Abstract
Carbon catabolite repression (CCR) allows bacteria to alter metabolism in response to the availability of specific sugar sources, and increasing evidence suggests that CCR is involved in regulating virulence gene expression in many pathogens. A scan of the M1 SF370 group A streptococcus (GAS) genome using a Bacillus subtilis consensus identified a number of potential catabolite-responsive elements (cre) important for binding by the catabolite control protein A (CcpA), a mediator of CCR in gram-positive bacteria. Intriguingly, a putative cre was identified in the promoter region of mga upstream of its distal P1 start of transcription. Electrophoretic mobility shift assays showed that a His-CcpA fusion protein was capable of binding specifically to the cre in Pmga in vitro. Deletion analysis of Pmga using single-copy Pmga-gusA reporter strains found that Pmga P1 and its upstream cre were not required for normal autoregulated mga expression from Pmga P2 as long as Mga was produced from its native locus. In fact, the Pmga P1 region appeared to show a negative influence on Pmga P2 in these studies. However, deletion of the cre at the native Pmga resulted in a reduction of total mga transcripts as determined by real-time reverse transcription-PCR, supporting a role for CcpA in initial expression. Furthermore, normal transcriptional initiation from the Pmga P1 start site alone was dependent on the presence of the cre. Importantly, inactivation of ccpA in the M6 GAS strain JRS4 resulted in a reduction in Pmga expression and Mga protein levels in late-logarithmic-phase cell growth. These data support a role for CcpA in the early activation of the mga promoter and establish a link between CCR and Mga regulation in the GAS.
Glucose is the preferred source of energy in many microorganisms, whereby the conversion of glucose into two molecules of pyruvate in the Embden-Meyerhof pathway generates two molecules of ATP. In order for another carbohydrate to be metabolized via this pathway, it first must be converted to glucose, an energy-consuming process. Therefore, bacteria have developed means to ensure that glucose is utilized before other energy sources and to ensure that enzymes necessary for the metabolism of alternative energy sources are available only when glucose is depleted (4). Carbon catabolite repression (CCR) is a process in which enzymes necessary for the metabolism of alternative sugars are inhibited in the presence of glucose.
In gram-positive organisms, regulation of carbon catabolism centers on a component of the phosphoenolpyruvate phosphotransferase system (PTS) (4). The primary purpose of the PTS is to regulate sugar uptake through phosphorylation, which is achieved by shuttling a phosphate from phosphoenolpyruvate to a cytosolic enzyme called EI, to HPr (heat-stable protein), and then to a sugar-specific membrane-bound enzyme, EII, before being attached to an incoming sugar (18). Unlike gram-negative organisms, which use cyclic AMP levels to detect the energy level of the cell, gram-positive bacteria use HPr (6). Therefore, the PTS is multifunctional in gram-positive bacteria, being involved in sugar transport as well as signal transduction in response to sugar availability.
During sugar transport, HPr is phosphorylated on a conserved histidine residue (H15). However, in the presence of glucose, an enzyme called HPr kinase phosphorylates HPr on a serine residue (S46) in response to products of glycolysis (7). This phosphorylation event allows HPr-Ser to bind to the catabolite control protein (CcpA), which belongs to the LacI/GalR family of transcription factors (5). CcpA then is capable of binding with specificity to a catabolite-responsive element (cre) located in promoters or coding sequences of genes with products that are involved in the metabolism of alternative sugars. Although CcpA primarily acts to repress expression of these operons, it also is known to activate transcription of genes important for growth in glucose (11, 14).
There is accumulating evidence that CCR is important for virulence in several low-G+C gram-positive pathogens. For instance, CCR has been linked to virulence in Clostridium perfringens (42), Staphylococcus aureus (29), and Listeria monocytogenes (26). In Streptococcus pneumoniae, the inactivation of CcpA, also called RegM, significantly attenuated virulence in various mouse models of pneumococcal colonization and infection (13, 16). CcpA (RegM) may be affecting systemic infection through the control of capsular gene expression (13), whereas its influence on mucosal colonization and pneumoniae could reflect alterations in basic cellular metabolism (16).
The group A streptococcus (GAS; Streptococcus pyogenes) is an important gram-positive pathogen capable of eliciting a wide array of diseases in humans. Surface M protein is a major virulence determinant that is capable of such functions as inhibiting phagocytosis, binding fibrinogen, and facilitating host cell invasion. Previous studies on the GAS have shown that M protein production is affected by the sugar source (33), suggesting that CCR plays a role in its expression. Transcription of the gene encoding M protein (emm) is activated by the stand-alone response regulator Mga, which also is responsible for transcriptional activation of other virulence genes involved in adhesion, invasion, and immune evasion. Expression of mga is autoregulated, and its promoter (Pmga) has two Mga-binding sites (MBSs) as well as two transcriptional start sites, named P1 (distal) and P2 (proximal) based on their proximity to the translational start site of mga (25, 30). Mga is known to be responsive to environmental signals such as growth phase, and the entire promoter, including 84 bp of sequence upstream of P1, has been reported to be necessary for full activity (3, 21, 23, 30, 34). Given this information, it was hypothesized that CCR could be involved in the control of M protein expression through the regulation of mga. In the present study, the GAS genome was scanned for cre based on a published Bacillus subtilis consensus. A functional cre involved in activation at the P1 start of transcription was identified within Pmga, suggesting a direct link between carbon metabolism and Mga regulation in S. pyogenes.
MATERIALS AND METHODS
Bacterial strains and media.
Bacterial strains used in this study are listed in Table 1. The GAS was cultured static in Todd-Hewitt medium supplemented with 0.2% (wt/vol) yeast extract (THY) at 37°C except where noted, and growth was monitored by optical density (OD) using a Klett-Summerson photoelectric colorimeter with the A filter. Escherichia coli was grown shaking in Luria-Bertani medium (LB) at 37°C, and growth was monitored by OD. Antibiotics were used at the following concentrations: ampicillin at 100 μg/ml for E. coli; chloramphenicol at 5 μg/ml for E. coli and 1.5 μg/ml for the GAS; erythromycin at 500 μg/ml for E. coli and 1.0 μg/ml for the GAS; kanamycin at 50 μg/ml for E. coli and 300 μg/ml for the GAS; and spectinomycin at 100 μg/ml for both E. coli and the GAS.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| E. coli | ||
