Abstract
General-diffusion porins form large β-barrel channels that control the permeability of the outer membrane of gram-negative bacteria to nutrients, some antibiotics, and external signals. Here, we have analyzed the effects of mutations in the OmpU porin of Vibrio cholerae at conserved residues that are known to affect pore properties in the Escherichia coli porins OmpF and OmpC. Various phenotypes were investigated, including sensitivity to β-lactam antibiotics, growth on large sugars, and sensitivity to and biofilm induction by sodium deoxycholate, a major bile component that acts as an external signal for multiple cellular responses of this intestinal pathogen. Overall, our results indicate that specific residues play different roles in controlling the passage of various compounds. Mutations of barrel wall arginine residues that protrude in the pore affect pore size and growth in the presence of large sugars or sodium deoxycholate. Sensitivity to large cephalosporins is mostly affected by D116, located on the L3 loop, whose homolog in E. coli, OmpF, is a known binding determinant for these drugs. L3 loop residues also affect biofilm induction. The results are interpreted in terms of a homology model based on the structures of E. coli porins.
General-diffusion porins form a large class of membrane proteins that play a major role in controlling the permeability of the outer membrane of gram-negative bacteria (33). The porins from Escherichia coli have been investigated extensively at the structural and functional levels (1, 3, 11, 12, 17, 33). They assemble as trimers in the outer membrane and form triple-barrel channels, as each β-barrel subunit contains an individual pore. The narrowest part of each pore, called the constriction zone or eyelet, is delimited by the barrel wall and an inwardly folded loop, the L3 loop. The permeability properties of the channels have been studied with a variety of assays, such as antibiotic flux assays, antibiotic sensitivity assays, liposome swelling assays, and electrophysiology. As the porins present the major conduit for hydrophilic solutes inside the cell, several cellular phenotypes have been found to be highly dependent on the presence and the functional properties of porins. One example is porin mutants with enlarged pores, which were isolated on the basis of their ability to allow the growth of cells on maltodextrins as the sole carbon source in the absence of the LamB maltoporin that is specific for this transport. Prior to the publication of the solved structure of OmpF, Benson and colleagues (2) had identified residues essential in determining pore size by using this approach. Later, the structure revealed that these residues indeed decorate the constriction zone of the channel, where the size exclusion limit is determined. Some of the important players in these studies are arginine residues (R42 and R82 in OmpF [2] and R37 and R74 in OmpC [31]) that belong to a cluster of positively charged amino acids of the barrel wall opposite to constriction loop L3.
Sensitivity to antibiotics is also greatly influenced by the porin composition of the outer membrane, particularly for β-lactams (33). Resistance to such drugs is conferred, in some cases, by the loss of the major porins or by mutations that functionally impact the porin (5, 10, 15, 29). For example, Dé and coworkers characterized a variant of porin from a clinical isolate of Enterobacter aerogenes that showed profound alterations in channel properties, as well as multidrug resistance (15). Various effects have been observed, depending on the type of porins and the antibiotics. Increased sensitivity to ampicillin and penicillin was described for OmpC mutated at the R37, R74, and D105 residues (homologous to R42, R82, and D113, respectively, in OmpF; see Fig. 2A), originally isolated by Misra and Benson (31). Similar mutations in the homologous OmpF residues generally produced an increase in sensitivity to a variety of β-lactams, albeit less drastic and more variable than for OmpC (2). The exception is D113A, which has been shown to be a major determinant in the sensitivity of OmpF to several β-lactams (42). However, other mutations can reduce antibiotic diffusion and confer resistance. For example, mutating G119 to aspartate or glutamate diminished sensitivity to cephalosporins (40). The diffusion of cefepime was substantially decreased by mutation G119D or G119E (40) and mutation K16A (5). Interestingly, most of these residues have also been shown to play a role in the permeability properties of the porins, as studied by electrophysiology (17). They were found to influence the rate of ion flux, as determined by conductance, and the selectivity of the channels. In addition, D113 appears to be part of a binding site for the polyamine spermine (23), which induces channel closure and interferes with antibiotic flux through the pores (9, 16).
FIG. 2.
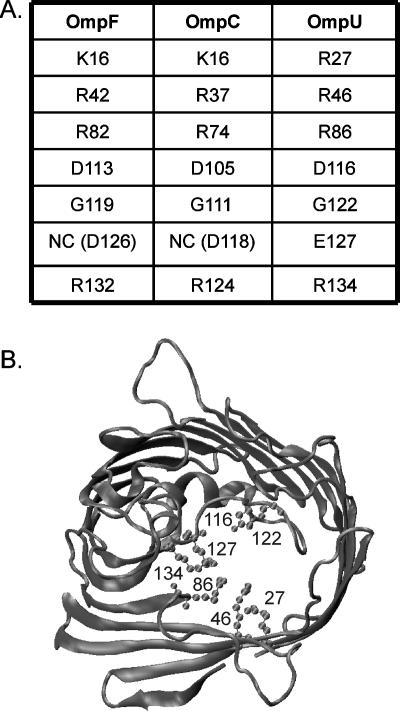
Homology model of OmpU. (A) Table of the homologous residues in the E. coli porins OmpF and OmpC and the V. cholerae porin OmpU that are tested here. NC, not conserved. (B) Graphic representation of the cross section of a monomer of OmpU, modeled on the structures of E. coli OmpF, OmpC, and PhoE. The numbers of the residues studied in this work are marked: R27, R46, R86, D116, G122, E127, and R134.
