lesion. Accordingly, the MMS sensitivity of yeast cells is greatly enhanced in the absence of Mag1, which is required for 3meA removal. Since the 3meA lesions would persist in mag1Δ cells, for replication to proceed through the lesion site would require the action of lesion bypass processes.
Here we provide genetic evidence that replication through the 3meA lesion in yeast cells can be effected by three Rad6-Rad18-dependent pathways that include TLS mediated by Pols η and ζ and an Mms2-Ubc13-Rad5-dependent pathway which presumably operates by template switching and involves a copy choice type of DNA synthesis. In addition to the inferred role of yeast Pols η and ζ in promoting TLS through the 3meA lesion, we show here that the expression of human Polι or Polκ in yeast cells confers a large increase in MMS resistance to mag1Δ and mag1Δ rad30Δ cells, indicating that both these human Pols are also able to support replication through the 3meA lesion. Our observations for the role of yeast Pols η and ζ and human Pols ι and κ in promoting TLS through the 3meA lesion when put into the context of the available information on their role in the bypass of various N2-dG adducts lead us to draw conclusions that are relevant to the variety of reactions these Pols can support opposite the minor-groove lesions of differing structural complexity.
Previously, we showed that yeast and human Polη and human Pols ι and κ can all replicate through the N2-dG adduct, the ring-opened analog of γ-HOPdG. Thus, in spite of the fact that this adduct would project into the minor groove, it presents no significant block to any of these polymerases at either the nucleotide insertion step or at the subsequent extension step. By contrast, the HNE adduct, which also conjugates at the N2-dG and which because of its increased size would be more blocking to replication than the γ-HOPdG adduct, is handled very differently by human Pols η, ι, and κ. Whereas Polη is inhibited by the HNE-dG adduct at both the insertion and extension steps, Polι can efficiently insert a C opposite from it but cannot carry out the subsequent extension reaction. Polκ, on the other hand, is unable to insert a nucleotide opposite from HNE-dG, but it can extend from a C inserted opposite it by Polι. Whereas the proficiency of Polι to insert a C opposite the HNE-dG adduct would derive from its ability to push the adduct into a syn conformation, where there would be no steric hindrance from the Polι active site, Polι will be unable to extend from the C inserted opposite the HNE-dG adduct because of the structural constraints in Polι at the templating side of the template-primer junction. Polκ, however, is able to extend from a C opposite HNE-dG, because the ring-open form of HNE-dG at the template-primer junction can be easily accommodated in the Polκ active site (29). Overall, then, it appears that whereas lesions such as the ring-open form of γ-HOPdG can be accommodated in the active sites of Pols η, ι, and κ at both the insertion and extension steps, for a structurally more complex lesion, such as HNE-dG, only the combined action of Pols ι and κ can promote replication through it. We presume this reflects the abilities of Polι to accommodate the lesion in its active site at the insertion step and of Polκ to accommodate the lesion in its active site for mediating the extension reaction.
The N2-dG adducts, such as γ-HOPdG and HNE, can block synthesis by replicative DNA polymerases, presumably because their presence at the site of either the templating residue or in the template at the template-primer junction introduces a steric hindrance into the active site. A minor-groove adduct such as 3meA would be additionally blocking to replicative polymerases because of the involvement of N-3 in hydrogen bonding with the residues in the polymerase. Since high-fidelity polymerases use this minor-groove hydrogen bonding to detect the correct Watson-Crick geometry of the postinsertion template primer base pair, a 3meA lesion would be particularly susceptible to detection by this mechanism at the extension step of lesion bypass. 3meA could additionally block replicative polymerases at both the insertion and extension steps because of the steric constraints in the active site.