| BL21(DE3) | F−ompT hsdSB(rB− mB−) gal dcm (DE3) | Novagen |
| DH5α | hsdR17 recA1 gyrA endA1 relA1 | 15 |
| M6 GAS | ||
| JRS4 | Streptomycin-resistant derivative of D471 | 37 |
| GA19681 | Clinical invasive isolate | 36 |
| KSM310 | Pmga (full)-gusA in VIT locus, Mga+ | This study |
| KSM310.150Lg | Pmga (full)-gusA in VIT locus, Mga− | This study |
| KSM310.700 | Pmga (full)-gusA in VIT locus, Mga+ CcpA− | This study |
| KSM427 | Pmga (ΔP1)-gusA in VIT locus, Mga+ | This study |
| KSM427.150Lg | Pmga (ΔP1)-gusA in VIT locus, Mga− | This study |
| KSM428 | Pmga (ΔMBS I)-gusA in VIT locus, Mga+ | This study |
| KSM428.150Lg | Pmga (ΔMBS I)-gusA in VIT locus, Mga− | This study |
| KSM429 | Pmga (ΔMBS I & II)-gusA in VIT locus, Mga+ | This study |
| KSM429.150Lg | Pmga (ΔMBS I & II)-gusA in VIT locus, Mga− | This study |
| KSM438 | Pmga (Δcre) in native locus | This study |
| KSM440 | Pmga (full length) in native locus | This study |
| KSM442 | Pmga (ΔP1) in native locus | This study |
| KSM435 | Pmga (Δcre)-gusA in VIT locus, Mga+ | This study |
| KSM435.150Lg | Pmga (Δcre)-gusA in VIT locus, Mga− | This study |
| KSM444 | Pmga (Δcre)-gusA in VIT locus, Mga+ | This study |
| KSM444.150Lg | Pmga (P1 only)-gusA in VIT locus, Mga− | This study |
| KSM445 | Pmga (P1 Δcre)-gusA in VIT locus, Mga+ | This study |
| KSM445.150Lg | Pmga (P1 Δcre)-gusA in VIT locus, Mga− | This study |
| RTG229 | VIT strain, Mga+ | 12 |
| RTG229.150Lg | VIT strain, Mga− | This study |
| VIT-GusA | Promoterless gusA in VIT locus, Mga+ | This study |
| VIT-GusA-586 | Promoterless gusA in VIT locus, Mga− | This study |
| Plasmids | ||
| pBluescript II KS− | ColE1 ori AmprlacZα | Stratagene |
| pBlue-cat194 | cat194 in pBluescript II KS− | This study |
| pJRS233 | Temperature-sensitive shuttle vector | 32 |
| pJRS586 | M6 mga suicide vector, pUC ori | 25 |
| pKSM148 | Pemm-gusA | 35 |
| pKSM150Lg | Fragment of M6 mga | This study |
| pKSM162 | Pspac-mga in pEU308 ΔlacIq | 22 |
| pKSM310 | Same as pPmga-gusA | 41 |
| pKSM427 | Pmga (ΔP1)-gusA | This study |
| pKSM428 | Pmga (ΔMBS I)-gusA | This study |
| pKSM429 | Pmga (ΔMBS I & II)-gusA | This study |
| pKSM435 | Pmga (Δcre)-gusA | This study |
| pKSM436 | Pmga upstream of cre in pJRS233 | This study |
| pKSM437 | Pmga downstream of cre cloned into pKSM436 | This study |
| pKSM438 | Pmga Δcre in pJRS233, ΩKm2 | This study |
| pKSM439 | Pmga (full) cloned into pKSM436 | This study |
| pKSM440 | Pmga (full) in pJRS233 | This study |
| pKSM441 | Pmga downstream of P1 cloned into pKSM436 | This study |
| pKSM442 | Pmga ΔP1 in pJRS233, ΩKm2 | This study |
| pKSM444 | Pmga (P1 only)-gusA | This study |
| pKSM445 | Pmga (P1 Δcre)-gusA | This study |
| pKSM540 | Promoterless gusA, pUC ori | This study |
| pKSM700 | Internal fragment of M6 ccpA in pJRS233 | This study |
| pKSM711 | M6 his-ccpA in pProEX-HTb | This study |
| pLZ12 | pSH71 origin, cat194 | 31 |
| pPmga-blue | Pmga in pBluescript II KS− | 41 |
| pPmga-gusA | Pmga (full)-gusA | 41 |
| pProEX-HTb | Expression vector N-terminal 6× His | Invitrogen |
| pUC4Km2 | pMB1 ori, ΩKm2 | 31 |
| pVIT164 | Plasmid vector for integration into Tn916 | 12 |
| pVIT-gusA | Promoterless gusA | This study |
DNA manipulations.
Plasmid DNA was isolated from E. coli by alkaline lysis using either the Wizard Miniprep system (Promega) or Maxi/Midi prep purification systems (Qiagen). Chromosomal DNA from the GAS was isolated using the FastDNA kit and a FastPrep cell disruptor (Bio101, Inc.). DNA fragments were isolated from agarose gels using the QIAquick gel extraction kit (Qiagen). PCR for cloning and promoter probes was performed using Platinum Pfx high-fidelity DNA polymerase (Invitrogen), and reactions were purified by using the QIAquick PCR purification system (Qiagen). PCR for diagnostic assays was performed using Taq DNA polymerase (New England Biolabs). DNA sequencing was performed either using the Excel II cycle sequencing kit (Epicenter, Inc.), through the McDermott Center at the University of Texas Southwestern Medical Center, or through Genewiz, Inc.
Construction of pKSM711 and purification of GAS His-CcpA.
An amino-terminal fusion of 6× His to CcpA from M6 GAS was constructed as follows. A 1,019-bp region containing the entire ccpA gene was PCR amplified from serotype M6 GA19681 (Table 1) genomic DNA (gDNA) using the primer pair M6ccpA_NcoI-L and M6ccpA_XhoI-R (Table 2). The resulting product was digested with NcoI/XhoI and ligated into NcoI/XhoI-digested pProEX-HTb to produce pKSM711 (Table 1). Following verification by PCR and DNA sequence analysis, pKSM711 was transformed into BL21[DE3] Gold (Stratagene) for protein expression.
TABLE 2.
PCR primers used in this study
| Target and primer | Sequencea (5′-3′) | Reference or source |
|---|---|---|
| ccpA | ||
| ccpAL | TTCAATGGCAACCGTTAG | This study |
| ccpAR | TCCTGACACAAAAGCGAT | This study |
| ccpAR1 | CCCTAAGGCTGATTTTAGTATT | This study |
| M6ccpA_NcoI-L | catccatggCTAATACAGATGATACCAT | This study |
| M6ccpA_XhoI-R | gcgctcgagTTACTTAGTTGTCCC | This study |
| gusA | ||
| gusA-PE | GTTGGGGTTTCTACAGGACG | 1 |
| Steph-gusA-PE | TTGTTTAAACAAATAGACGA | This study |
| gyrA RT | ||
| gyrA M1 RT L | CGACTTGTCTGAACGCCAAAG | This study |
| gyrA M1 RT L | ATCACGTTCCAAACCAGTCAAAC | This study |
| M13 | ||
| 1201 M13 Rev | AACAGCTATGACCATGATTACG | Clontech |
| 1211 M13 For | GTTGTAAAACGACAACCAGT | Clontech |
| mga | ||
| OYL-13 | GACGGCAGAGTATCCCTTGT | 23 |
| OYR-4 | GTACCATCAACATTGCG | 25 |
| mga P1 RT | ||
| mgaP1_L2 | TAAATAATGAACAAAAAGGAATAATTGCG | This study |
| mgaP1_R2 | AATACCTTTCAAATTCTTTCATTAAAATCC | This study |
| mga RT | ||
| mga M6 RT L | AGATGAATCCAGTTGGTCACTTTTC | This study |
| mga M6 RT R | AAATCGGTTATGCGTTTGATAGC | This study |
| PccpA | ||
| PccpA-L1 | GCCAATTCAGCTCCCTTT | This study |
| PccpA-R1 | CTTCACGGGCAACATCAT | This study |
| Pmga | ||
| Δcre-L | TTTTTGTGAACTGGTTAA | This study |
| Δcre-R_Bam | cgggatccAATATTGGAGTAAATTGAC | This study |
| ΔPmga_Bam | cgggatccATTTCTAATTGGTCATTAA | This study |
| MgaL3_Bam | caggatccGGATTTTAATGAAAGAATTT | 25 |
| OYL-1 | TATGCCATTTATGCTCT | 25 |
| OYL-14 | GTCACTAACTTAATTAGGT | 25 |
| OYR-1 | AGAGCATAAATGGCATA | 25 |
| OYR-14 | AATCTGCGAGATTAGAGTAAT | This study |
| OYR-17 | ATATGGTAGAAGACACTATC | This study |
| OYR-22 | TAGACCCCAAATTCCCGT | 25 |
| OYR-25 | GGTTGTACCATAACAGTC | 22 |
| OYL-1 | TATGCCATTTATGCTCT | 25 |
| rpsL | ||
| GAS-rpsL5 | GGTTGATATAGCACTTGGTGAC | This study |
| GAS-rpsL6 | GTGCGCCACGAACGATATG | 24 |
| Spn-rpsL1 | GAATGTAGATGCCTACAATTAACCA | 24 |
| VIT | ||
| VIT-R1 | TCAACGTCGCCATGAAGTAC | This study |
Underlined nucleotides are restriction sites, and lowercase nucleotides are anchor sequences introduced into the primer.