Since specific residues lining the constriction zone of the E. coli porins OmpF and OmpC are major determinants in the permeability properties of these porins, we sought to test whether the homologous residues of the Vibrio cholerae OmpU porin might play similar roles. OmpU shares about 25% identity and 40% similarity in amino acid sequence with OmpF, OmpC, and PhoE of E. coli. The ClustalW alignment of OmpU and these three E. coli porins is shown in Fig. 1. Since the structure of the OmpU porin is unknown, we constructed a homology model of OmpU structure based on sequence alignment and the deposited (Protein Data Bank) structures of OmpF, OmpC, and PhoE from E. coli (Fig. 2B). Previous studies on OmpU have shown that it forms a pore with an exclusion limit of approximately 700 Da, based on liposome swelling assays (7); that it is permeable to cephaloridine (43); and that it has the typical behavior of porins in electrophysiology experiments, i.e., a stable open state with occasional transient spontaneous closures and voltage-induced inactivation (4, 41). OmpU and OmpT are the major general diffusion porins of V. cholerae and are differentially expressed, such that OmpU is believed to be expressed while OmpT is repressed, when the cells are colonizing a human host (13, 27). Interestingly, OmpU confers resistance to sodium deoxycholate (DC), a major bile component, when solely expressed in V. cholerae cells, while OmpT makes the cells more sensitive to this compound (36, 37). The characterization of the channel properties by electrophysiology has shown that OmpU and OmpT are functionally distinct and differ markedly in their ionic selectivity, spontaneous activity, and sensitivity to voltage (41), to deoxycholic acid (18), and to acidic pH (19).
FIG. 1.
Sequence alignments. The OmpU sequence was aligned with those of the E. coli porins OmpF, PhoE, and OmpC by using the ClustalW program. *, identical residue in all sequences; :, conserved substitutions; ., semiconserved substitutions; -, gap.
In the work presented here, we have tested the hypothesis that conserved residues known to affect the pore properties of OmpF and OmpC also influence the pore characteristics of OmpU. For this, we have analyzed several phenotypes of OmpU mutants that relate to pore properties, such as antibiotic sensitivity, growth on maltodextrins or DC, and biofilm induction by DC. Effects on these functions are observed with specific mutations, and the results are discussed in terms of the nature and the presumed location of the substituted residues based on their homology with E. coli porins.
MATERIALS AND METHODS
Strains and plasmids.
The wild-type (WT) and mutant ompU genes were cloned into the EcoRI and KpnI sites of the pBAD24 plasmid and the SalI and BamHI sites of the pNL9 plasmid. The site-directed mutations were constructed with a quick change kit from Stratagene, except for the R46A mutation, which was made by using ProteinOne. After the mutated sequences were confirmed, the mutant genes were recloned into a clean vector. For the experiments testing growth on maltodextrin, we expressed the WT and mutated genes from the pBAD plasmid into the E. coli strain BZB1107 (35), because these experiments require that the maltoporin be absent and we do not have a maltoporin-deficient V. cholerae strain (ompS null). For all other experiments, the genes were expressed in the V. cholerae porin-deficient strain KKV884 (37) transformed with the appropriate pNL9 plasmids. We have found that V. cholerae retains the pNL plasmids more efficiently than the pBAD plasmids.
Media and chemicals.
Luria-Bertani (LB) medium contained 1% tryptone, 1% NaCl, and 0.5% yeast extract. Tryptone, yeast extract, and Mueller-Hinton broth were from Difco Laboratories. All chemicals, including antibiotics, inducers, and sugars, were from Sigma.
Antibiotic disc assays.
Single colonies of KKV884 freshly transformed with each of the ompU gene-containing plasmids were toothpicked and grown in LB broth in the presence of 50 μg/ml kanamycin, 100 μg/ml streptomycin, and 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) until the optical density at 600 nm (OD600) reached 0.7. The cultures were then diluted to an OD600 of 0.14 and swabbed onto a Mueller-Hinton agar plate containing 50 μg/ml kanamycin, 100 μg/ml streptomycin, and 1 mM IPTG. Six-millimeter-diameter sterile, blank filter disks (VWR) were placed onto the plate and spotted with 20 μl of antibiotic stock solution such that the final amount of antibiotic was 30 μg (10 μg for ceftazidime). The plates were incubated for 16 to 18 h at 37°C, after which the diameter of the whole zone of inhibition was measured. E. coli strain ATCC 29522 was used as a control.
Biofilm assay.