The involvement of yeast Pols η and ζ and of human Pols κ and ι in 3meA bypass suggest that these Pols do not form hydrogen bonds with the N-3 of an A present at the templating position or at the template-primer junction and, therefore, they lack the sensing mechanism for the correct Watson-Crick base-pairing geometry at either of these positions. In keeping with this inference, we have shown previously that the replacement of a guanine with 3-deazaguanine at the templating position or at the postinsertion template site has no adverse effect on synthesis by yeast Polη or human Polκ (53, 56; unpublished observations).
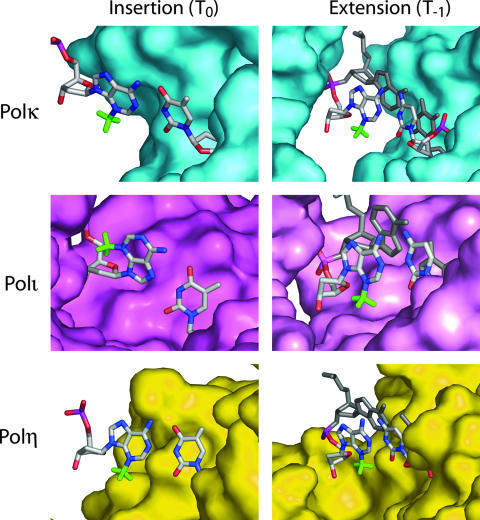
To examine whether a 3meA present on the templating residue or on the template residue at the template-primer junction can be accommodated into the active sites of Pols η, ι, and κ, we modeled the 3meA lesion into the active sites of these Pols at either the templating base (T0) or the preceding template residue (T−1), at the postinsertion site. The ternary structures of Polκ and Polι with DNA and incoming deoxynucleoside triphosphate have been solved (29, 35-38, 47). Modeling of 3meA in Polκ shows that there is ample room in the active site to accommodate the methyl group when positioned at either the templating base or the postinsertion site (Fig. 4). Thus, 3meA is not expected to be a block at either the insertion step or the subsequent extension from 3meA by Polκ. In fact, the 3meA would not impart any steric impediment to Polκ as the DNA passes through the protein during replication. This scenario also holds for Polη. We modeled the DNA from Polκ into the active site of Polη, and again, there is no contact between the protein and the minor-groove perturbation of 3meA when present at either the templating base or opposite the primer terminus (Fig. 4). Although Polι is able to rescue the MMS sensitivity of the yeast mag1Δ and mag1Δ rad30Δ strains, and thus is predicted to bypass 3meA, the structural modeling suggests steric constraints. Polι inserts nucleotides opposite purine templates by rotating the purine residue into the syn conformation and forming a Hoogsteen base pair with the incoming pyrimidine. When 3meA is modeled into the Polι active site, we find that the methyl group clashes with the 5′ oxygen in the DNA backbone, which would inhibit the syn conformation and thus make Hoogsteen base pair formation unlikely. However, it may be that the complete rotation of the 3meA residue into the syn conformation is somehow prevented in the Polι active site, and a single hydrogen bond can still form between the N-7 of 3meA and the N-3 of the incoming T. As we have shown previously, for proficient T incorporation opposite template A, only N-7 hydrogen bonding is needed (13). Polι is also less open to minor-groove disturbances at the template-primer junction than is Polκ or Polη. When 3meA is modeled into Polι at the T−1 position, there is substantially less room, and the methyl group comes in close proximity to the active site floor. However, there is no severe clash, suggesting that Polι would be able to extend from 3meA paired with T.
FIG. 4.
Modeling of 3meA in the active sites of Polκ, Polι, and Polη. 3meA was modeled in place of A at the templating base (insertion) or in place of G at the postinsertion template residue (extension) for each polymerase. Surface representations of Polκ (upper panels; PDB accession no. 2OH2), Polι (middle panels; PDB accession no. 2FLL), and Polη (lower panels; PDB accession no. 1JIH) are shown in cyan, magenta, and yellow, respectively. The methyl group of 3meA is shown in green. The DNA from the Polκ structure was modeled into Polη based on the alignment of the Polκ and Polη catalytic residues. The incoming dTTP is shown opposite the 3meA at position T0. For modeling 3meA at the T−1 position, the G residue present in the structures was replaced by 3meA; the original cognate C residue is shown to provide a reference point for the base pair; the next template residue and incoming dTTP are shown in gray. The thumb region of each polymerase was removed to provide a clear view of the active sites.