GAS His-CcpA was purified via Ni-nitrilotriacetic acid resin (Qiagen) based on the manufacturer's protocol. Briefly, expression of protein was induced at an OD at 600 nm (OD600) of 0.6 nm for 4 h with 1 mM IPTG, and cell pellets were stored at −80°C. The frozen pellet was lysed in the presence of 1 mg/ml lysozyme and 1× Complete Protease inhibitors (Roche) using a Branson sonicator (5 cycles of 30-s pulses at a 50% duty cycle, output of 7.5). His-CcpA was purified from the resulting lysate over Ni-nitrilotriacetic acid resin under native conditions, and the protein concentration was determined for each fraction using protein assay reagent (Bio-Rad) with an Ultrospec 2100 spectrophotometer (GE Healthcare). Chosen fractions were dialyzed with two buffer changes in 4 liters of TKED buffer (100 mM Tris-HCl, 150 mM KCl, 1 mM EDTA, 0.1 mM dithiothreitol), and glycerol was added to 10% prior to storage of protein aliquots at −20°C.
Electrophoretic mobility shift assay (EMSA).
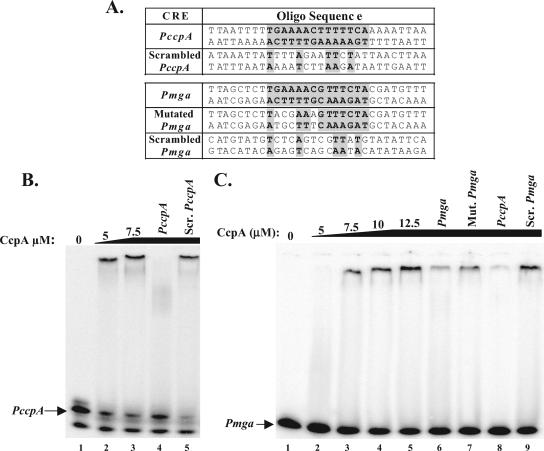
Double-stranded DNA probes were generated by annealing 30-bp sense and antisense oligonucleotide pairs representing PmgaCRE, PccpACRE, a mutated PmgaCRE, and randomly rearranged PmgaCRE or PccpACRE, termed scrambled (see Fig. 2A). Briefly, gel-purified oligonucleotide pairs were annealed by being heated to 85°C for 5 min in 12.5 μg of each pair in 10 mM Tris-HCl, pH 8.0, 5 mM MgCl2 and slowly being cooled to room temperature for 30 min. Annealed oligonucleotides were end labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs), and the resulting radiolabeled probes were separated on a 5% polyacrylamide gel and extracted by crush-and-soak elution.
FIG. 2.
EMSA of Pmga cre using His-CcpA. (A) Sequence of annealed oligonucleotide (Oligo) probes used in EMSAs. Putative cre are indicated by shaded boxes. Base pairs matching Pmga cre are shaded and in boldface. EMSA was performed on radiolabeled PccpA-annealed (B) or (C) Pmga-annealed oligonucleotide probes. A constant amount (1 to 2 ng) of [γ-32P]ATP-labeled probe was incubated with increasing amounts (5 to 12.5 μM) of purified GAS His-CcpA for 30 min at 30°C prior to separation on a 5% polyacrylamide, 10% glycerol gel. The specificity of His-CcpA binding to Pmga was assayed by addition of 700 ng of unlabeled competitor annealed oligonucleotide probes corresponding to Pmga (lane 6), mutated (Mut.) Pmga (lane 7), PccpA (lanes 4 and 8), and scrambled (Scr.) PccpA (lane 5) or Pmga (lane 9) using the oligonucleotide pairs listed in Table 2.
EMSA was performed as described previously (20). Briefly, a constant amount of labeled double-stranded oligonucleotide probe (ca. 1 to 5 ng) and increasing amounts of purified GAS His-CcpA (5 to 20 μM) were used in each reaction. Competition assays were performed by the addition of 700 ng unlabeled double-stranded oligonucleotide probes to binding reaction mixtures. After incubating for 30 min at 30°C, reactions were mixed in 1% vol/vol Ficoll, 0.02% (wt/vol) bromophenol blue and were separated on a 5% polyacrylamide, 10% (vol/vol) glycerol gel at room temperature. Gels were dried under vacuum at 80°C for 1 h and were exposed overnight to a phosphorimaging screen. Screens were scanned using a Storm860 (Amersham Biosciences), and resulting data were analyzed with the ImageQuant analysis software (version 5.0).
Construction of the Mga− M6 GAS strain RTG229.150Lg.
A chloramphenicol-resistant GAS suicide plasmid for inactivation of mga was constructed by ligation of a 1.5-kb cat194 fragment of HincII-digested pLZ12 (31) into XmnI/BsaI-digested pBluescript II KS− to form pBlue-cat194 (Table 1). A 454-bp internal fragment of mga was PCR amplified from M6 JRS4 gDNA using OYR-4/OYL-13 (Table 2) and was ligated into EcoRV-digested pBlue-cat194 to form pKSM150Lg (Table 1). This plasmid was introduced into RTG229 by electroporation, and chloramphenicol-resistant insertion mutants (RTG229.150Lg) were selected and verified by PCR.
Chromosomal GusA-based transcriptional reporters at the VIT locus.
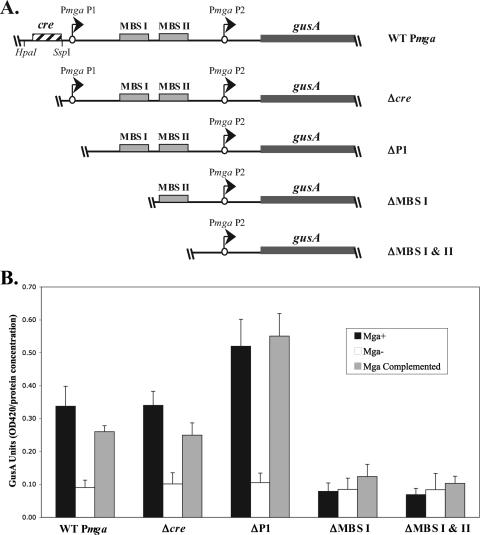
A 706-bp fragment of M6 Pmga containing sequence downstream of the putative cre (see Fig. 3) was PCR amplified from pPmga-blue using the 1201/1211 primer pair (Table 2), digested with SspI/EcoRI, and ligated into the HpaI/EcoRI-digested pPmga-gusA to form pKSM435 (Table 1). A 520-bp fragment of M6 Pmga (see Fig. 3) containing sequence downstream of P1 was amplified from pPmga-blue (41) using the 1201/OYR-25 primer pair (Table 2), digested with EcoRI, and ligated into the HpaI/EcoRI-digested pPmga-gusA (41) to form pKSM427 (Table 1). A 371-bp fragment of M6 Pmga containing sequences downstream of MBS I (see Fig. 3) was PCR amplified from pPmga-blue using the 1201/OYR-1 primer pair (Table 2), digested with EcoRI, and ligated into the HpaI/EcoRI-digested pPmga-gusA to form pKSM428 (Table 1). A 265-bp fragment of M6 Pmga containing sequence downstream of MBS II (see Fig. 3) was PCR amplified from pPmga-blue using the 1201/MgaL3_Bam primer pair (Table 2), digested with EcoRI, and ligated into the HpaI/EcoRI-digested pPmga-gusA to form pKSM429 (Table 1). A PstI fragment of pPmga-gusA containing the gusA gene was ligated into the PstI-digested pVIT164 to form pKSM540 (Table 1).
FIG. 3.
VIT GusA transcriptional reporter assay for deletion analysis of Pmga. (A) Schematic representation of gusA transcriptional reporters in the chromosomal VIT locus of the M6 GAS corresponding to wild-type Pmga (KSM310), Δcre (KSM435), ΔP1 (KSM427), ΔMBS I (KSM428), and ΔMBS I & II (KSM429). The starts of transcription for mga (circles with arrows), the gusA gene (thick line), MBSs (solid boxes), putative cre (striped box), and relevant restriction sites are shown. (B) GusA assays for the Pmga-gusA constructs depicted in panel A inserted into the VIT locus of the wild-type JRS4-derived M6 GAS strain RTG229 (black bars), isogenic mga-inactivated strain KSM150Lg (open bars), and mga-inactivated KSM150Lg complemented with the Pspac-mga plasmid pKSM162 (shaded bars). Data are reported in GusA units (OD420/concentration of total protein [in micrograms per microliter]) and represent an average of the results from at least three independent experiments. The error bars express the standard deviations for each strain measured.