Finally, the observation that human Pols ι and κ are able to promote TLS opposite the 3meA lesion in yeast cells points to a high degree of evolutionary conservation of the mechanisms that control TLS in yeast and human cells. The ability of Pols ι and κ to function in TLS in yeast cells indicates that they can access the replication ensemble stalled at the lesion site, which we presume involves their binding to PCNA since that is a necessary precondition for TLS to occur in both yeast and human cells (40). Furthermore, we consider it quite likely that the TLS Pols are additionally involved in physical interactions with many of the other components of the replication ensemble and suspect that they too have been conserved between yeast and humans.
Acknowledgments
This work was supported by National Institutes of Health grants CA107650 and ES012411.
Footnotes
Published ahead of print on 13 August 2007.
REFERENCES
- 1.Beranek, D. T., C. C. Weis, and D. H. Swenson. 1980. A comprehensive quantitative analysis of methylated and ethylated DNA using high pressure liquid chromatography. Carcinogenesis 1:595-606. [DOI] [PubMed] [Google Scholar]
- 2.Bjoras, M., A. Klungland, R. F. Johansen, and E. Seeberg. 1995. Purification and properties of the alkylation repair DNA glycosylase encoded MAG gene from Saccharomyces cerevisiae. Biochemistry 34:4577-4582. [DOI] [PubMed] [Google Scholar]
- 3.de los Santos, C., T. Zaliznyak, and F. Johnson. 2001. NMR characterization of a DNA duplex containing the major acrolein-derived deoxyguanosine adduct γ-OH-1,-N2-propano-2′-deoxyguanosine. J. Biol. Chem. 276:9077-9082. [DOI] [PubMed] [Google Scholar]
- 4.Engelward, B. P., J. M. Allan, A. J. Dreslin, J. D. Kelly, M. M. Wu, B. Gold, and L. D. Samson. 1998. A chemical and genetic approach together define the biological consequences of 3-methylanine lesions in the mammalian genome. J. Biol. Chem. 273:5412-5418. [DOI] [PubMed] [Google Scholar]
- 5.Gangavarapu, V., L. Haracska, I. Unk, R. E. Johnson, S. Prakash, and L. Prakash. 2006. Mms-Ubc13-dependent and -independent roles of Rad5 ubiquitin ligase in postreplication repair and translesion DNA synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 26:7783-7790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haracska, L., R. E. Johnson, I. Unk, B. B. Phillips, J. Hurwitz, L. Prakash, and S. Prakash. 2001. Targeting of human DNA polymerase ι to the replication machinery via interaction with PCNA. Proc. Natl. Acad. Sci. USA 98:14256-14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haracska, L., C. A. Torres-Ramos, R. E. Johnson, S. Prakash, and L. Prakash. 2004. Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Mol. Cell. Biol. 24:4267-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haracska, L., I. Unk, R. E. Johnson, E. Johansson, P. M. J. Burgers, S. Prakash, and L. Prakash. 2001. Roles of yeast DNA polymerases δ and ζ and of Rev1 in the bypass of abasic sites. Genes Dev. 15:945-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haracska, L., M. T. Washington, S. Prakash, and L. Prakash. 2001. Inefficient bypass of an abasic site by DNA polymerase η. J. Biol. Chem. 276:6861-6866. [DOI] [PubMed] [Google Scholar]
- 10.Higgins, N. P., K. Kato, and B. Strauss. 1976. A model for replication repair in mammalian cells. J. Mol. Biol. 101:417-425. [DOI] [PubMed] [Google Scholar]
- 11.Hoege, C., B. Pfander, G.-L. Moldovan, G. Pyrowolakis, and S. Jentsch. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419:135-141. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann, R. M., and C. M. Pickart. 1999. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96:645-653. [DOI] [PubMed] [Google Scholar]
- 13.Johnson, R. E., L. Haracksa, L. Prakash, and S. Prakash. 2006. Role of Hoogsteen edge hydrogen bonding at tempate purines in nucleotide incorporation by human DNA polymerase ι. Mol. Cell. Biol. 26:6435-6441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson, R. E., C. M. Kondratick, S. Prakash, and L. Prakash. 1999. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science 285:263-265. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, R. E., S. Prakash, and L. Prakash. 1999. Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Polη. Science 283:1001-1004. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, R. E., S. Prakash, and L. Prakash. 2000. The human DINB1 gene encodes the DNA polymerase Polθ. Proc. Natl. Acad. Sci. USA 97:3838-3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, R. E., S. Prakash, and L. Prakash. 1994. Yeast DNA repair protein RAD5 that promotes instability of simple repetitive sequences is a DNA-dependent ATPase. J. Biol. Chem. 269:28259-28262. [PubMed] [Google Scholar]
- 18.Johnson, R. E., C. A. Torres-Ramos, T. Izumi, S. Mitra, S. Prakash, and L. Prakash. 1998. Identification of APN2, the Saccharomyces cerevisiae homolog of the major human AP endonuclease HAP1, and its role in the repair of abasic sites. Genes Dev. 12:3137-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, R. E., M. T. Washington, L. Haracska, S. Prakash, and L. Prakash. 2000. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature 406:1015-1019. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, R. E., M. T. Washington, S. Prakash, and L. Prakash. 2000. Fidelity of human DNA polymerase η. J. Biol. Chem. 275:7447-7450. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, R. E., S.-L. Yu, S. Prakash, and L. Prakash. 2003. Yeast DNA polymerase zeta (ζ) is essential for error-free replication past thymine glycol. Genes Dev. 17:77-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, S. J., and L. S. Beese. 2004. Structures of mismatch replication errors observed in a DNA polymerase. Cell 116:803-816. [DOI] [PubMed] [Google Scholar]
- 23.Kim, H.-Y. H., M. Voehler, T. M. Harris, and M. P. Stone. 2002. Detection of an interchain carbinolamine cross-link formed in a CpG sequence by the acrolein DNA adduct γ-OH-1,N2-propano-2′-deoxyguanosine. J. Am. Chem. Soc. 124:9324-9325. [DOI] [PubMed] [Google Scholar]
- 24.Kozekov, I. D., L. V. Nechev, M. S. Moseley, C. M. Harris, C. J. Rizzo, M. P. Stone, and T. M. Harris. 2003. DNA interchain cross-links formed by acrolein and crotonaldehyde. J. Am. Chem. Soc. 125:50-61. [DOI] [PubMed] [Google Scholar]
- 25.Larson, K., J. Sahm, R. Shenkar, and B. Strauss. 1985. Methylation-induced blocks to in vitro DNA replication. Mutat. Res. 150:77-84. [DOI] [PubMed] [Google Scholar]
- 26.Lawley, P. D., D. J. Orr, and M. Jarman. 1975. Isolation and identification of products from alkylation of nucleic acids: ethyl- and isoproyl-purines. Biochem. J. 145:73-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawrence, C. W., and R. B. Christensen. 1979. Ultraviolet-induced reversion of cyc1 alleles in radiation-sensitive strains of yeast. III. rev3 mutant strains. Genetics 92:397-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lawrence, C. W., P. E. Nisson, and R. B. Christensen. 1985. UV and chemical mutagenesis in rev7 mutants of yeast. Mol. Gen. Genet. 200:86-91. [DOI] [PubMed] [Google Scholar]