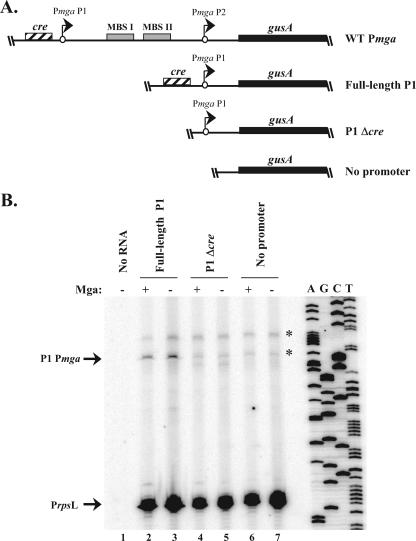
A 381-bp fragment of Pmga P1 (see Fig. 5) was amplified from the M6 GAS strain JRS4 using the OYR-14/OYL-14 primer pair (Table 2) and ligated into the HpaI-digested pKSM540 to form pKSM444 (Table 1). A 135-bp fragment of Pmga P1 downstream of the putative cre (see Fig. 5) was PCR amplified from the M6 GAS strain JRS4 using the Δcre-R_Bam/OYL-14 primer pair (Table 2) and was ligated into the HpaI-digested pKSM540 to form pKSM445 (Table 1). A promoterless GusA transcriptional reporter plasmid (see Fig. 5) was constructed by ligation of the 1.9-kb BamHI gusA fragment from pKSM148 (35) into BamHI-digested pVIT164 (12) to form pVIT-gusA (Table 1).
FIG. 5.
Deletion analysis of the Pmga P1 promoter region. (A) Schematic representation of gusA transcriptional reporters in the chromosomal VIT locus of the M6 GAS strains KSM310 (WT Pmga), KSM444 (full-length P1), KSM445 (P1 Δcre), and VIT-GusA (no promoter). The starts of transcription for mga (circles with arrows), the gusA gene (thick line), MBSs (solid boxes), and the putative cre (striped box) are shown. (B) Semiquantitative primer extension analysis was performed on total RNA from the full-length P1, P1 Δcre, and no-promoter reporter strains using the radiolabeled antisense primers Steph_gusA-PE for gusA and GAS-rpsL5 for rpsL (Table 2) in the same reaction. The starts of transcription for gusA (P1 Pmga) and rpsL (PrpsL) are shown at the left, and a Pmga P1 sequence ladder is provided at the right. Nonspecific background bands are indicated with an asterisk.
Plasmids pKSM310, pKSM427, pKSM428, pKSM429, pKSM435, pKSM444, and pKSM445 were linearized with PvuII and were introduced into RTG229 (Mga+) and RTG229.150Lg (Mga−) by electroporation. Exchange of DNA at the VIT locus was verified by the presence of kanamycin resistance and erythromycin sensitivity. Additionally, PCR amplification with the primer pair VIT-R1/gusA-PE was used to verify the presence of the constructs in the chromosome. Plasmid pVIT-gusA was linearized with PvuII and introduced into RTG229 by electroporation to form the control strain VIT-gusA (Table 1). Mga was inactivated in this strain by transformation with the mga suicide plasmid pJRS586 (25) to form strain VIT-gusA-586 (Table 1). Complementation of the Mga− GusA reporter strains with constitutively expressed mga (Pspac-mga) was performed by introduction of pKSM162 (22).
GusA assays were performed as previously described (10). Briefly, cells were grown to late logarithmic phase, lysed using a FastPrep cell disruptor (Bio101, Inc.), and assayed for GusA activity. Results are reported in GusA units, which are equivalent to the A420 of the lysate divided by the concentration of total lysate protein (in micrograms/microliter).
Construction of Pmga deletions at the native locus.
A 1,196-bp fragment upstream of the putative Pmga cre was amplified from M6 JRS4 gDNA using the primer pair OYR-17/Δcre-L (Table 2) and was cloned blunt into EcoRV-digested pJRS233 (32) to form pKSM436 (Table 1). A fragment of Pmga downstream of the cre was PCR amplified from JRS4 using the primer pair Δcre-R_Bam/ΔPmga_Bam (Table 2), digested with BamHI, and cloned into BamHI-digested pKSM436 to form pKSM437 (Table 1). Similarly, the primer pair OYR-22/ΔPmga_Bam (Table 2) was used to amplify wild-type Pmga from JRS4, digested with BamHI, and cloned into SmaI/BamHI-digested pKSM436 to form pKSM439 (Table 1). Finally, Pmga ΔP1 was amplified from JRS4 using the primer pair OYR-25/ΔPmga_Bam (Table 2), digested with BamHI, and cloned into SmaI/BamHI-digested pKSM436 to form pKSM441 (Table 1). The 2.1-kb SmaI-digested ΩKm2 from pUC4ΩKm2 (31) was cloned into either SmaI-digested pKSM437 or PstI-digested and -blunted plasmids pKSM439 and pKSM441 to form pKSM438 (Δcre), pKSM440 (Full), and pKSM442 (ΔP1), respectively (Table 1). Plasmids were introduced into JRS4 by electroporation at 30°C, and plasmid integrants were selected by passage of cells at 37°C with screening for kanamycin resistance and erythromycin sensitivity as described previously (35). Strains constructed from plasmids pKSM438, pKSM440, and pKSM442 were named KSM438 (Δcre), KSM440 (Full), and KSM442 (ΔP1), respectively (Table 1; also see Fig. 4).
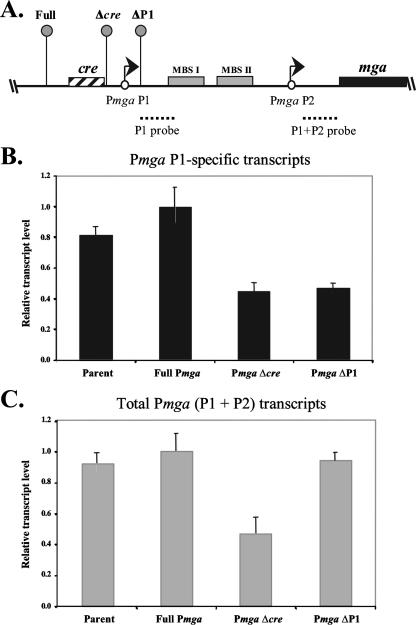
FIG. 4.
Real-time RT-PCR analysis of native Pmga P1 and mga transcripts. (A) Real-time RT-PCR was performed on total RNA isolated from JRS4-derived strains KSM440 (Full), KSM438 (Δcre), and KSM442 (ΔP1) as shown in the schematic. The ΩKm2 cassette (lollipops) and all relevant Pmga elements are shown. The location of the P1 (mga P1 RT) and the P1 plus P2 (mga RT) probes are indicated (dotted lines). (B) Levels of mga transcribed from the Pmga P1 start site only (black bars) were assessed using the P1 probe. (C) Total Pmga transcript levels (gray bars) were assessed using the P1 plus P2 probe. Transcript levels are shown as the fold transcript level above that of the full-length promoter (KSM440) that had been normalized to levels of the gyrA control. Reactions were performed in triplicate for three independent experiments. Error bars represent the minimum and maximum relative transcript levels based on the standard errors for the samples.
Primer extension analysis.
Total RNA was extracted from samples in late-logarithmic phase as described previously (22) using the FastRNA kit and a FastPrep cell disruptor (Bio101, Inc.). Primer extensions were performed on 10 to 20 μg of total RNA as described previously (25) using the primers Steph-gusA-PE and GAS-rpsL5 (Table 2). Primer extension products were run on a 6% denaturing polyacrylamide gel (Amresco). Gels were dried under vacuum at 80°C for 1 h and were exposed overnight to a phosphorimaging screen. Sequence was generated using a labeled Steph-gusA-PE primer on the pKSM444 plasmid.