- 29.Lone, S., S. A. Townson, S. N. Uljon, R. E. Johnson, A. Brahma, D. T. Nair, S. Prakash, L. Prakash, and A. K. Aggarwal. 2007. Human DNA polymerase κ encircles DNA: implications for mismatch extension and lesion bypass. Mol. Cell 25:601-614. [DOI] [PubMed] [Google Scholar]
- 30.Masutani, C., R. Kusumoto, A. Yamada, N. Dohmae, M. Yokoi, M. Yuasa, M. Araki, S. Iwai, K. Takio, and F. Hanaoka. 1999. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase η. Nature 399:700-704. [DOI] [PubMed] [Google Scholar]
- 31.McDonald, J. P., A. S. Levine, and R. Woodgate. 1997. The Saccharomyces cerevisiae RAD30 gene, a homologue of Escherichia coli dinB and umuC, is DNA damage inducible and functions in a novel error-free postreplication repair mechanism. Genetics 147:1557-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mellor, J., M. J. Dobson, N. A. Roberts, M. F. Tuite, J. S. Emtage, S. White, P. A. Lowe, T. Patel, A. J. Kingsman, and S. M. Kingsman. 1983. Efficient synthesis of enzymatically active calf chymosin in Saccharomyces cerevisiae. Gene 24:1-14. [DOI] [PubMed] [Google Scholar]
- 33.Minko, I. G., M. T. Washington, M. Kanuri, L. Prakash, S. Prakash, and R. S. Lloyd. 2003. Translesion synthesis past acrolein-derived DNA adduct, γ-hydroxypropanodeoxyguanosine, by yeast and human DNA polymerase η. J. Biol. Chem. 278:784-790. [DOI] [PubMed] [Google Scholar]
- 34.Monti, P., R. Iannone, P. Campomenosi, Y. Ciribilli, S. Varadarajan, D. Shah, P. Menichini, B. Gold, and G. Fronza. 2004. Nucleotide excision repair defect influences lethality and mutagenicity induced by Me-lex, a sequence-selective N3-adenine methylating agent in the absence of base excision repair. Biochemistry 43:5592-5599. [DOI] [PubMed] [Google Scholar]
- 35.Nair, D. T., R. E. Johnson, L. Prakash, S. Prakash, and A. K. Aggarwal. 2006. Hoogsteen base pair formation promotes synthesis opposite the 1,N6-ehthenodeoxyadenosine lesion by human DNA polymerase ι. Nat. Struct. Mol. Biol. 13:619-625. [DOI] [PubMed] [Google Scholar]
- 36.Nair, D. T., R. E. Johnson, L. Prakash, S. Prakash, and A. K. Aggarwal. 2005. Human DNA polymerase ι incorporates dCTP opposite template G via a G · C+ Hoogsteen base pair. Structure 13:1569-1577. [DOI] [PubMed] [Google Scholar]
- 37.Nair, D. T., R. E. Johnson, S. Prakash, L. Prakash, and A. K. Aggarwal. 2006. An incoming nucleotide imposes an anti to syn conformational change on the templating purine in the human DNA polymerase-ι active site. Structure 14:749-755. [DOI] [PubMed] [Google Scholar]
- 38.Nair, D. T., R. E. Johnson, S. Prakash, L. Prakash, and A. K. Aggarwal. 2004. Replication by human DNA polymerase ι occurs via Hoogsteen base-pairing. Nature 430:377-380. [DOI] [PubMed] [Google Scholar]
- 39.Nelson, J. R., C. W. Lawrence, and D. C. Hinkle. 1996. Thymine-thymine dimer bypass by yeast DNA polymerase ζ. Science 272:1646-1649. [DOI] [PubMed] [Google Scholar]
- 40.Prakash, S., R. E. Johnson, and L. Prakash. 2005. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 74:317-353. [DOI] [PubMed] [Google Scholar]
- 41.Prakash, S., and L. Prakash. 2002. Translesion DNA synthesis in eukaryotes: a one- or two-polymerase affair. Genes Dev. 16:1872-1883. [DOI] [PubMed] [Google Scholar]
- 42.Rothstein, R. J. 1983. One-step gene disruption in yeast. Methods Enzymol. 101:202-211. [DOI] [PubMed] [Google Scholar]
- 43.Stelter, P., and H. D. Ulrich. 2003. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425:188-191. [DOI] [PubMed] [Google Scholar]