Real-time RT-PCR.
gDNA was removed from total RNA by use of the MessageClean kit (GenHunter Corp.). Real-time reverse transcription-PCR (RT-PCR) was performed on 25 ng of RNA mixed with 5 pmol of each primer, 6.25 U of MultiScribe RT (Applied Biosystems), and 1× SYBR green PCR master mix (Applied Biosystems). Reaction mixtures were transferred in triplicate into a 96-well optical reaction plate (Applied Biosystems), and the plate was covered with optical adhesive covers (Applied Biosystems). An Applied Biosystems 7500 real-time PCR system was used to detect transcript levels in the absolute quantification mode with reaction conditions of 48°C for 30 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s followed by 60°C for 1 min. Analysis of data was performed using Sequence Detection Software, version 1.3 (Applied Biosystems). The mga RT and mga P1 RT primer pairs (Table 2) were used to detect levels of total mga and mga P1 transcripts, respectively, in relation to gyrA transcript levels (detected with the gyrA RT primer pair; Table 2) in RNA isolated from JRS4 (wild type), KSM440 (Full), KSM438 (Δcre), and KSM442 (ΔP1). A standard curve using total RNA was used to quantify the levels of each transcript.
Inactivation of ccpA in the GAS.
To produce the ccpA mutant strain KSM310.700, a 511-bp internal region of ccpA was amplified from JRS4 gDNA using the primer pair ccpAL/ccpAR (Table 2). The resulting fragment was ligated into EcoRV-digested pJRS233 to form pKSM700 (Table 1) and was verified by PCR using the primer pair ccpAL/ccpAR (Table 2). pKSM700 was transformed into KSM310 at 30°C, and erythromycin-resistant integrants were isolated at 37°C by following the protocol described previously (35). Mutants were verified by PCR using the primer pairs 1201/ccpAR1 and 1211/PccpA-L1 (Table 2). GusA assays were performed as described above.
GAS protein extracts and Western blot analyses.
Whole-cell GAS protein extractions, separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blot analysis, were performed as previously described (22). Blots were incubated with a 1:1,000 dilution of α-Mga-pep2 antiserum (22), incubated with a 1:25,000 dilution of α-rat (Santa Cruz Biotechnologies) horseradish peroxidase-conjugated secondary antibody, and visualized using the Western Lightning chemiluminescence system (Perkin Elmer).
RESULTS
Identification of putative cre in the GAS genome.
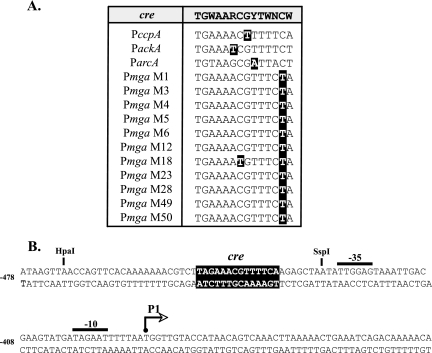
As an initial step to find GAS genes under CcpA-mediated CCR, the genome of the serotype M1 strain SF370 was scanned for putative cre based on similarity to the published B. subtilis consensus sequence TGWAARCGYTWNCW (39). Allowing for a single mismatch from the consensus, 60 cre were identified on the direct strand, and 58 were identified on the complementary strand, resulting in 98 unique sites scattered throughout the genome (see Table S1 in the supplemental material). Many of the putative cre in the M1 genome were located upstream or within annotated open reading frames similar to genes regulated by CcpA in B. subtilis (27, 28), including ndk, lctE, and numerous sugar transport operons (Fig. 1; also see Table S1 in the supplemental material). The promoter of ccpA, which can be autoregulated (9), also was found to have a cre similar to that of the B. subtilis consensus, with a single mismatch (Fig. 1A). Likewise, putative cre were identified in the promoters of ackA, encoding acetate kinase, and arcA, encoding arginine deiminase (Fig. 1A), which are positively and negatively regulated by CcpA, respectively, in other gram-positive bacteria (8, 40). Thus, the location of cre identified in the GAS genome corresponds to known CcpA-regulated genes.
FIG. 1.
Alignment of putative cre identified in GAS genomes. (A) Possible cre from the Pmga region in 11 different serotypes of the GAS as well as those identified in the promoters of known CcpA-regulated genes PccpA, PackA, and ParcA were aligned to the consensus B. subtilis cre (shaded at the top) used to identify them. Nucleotides that did not match the consensus are in black boxes. (B) Location of the putative Pmga cre (black box) relative to the P1 start of transcription in the M1 SF370 GAS genome. The P1 start of transcription (arrow), −10 and −35 hexamers (overlines), and relevant restriction sites are shown. The numbers at left reflect the position relative to the start codon for mga.
One such cre was identified within the mga promoter (Pmga) upstream of the distal P1 start of transcription (Fig. 1B), and the site was highly conserved among all serotypes of the GAS for which genome sequence is available (Fig. 1A). Based on CcpA studies with Lactococcus lactis, the position of the Pmga P1 cre centered at −54.5 bp from the start of transcription strongly suggests that it plays a role in activating Pmga activity (44).
The catabolite control protein, CcpA, specifically binds to PccpA and Pmga in vitro.
To determine if CcpA interacts with the cre identified in our bioinformatic screen of the GAS M1 genome, EMSAs were performed on PccpA and Pmga. Double-stranded oligonucleotide probes (30 bp) were generated that contained either the PccpA (positive control) or the Pmga cre (14 bp each) centered within the sequence (Fig. 2A). To address the specificity of CcpA binding, probes consisting of a random rearrangement of the respective nucleotides (scrambled PccpA and Pmga) or a probe containing four specific mutations in the Pmga cre (mutated Pmga) were generated (Fig. 2A). Since CcpA is capable of binding DNA in the absence of phosphorylated HPr-Ser in vitro (2), assays were performed using purified GAS His-CcpA alone (see Materials and Methods).
Studies with other gram-positive bacteria predict that CcpA will bind to a cre located within its own promoter (19, 44); therefore, we first tested the ability of purified His-CcpA to bind to the identified GAS PccpA cre probe (Fig. 2A). Increasing amounts of His-CcpA (5.0 to 7.5 μM) resulted in a mobility shift of labeled PccpA, indicating DNA binding (Fig. 2B, lanes 1 to 3). The addition of 700 ng of cold PccpA to the reaction was able to compete for His-CcpA interaction, whereas the addition of the same amount of a cold scrambled PccpA probe had no effect (Fig. 2B, lanes 4 and 5). Thus, GAS His-CcpA is able to bind specifically to the PccpA cre in vitro.
The Pmga cre probe also demonstrated slower migration upon addition of increasing amounts of purified His-CcpA (7.5 to 12.5 μM), indicating protein-DNA interaction (Fig. 2C). The 700 ng of cold Pmga or PccpA cre probe that was added to the reaction was able to compete for binding of His-CcpA to the labeled Pmga probe to various degrees (Fig. 2C, lanes 6 and 8). In contrast, a scrambled Pmga cre probe (9/14 mismatches) was not able to compete for His-CcpA (Fig. 2B, lane 9), suggesting that the interaction with Pmga cre is specific. In support of this conclusion, mutation of only 4/14 nucleotides in the predicted Pmga cre exhibited an intermediate level of competition (Fig. 2B, lane 7). Thus, GAS His-CcpA is capable of binding directly to the predicted cre sequences located upstream of PccpA and Pmga P1 promoters in vitro.
P1 and cre are not essential for Pmga activity in the presence of existing Mga.