- 44.Tissier, A., J. P. McDonald, E. G. Frank, and R. Woodgate. 2000. Polι, a remarkably error-prone human DNA polymerase. Genes Dev. 14:1642-1650. [PMC free article] [PubMed] [Google Scholar]
- 45.Torres-Ramos, C., S. Prakash, and L. Prakash. 2002. Requirement of RAD5 and MMS2 for post replication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 22:2419-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Torres-Ramos, C. A., R. E. Johnson, L. Prakash, and S. Prakash. 2000. Evidence for the involvement of nucleotide excision repair in the removal of abasic sites in yeast. Mol. Cell. Biol. 20:3522-3528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trincao, J., R. E. Johnson, C. R. Escalante, S. Prakash, L. Prakash, and A. K. Aggarwal. 2001. Structure of the catalytic core of S. cerevisiae DNA polymerase η: implications for translesion DNA synthesis. Mol. Cell 8:417-426. [DOI] [PubMed] [Google Scholar]
- 48.Ulrich, H. D., and S. Jentsch. 2000. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 19:3388-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Washington, M. T., R. E. Johnson, L. Prakash, and S. Prakash. 2004. Human DNA polymerase ι utilizes different nucleotide incorporation mechanisms dependent upon the template base. Mol. Cell. Biol. 24:936-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Washington, M. T., R. E. Johnson, S. Prakash, and L. Prakash. 2000. Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc. Natl. Acad. Sci. USA 97:3094-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Washington, M. T., I. G. Minko, R. E. Johnson, W. T. Wolfle, T. M. Harris, R. S. Lloyd, S. Prakash, and L. Prakash. 2004. Efficient and error-free replication past a minor groove DNA adduct by the sequential action of human DNA polymerases ι and κ. Mol. Cell. Biol. 24:5687-5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Washington, M. T., L. Prakash, and S. Prakash. 2003. Mechanism of nucleotide incorporation opposite a thymine-thymine dimer by yeast DNA polymerase η. Proc. Natl. Acad. Sci. USA 100:12093-12098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Washington, M. T., W. T. Wolfle, T. E. Spratt, L. Prakash, and S. Prakash. 2003. Yeast DNA polymerase η makes functional contacts with the DNA minor groove only at the incoming nucleoside triphosphate. Proc. Natl. Acad. Sci. USA 100:5113-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolfle, W. T., R. E. Johnson, I. G. Minko, R. S. Lloyd, S. Prakash, and L. Prakash. 2005. Human DNA polymerase ι promotes replication through a ring-closed minor-groove adduct that adopts a syn conformation in DNA. Mol. Cell. Biol. 25:8748-8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolfle, W. T., R. E. Johnson, I. G. Minko, R. S. Lloyd, S. Prakash, and L. Prakash. 2006. Replication past a trans-4-hydroxynonenal minor-groove adduct by the sequential action of human DNA polymerase ι and κ. Mol. Cell. Biol. 26:381-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolfle, W. T., M. T. Washington, E. T. Kool, T. E. Spratt, S. A. Helquist, L. Prakash, and S. Prakash. 2005. Evidence for a Watson-Crick hydrogen bonding requirement in DNA synthesis by human DNA polymerase κ. Mol. Cell. Biol. 25:7137-7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu, S.-L., R. E. Johnson, S. Prakash, and L. Prakash. 2001. Requirement of DNA polymerase η for error-free bypass of UV-induced CC and TC photoproducts. Mol. Cell. Biol. 21:185-188. [DOI] [PMC free article] [PubMed] [Google Scholar]