To address whether binding of CcpA to the Pmga cre is important for transcriptional regulation of mga, β-glucuronidase (GusA) transcriptional reporter assays were performed. Various single-copy Pmga-gusA alleles were constructed in the JRS4-derived M6 GAS strain RTG229 (12) genome at an ectopic site (VIT) separate from the native mga locus. Promoter fragments corresponding to the full-length (wild type; KSM310) Pmga, a deletion of the cre only (Δcre; KSM435), a deletion of the entire P1 with cre (ΔP1; KSM427), a deletion of P1/cre and Mga-binding site 1 (ΔMBS I; KSM428), and a deletion of P1/cre and both Mga-binding sites (ΔMBS I & II; KSM429) were introduced into the VIT locus of both a wild-type and Mga− M6 GAS (Fig. 3A and Table 1). The Mga− strains then were complemented by introduction of a multicopy Pspac-mga plasmid for constitutive expression of mga (22). Strains were grown to late logarithmic phase, and the levels of GusA activity were quantified (see Materials and Methods).
The wild-type Pmga, Δcre, and ΔP1 strains all showed Mga-regulated GusA activity, which could be restored upon complementation (Fig. 3B). As predicted from previous studies (25), deletion of either MBS I (ΔMBS I) or both MBSs (ΔMBS I & II), in addition to the upstream P1 promoter and cre, eliminated all Mga-dependent regulation of Pmga (Fig. 3B). Thus, deletion of the cre alone or the entire P1/cre had little effect on wild-type activity (Fig. 3B), suggesting that the Mga produced from its native locus was sufficient to activate Pmga at the VIT locus from the P2 promoter alone. Of note, GusA levels appeared to increase slightly upon deletion of P1 (Fig. 3B), indicating that the Pmga P1 region also may contain sequences that repress mga expression.
The Pmga cre is necessary for full transcriptional activity of Pmga at its native locus.
To determine whether the cre is important for Pmga activity in the absence of exogenously produced Mga, Pmga mutations were constructed using the ΩKm2 cassette at the native mga locus in the serotype M6 strain JRS4 chromosome. The ΩKm2 cassette has T4 terminators that flank the antibiotic resistance marker to prevent upstream transcription through the element (31). As a wild-type control, the ΩKm2 cassette was inserted by allelic exchange upstream of the wild-type Pmga promoter (Fig. 4A). To generate Pmga alleles lacking either cre or the entire P1 promoter, the ΩKm2 cassette was inserted either immediately downstream of cre (Δcre) or downstream of the P1 transcriptional start site (ΔP1), respectively (Fig. 4A) (see Materials and Methods).
Given that transcript levels from Pmga P1 often are quite low (25), quantitative real-time RT-PCR was utilized to assess the effects of promoter deletions on the expression of mga. Total RNA was isolated from the wild-type JRS4, full Pmga, Δcre, and ΔP1 strains grown to late logarithmic phase. Transcript levels of both Pmga P1 alone (Fig. 4B) and total Pmga (Fig. 4C) were detected using the mga P1 RT and mga RT primer pairs (Table 2), respectively, and results were normalized to transcript levels of the housekeeping gene gyrA.
As expected, relative levels of mga transcribed from Pmga P1 alone and total Pmga were similar between the parent strain, JRS4, and the full Pmga control strain (Fig. 4B and C), indicating that the ΩKm2 cassette inserted upstream of Pmga had little effect on the transcription of mga (Fig. 4B and C). Importantly, deletion of the cre in Pmga caused an approximately twofold reduction in the levels of both Pmga P1 and total Pmga, suggesting that the cre is important in the activation of mga transcription. Unexpectedly, the deletion of the P1 promoter resulted in increased transcript levels from P2 (mga P1-specific transcript levels remained low) (Fig. 4B and C). As seen with the GusA reporter assays described above (Fig. 3B), this result supports previous reports of a repressor region in Pmga near P1 (25).
cre is necessary for activation of Pmga P1.
In order to further assess the effect of the cre on the activity of Pmga P1 alone, a gusA transcriptional fusion was made to Pmga P1 with and without the cre in the VIT locus of wild-type and Mga− M6 GAS to form full-length (cre+) and P1Δcre (cre mutant) strains, respectively (Fig. 5A). Since the level of GusA activity from the reporter strains was too low to detect, transcript levels were assessed directly using semiquantitative primer extension. Total RNA was extracted from late-logarithmic-phase cells representing the full-length Pmga P1, the P1 Δcre, and the no-promoter control gusA reporter strains either in the presence or absence of a functional mga. Primer extensions were performed simultaneously for both gusA and the constitutive rpsL for each of the strains.
Promoter-specific products were not observed in the absence of RNA (Fig. 5B, lane 1) or in the no-promoter control strains (Fig. 5B, lanes 6 and 7), with the exception of two light background bands (Fig. 5B). However, a product of the predicted size for Pmga P1 was detected in the cre+ strains (Fig. 5B, lanes 2 and 3), while it was reduced approximately 4.2-fold in the cre mutant strains, as determined by densitometry (Fig. 5B, lanes 4 and 5). This correlates with the Δcre results from the real-time RT-PCR analysis of Pmga P1 at its native locus (Fig. 4B). As expected from the absence of Mga-binding sites in P1, Mga had no detectable effect on transcription from Pmga P1 (Fig. 5B, lanes 2 to 5) when its transcript levels were normalized to rpsL. Thus, the cre is necessary for transcriptional activation from the Pmga P1 start site.
Inactivation of ccpA affects mga expression.
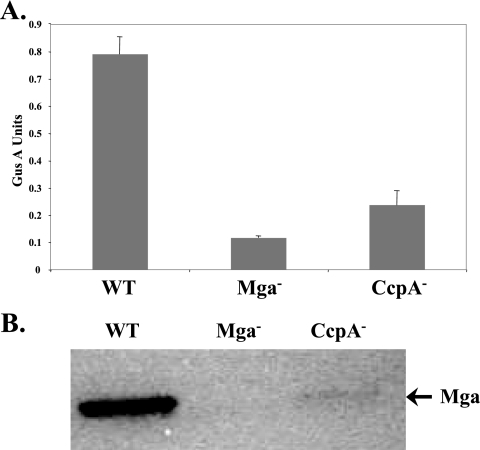
The role of CcpA-mediated activation of Pmga P1 on mga expression in vivo was assessed using an insertion-inactivation mutant of ccpA in the M6 Pmga-gusA VIT reporter strain KSM310 (KSM310.700; Table 2). The ccpA-defective strain did not exhibit any significant growth defects compared to growth of the wild-type KSM310 when grown in rich THY medium (data not shown). Inactivation of ccpA resulted in a greater than threefold reduction in Pmga-specific GusA activity compared to the activity of the parental KSM310 samples grown to late-logarithmic phase in THY medium (Fig. 6A). The resulting GusA activity was slightly higher than background levels observed in the Mga− control strain KSM310.150 (Fig. 6A). In addition, Western analysis of whole-cell extracts found that steady-state levels of Mga also were reduced in the CcpA− mutant at the same point in growth (Fig. 6B). These data indicate that CcpA is necessary for wild-type production of Mga during logarithmic phase, a point in growth when the Mga virulence regulon shows maximal expression (23).
FIG. 6.
Effect of a CcpA− mutant on mga expression. (A) GusA reporter assay for the M6 GAS Pmga-gusA reporter strain KSM310 (WT), the mga-inactivated derivative KSM310.150Lg (Mga−), and the ccpA-inactivated derivative KSM310.700 (CcpA−). Data are reported in GusA units (OD420/concentration total protein [in micrograms per microliter]) and represent an average of the results from at least three independent experiments. The error bars express the standard deviations for each strain measured. (B) Mga protein production was assessed using Western analysis on whole-cell protein extracts derived from the same samples used for panel A and probed with an anti-M6 Mga antibody.
DISCUSSION
CcpA and cre in the GAS.
CcpA-mediated CCR of metabolic gene expression through direct binding to catabolite control elements (cre) is a highly conserved process in low-G+C gram-positive bacteria (4, 38, 43). Much of our knowledge on this topic comes from extensive studies on B. subtilis that have resulted in the determination of a consensus cre-binding site as well as the residues of CcpA essential for interaction with cre (17, 39). Using the B. subtilis consensus cre (TGWAARCGYTWNCW) to scan the genome of the serotype M1 SF370 with one mismatch, 98 potential cre were identified (Fig. 1; also see Table S1 in the supplemental material). This number is in accordance with the 126 putative cre found in a recent survey of the larger B. subtilis genome using a slightly more stringent version of the same consensus sequence (27) and with the 82 cre found in the L. lactis genome based on an L. lactis consensus (44). The fact that the majority of the potential GAS cre were found near genes regulated by CCR in other gram-positive organisms (data not shown) provides confidence that the overall approach identifies such genes in a heterologous genome. Of particular interest, several cre were found associated with genes encoding established virulence factors (e.g., SpeB and Ska) as well as the virulence regulator Mga (Fig. 1A; also see Table S1 in the supplemental material).
In the case of two identified GAS cre located in Pmga and PccpA, specific binding by a purified GAS His-CcpA was demonstrated in vitro (Fig. 2), demonstrating that these are functional CcpA-binding sites. An alignment of a subset of GAS cre found that the single-nucleotide mismatches to the B. subtilis consensus were located in variable positions in the site (Fig. 1A), suggesting that a GAS-derived consensus cre might be somewhat divergent. Furthermore, this would strongly indicate that these nucleotides, although important for B. subtilis, likely fall in positions that are not essential for CcpA-cre interactions in GAS. Regardless, a more comprehensive analysis utilizing CcpA-binding studies and growth in defined sugar sources will be required to determine whether these cre are important for CCR in the GAS.
What is the role of Pmga P1 in transcription of mga?
The Pmga region is quite complex and contains two starts of transcription, a strong gene-proximal P2 start site that is autoactivated by Mga and a weaker distal P1 start site that has been thought to be constitutive. Our studies show that CcpA was able to bind specifically to the putative cre located upstream of the P1 −35 region (Fig. 1B and 2) in a position that suggests an activating function based on recent studies on CcpA in L. lactis (44). Furthermore, deletion of the cre at the native mga locus or in the context of Pmga P1 alone leads to decreased mga transcription (Fig. 4 and 5B). Inactivation of ccpA in the serotype M6 JRS4 also shows a reduction in both Pmga activity and Mga levels in the cell (Fig. 6). Taken together, these results indicate that CcpA can activate the Pmga P1 start site through the upstream cre and that this interaction is important for normal expression of mga in the GAS during exponential growth. Previously, Pmga P1 was thought to provide low-level constitutive expression of mga early in growth that, when translated, would amplify its expression from Pmga P2 (30). Our results suggest a modification of this model, whereby CcpA may activate Pmga P1 in response to glucose levels in the cell early in growth and provide the initial trigger that leads to high production of Mga and autoactivation at Pmga P2. Experiments are currently under way to test whether Pmga P1 expression can be influenced by glucose and CCR via its cre.
Earlier Pmga deletion studies indicated that Pmga P1 was essential for the expression of mga (30). However, deletion of the entire Pmga P1 and cre did not prevent Mga-regulated expression from Pmga P2 as long as Mga was produced from its native locus (Fig. 3B). Although these data appear to contradict the published findings, the Pmga P1 deletion constructs used in this study included approximately 20 bp of extra sequence not found in the previous study and used a different reporter gene (30). Therefore, it is possible that this extra sequence is necessary for the normal expression of mga.
Rather than being essential to the activity of mga, Pmga P1 may actually play a role in repression. An increase in Pmga P2 expression is observed upon deletion of the P1 promoter at both its native locus (Fig. 4C) and at an ectopic locus (Fig. 3), suggesting that sequences in this region actually function to repress transcription initiated from Pmga P2. A repressor has been suggested to function at the MBS (MBS I) in Pmga (25); however, no such regulator has been identified to date. One possible model is that Pmga P1 repression is important for fine-tuning mga expression later in growth. However, this repression would be neutralized early on in the presence of CcpA and Pmga P1 cre (Fig. 4B). In theory, Pmga P1 functions to activate mga expression levels in conjunction with CCR and suppresses its activity later in growth in the absence of a preferred energy source. Overall, the regulation of mga requires a complex interplay between P1, P2, Mga, and other unknown regulators.
CcpA and Mga virulence regulation.
In this study, a link between carbon catabolite repression and mga regulation in the GAS has been demonstrated. The significance of this regulation may be that Mga virulence gene expression is influenced by the availability of a preferred carbon source during infection. Presumably, this would enable the bacterium to sense its environment and express the genes it requires to survive during early time points in growth. The importance of CCR in the regulation of mga may be observed by the strict conservation of the cre in an activating position in the mga promoter in all sequenced serotypes of the GAS (Fig. 1A). However, this may be unique to S. pyogenes, since an examination of the promoters of mga orthologs found that this conservation is not consistent across other pathogenic streptococcal species (data not shown). In the GAS, Mga originally may have functioned to regulate genes important in carbon metabolism and later evolved to regulate virulence genes. This hypothesis is supported by a recent microarray analysis of Mga-regulated genes from different serotypes of GAS showing that inactivation of mga alters the ability of the GAS to grow in different sugars (36).
It has been known for some time that the expression of Mga is regulated by growth phase, with maximal expression occurring during exponential phase (23). The results presented here may provide a mechanism for this connection. As the GAS colonize a new tissue site with available glucose, the pathogen would enter into exponential-phase growth. CCR would be active at this time, and the expression of mga from Pmga P1 is activated. Once the mga transcripts are translated, Mga autoactivates itself from Pmga P2. Upon depletion of glucose, the bacteria enter stationary-phase growth, and CcpA would no longer activate expression of Pmga P1. However, since Mga still may be able to activate transcription from Pmga P2, an unidentified factor that suppresses Mga activity during stationary phase must come into play, either by modifying Mga to inactivate it or through competition for binding at Pmga. A complete understanding of the events that occur at Pmga may provide insights into how the GAS is able to coordinate Mga regulation in response to growth-phase-dependent cues.
Supplementary Material
Acknowledgments
We thank Steve Melville for providing reagents important for the initiation of these studies.
This work was supported by a grant from the National Institutes of Health (NIH/NIAID AI47928 to K.S.M.). A.C.A. was supported in part by an NIH/NIAID Molecular Microbiology training grant (5T32 AI07520) and an NIH/NIAID research supplement for underrepresented minorities (RSUM AI-47928-S).
Footnotes
Published ahead of print on 28 September 2007.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Almengor, A. C., and K. S. McIver. 2004. Transcriptional activation of sclA by Mga requires a distal binding site in Streptococcus pyogenes. J. Bacteriol. 186:7847-7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aung-Hilbrich, L. M., G. Seidel, A. Wagner, and W. Hillen. 2002. Quantification of the influence of HPrSer46P on CcpA-cre interaction. J. Mol. Biol. 319:77-85. [DOI] [PubMed] [Google Scholar]
- 3.Caparon, M. G., R. T. Geist, J. Perez-Casal, and J. R. Scott. 1992. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J. Bacteriol. 174:5693-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deutscher, J., C. Francke, and P. W. Postma. 2006. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol. Mol. Biol. Rev. 70:939-1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deutscher, J., E. Kuster, U. Bergstedt, V. Charrier, and W. Hillen. 1995. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in gram-positive bacteria. Mol. Microbiol. 15:1049-1053. [DOI] [PubMed] [Google Scholar]
- 6.Deutscher, J., J. Reizer, C. Fischer, A. Galinier, M. H. Saier, Jr., and M. Steinmetz. 1994. Loss of protein kinase-catalyzed phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, by mutation of the ptsH gene confers catabolite repression resistance to several catabolic genes of Bacillus subtilis. J. Bacteriol. 176:3336-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deutscher, J., and M. H. Saier, Jr. 1983. ATP-dependent protein kinase-catalyzed phosphorylation of a seryl residue in HPr, a phosphate carrier protein of the phosphotransferase system in Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 80:6790-6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dong, Y., Y. Y. Chen, and R. A. Burne. 2004. Control of expression of the arginine deiminase operon of Streptococcus gordonii by CcpA and Flp. J. Bacteriol. 186:2511-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egeter, O., and R. Bruckner. 1996. Catabolite repression mediated by the catabolite control protein CcpA in Staphylococcus xylosus. Mol. Microbiol. 21:739-749. [DOI] [PubMed] [Google Scholar]
- 10.Eichenbaum, Z., M. J. Federle, D. Marra, W. M. de Vos, O. P. Kuipers, M. Kleerebezem, and J. R. Scott. 1998. Use of the lactococcal nisA promoter to regulate gene expression in gram-positive bacteria: comparison of induction level and promoter strength. Appl. Environ. Microbiol. 64:2763-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujita, Y., Y. Miwa, A. Galinier, and J. Deutscher. 1995. Specific recognition of the Bacillus subtilis gnt cis-acting catabolite-responsive element by a protein complex formed between CcpA and seryl-phosphorylated HPr. Mol. Microbiol. 17:953-960. [DOI] [PubMed] [Google Scholar]
- 12.Geist, R. T., N. Okada, and M. G. Caparon. 1993. Analysis of Streptococcus pyogenes promoters by using novel Tn916-based shuttle vectors for the construction of transcriptional fusions to chloramphenicol acetyltransferase. J. Bacteriol. 175:7561-7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giammarinaro, P., and J. C. Paton. 2002. Role of RegM, a homologue of the catabolite repressor protein CcpA, in the virulence of Streptococcus pneumoniae. Infect. Immun. 70:5454-5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grundy, F. J., D. A. Waters, S. H. Allen, and T. M. Henkin. 1993. Regulation of the Bacillus subtilis acetate kinase gene by CcpA. J. Bacteriol. 175:7348-7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanahan, D., and M. Meselson. 1983. Plasmid screening at high colony density. Methods Enzymol. 100:333-342. [DOI] [PubMed] [Google Scholar]
- 16.Iyer, R., N. S. Baliga, and A. Camilli. 2005. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J. Bacteriol. 187:8340-8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim, J. H., and G. H. Chambliss. 1997. Contacts between Bacillus subtilis catabolite regulatory protein CcpA and amyO target site. Nucleic Acids Res. 25:3490-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kundig, W., S. Ghosh, and S. Roseman. 1964. Phosphate bound to histidine in a protein as an intermediate in a novel phospho-transferase system. Proc. Natl. Acad. Sci. USA 52:1067-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahr, K., W. Hillen, and F. Titgemeyer. 2000. Carbon catabolite repression in Lactobacillus pentosus: analysis of the ccpA region. Appl. Environ. Microbiol. 66:277-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McIver, K. S., A. S. Heath, B. D. Green, and J. R. Scott. 1995. Specific binding of the activator Mga to promoter sequences of the emm and scpA genes in the group A streptococcus. J. Bacteriol. 177:6619-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McIver, K. S., A. S. Heath, and J. R. Scott. 1995. Regulation of virulence by environmental signals in group A streptococci: influence of osmolarity, temperature, gas exchange, and iron limitation on emm transcription. Infect. Immun. 63:4540-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McIver, K. S., and R. L. Myles. 2002. Two DNA-binding domains of Mga are required for virulence gene activation in the group A streptococcus. Mol. Microbiol. 43:1591-1602. [DOI] [PubMed] [Google Scholar]
- 23.McIver, K. S., and J. R. Scott. 1997. Role of mga in growth phase regulation of virulence genes of the group A streptococcus. J. Bacteriol. 179:5178-5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McIver, K. S., S. Subbarao, E. M. Kellner, A. S. Heath, and J. R. Scott. 1996. Identification of isp, a locus encoding an immunogenic secreted protein conserved among group A streptococci. Infect. Immun. 64:2548-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McIver, K. S., A. S. Thurman, and J. R. Scott. 1999. Regulation of mga transcription in the group A streptococcus: specific binding of Mga within its own promoter and evidence for a negative regulator. J. Bacteriol. 181:5373-5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Milenbachs, A. A., D. P. Brown, M. Moors, and P. Youngman. 1997. Carbon-source regulation of virulence gene expression in Listeria monocytogenes. Mol. Microbiol. 23:1075-1085. [DOI] [PubMed] [Google Scholar]
- 27.Miwa, Y., A. Nakata, A. Ogiwara, M. Yamamoto, and Y. Fujita. 2000. Evaluation and characterization of catabolite-responsive elements (cre) of Bacillus subtilis. Nucleic Acids Res. 28:1206-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moreno, M. S., B. L. Schneider, R. R. Maile, W. Weyler, and M. H. Saier, Jr. 2001. Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol. Microbiol. 39:1366-1381. [DOI] [PubMed] [Google Scholar]
- 29.Morse, S. A., R. A. Mah, and W. J. Dobrogosz. 1969. Regulation of staphylococcal enterotoxin B. J. Bacteriol. 98:4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okada, N., R. T. Geist, and M. G. Caparon. 1993. Positive transcriptional control of mry regulates virulence in the group A streptococcus. Mol. Microbiol. 7:893-903. [DOI] [PubMed] [Google Scholar]
- 31.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 173:2617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez-Casal, J., J. A. Price, E. Maguin, and J. R. Scott. 1993. An M protein with a single C repeat prevents phagocytosis of Streptococcus pyogenes: use of a temperature-sensitive shuttle vector to deliver homologous sequences to the chromosome of S. pyogenes. Mol. Microbiol. 8:809-819. [DOI] [PubMed] [Google Scholar]
- 33.Pine, L., and M. W. Reeves. 1978. Regulation of the synthesis of M protein by sugars, Todd Hewitt broth, and horse serum in growing cells of Streptococcus pyogenes. Microbios 21:185-212. [PubMed] [Google Scholar]
- 34.Podbielski, A., J. A. Peterson, and P. Cleary. 1992. Surface protein-CAT reporter fusions demonstrate differential gene expression in the vir regulon of Streptococcus pyogenes. Mol. Microbiol. 6:2253-2265. [DOI] [PubMed] [Google Scholar]
- 35.Ribardo, D. A., and K. S. McIver. 2003. amrA encodes a putative membrane protein necessary for maximal exponential phase expression of the Mga virulence regulon in Streptococcus pyogenes. Mol. Microbiol. 50:673-685. [DOI] [PubMed] [Google Scholar]
- 36.Ribardo, D. A., and K. S. McIver. 2006. Defining the Mga regulon: comparative transcriptome analysis reveals both direct and indirect regulation by Mga in the group A streptococcus. Mol. Microbiol. 62:491-508. [DOI] [PubMed] [Google Scholar]
- 37.Scott, J. R., P. C. Guenthner, L. M. Malone, and V. A. Fischetti. 1986. Conversion of an M− group A streptococcus to M+ by transfer of a plasmid containing an M6 gene. J. Exp. Med. 164:1641-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stülke, J., and W. Hillen. 1999. Carbon catabolite repression in bacteria. Curr. Opin. Microbiol. 2:195-201. [DOI] [PubMed] [Google Scholar]
- 39.Stülke, J., and W. Hillen. 2000. Regulation of carbon catabolism in Bacillus species. Annu. Rev. Microbiol. 54:849-880. [DOI] [PubMed] [Google Scholar]
- 40.Turinsky, A. J., F. J. Grundy, J. H. Kim, G. H. Chambliss, and T. M. Henkin. 1998. Transcriptional activation of the Bacillus subtilis ackA gene requires sequences upstream of the promoter. J. Bacteriol. 180:5961-5967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vahling, C. M., and K. S. McIver. 2005. Identification of residues responsible for the defective virulence gene regulator Mga produced by a natural mutant of Streptococcus pyogenes. J. Bacteriol. 187:5955-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Varga, J., V. L. Stirewalt, and S. B. Melville. 2004. The CcpA protein is necessary for efficient sporulation and enterotoxin gene (cpe) regulation in Clostridium perfringens. J. Bacteriol. 186:5221-5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Warner, J. B., and J. S. Lolkema. 2003. CcpA-dependent carbon catabolite repression in bacteria. Microbiol. Mol. Biol. Rev. 67:475-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zomer, A. L., G. Buist, R. Larsen, J. Kok, and O. P. Kuipers. 2007. Time-resolved determination of the CcpA regulon of Lactococcus lactis subsp. cremoris MG1363. J. Bacteriol. 189:1366-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.