Overnight cultures of KKV884 transformed with the pNL9 plasmid containing WT ompU or the mutated ompU genes were grown in LB medium with the appropriate antibiotics and inducer. Three-hundred-microliter static cultures of each strain were set up by diluting the overnight culture 100-fold in an 8-ml test tube in the absence or the presence of 0.01% DC. The cultures were grown for 24 h at room temperature in the absence of shaking. Detection of the biofilm was performed by adding 30 μl of 0.5% crystal violet solution in water, followed by a 30-minute incubation at room temperature, five washes with water, and a final addition of 400 μl of 95% ethanol. After 20 min of incubation at room temperature, 600 μl of water was added, and the A540 of the solution was read in an Ultrospec 2100 Pro spectrophotometer.
Homology model.
A homology model of OmpU was generated based upon the published structures of OmpF (PDB ID 2OMF), OmpC (PDB ID 2J1N), and PhoE (PDB ID 1PHO). The ClustalW program hosted at the European Bioinformatics Institute (http://www.ebi.ac.uk/clustalw/) was used to generate an alignment between OmpU, OmpF, OmpC, and PhoE. The primary sequence of OmpU was then loaded into the Deep-Viewer program along with the published structures of OmpF, OmpC, and PhoE. The alignment prepared by ClustalW was reproduced within this program and the project file sent to the Swiss-Model website (http://swissmodel.expasy.org/) for automatic model generation using project mode. The structure of the modeled OmpU was visualized with Visual Molecular Dynamics (VMD), which was developed by the theoretical and computational biophysics group in the Beckman Institute for Advanced Science and Technology at the University of Illinois at Urbana-Champaign (20).
RESULTS
The goal of this study was to test the effects of mutations in OmpU at conserved residues (Fig. 2A) that are pore exposed, based on the homology with E. coli porins, and that are known to alter pore properties in OmpF and OmpC. As in OmpF, a cluster of positively charged residues (in this case, all arginines—R27, R46, R86, and R134) is found in OmpU in the β-barrel wall opposite to constriction loop L3. As described in the introduction, mutations of the homologous residues have a profound influence on the pore properties of OmpF (2, 5, 17, 42). Negatively charged residues are located on L3: D116 is homologous to D113 in OmpF, a site that has been repeatedly identified as influencing the permeation and binding of various hydrophilic compounds (2, 6, 23, 34, 39, 42); and E127 does not have strict homology to a negatively charged residue in OmpF, but it protrudes into the pore lumen and is shifted by 1 residue relative to a conserved aspartate in OmpF (D126) and OmpC (D118) with similar orientation. A mutation at D118 of OmpC has been shown to affect channel properties (28). G122 is homologous to G119 in OmpF, a residue that was shown to play a role in colicin A and antibiotic sensitivities (6, 24, 40). The mutations were engineered on ompU cloned in the pBAD24 or pNL9 plasmids. Western blots show that the expression levels of the mutants were the same as the expression level for the WT (Fig. 3A and 5A).
FIG. 3.
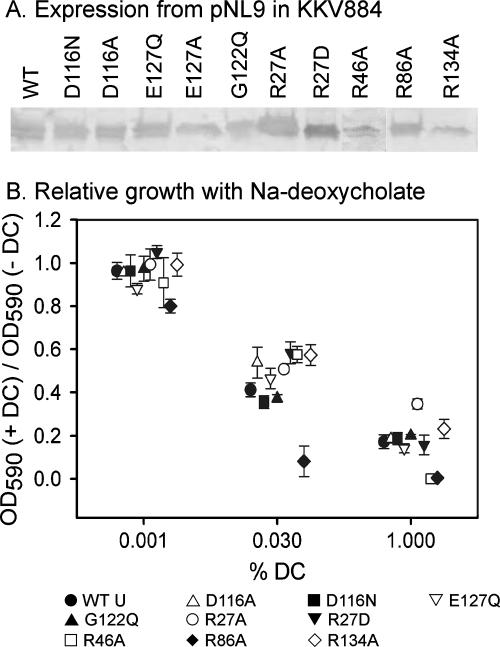
Growth on DC (Na deoxycholate). (A) Western blot of envelope preparations isolated from KKV884 cells expressing the mutant porins from the pNL9 plasmid. (B) Growth of KKV884 cells expressing WT or mutant OmpU porins in the presence of the indicated concentrations of DC (+ DC) relative to that in the absence of DC (− DC). The symbols and error bars show the averages ± standard errors of the means of the ratios of the OD590 values obtained from three independent colonies at the 4-h time point.
FIG. 5.
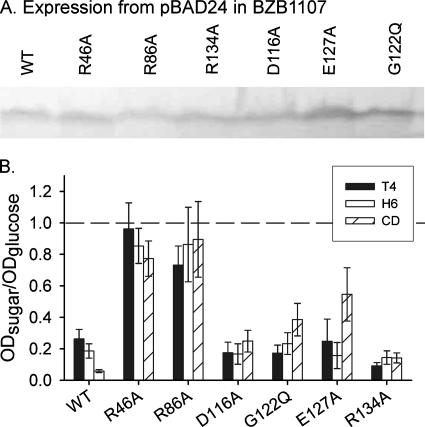
Growth on maltodextrins and cyclodextrin (CD). Relative growth of KKV884 cells expressing WT or mutant OmpU porins in the presence of 1 mM of the test sugar: maltotetraose (T4), maltohexaose (H6), or CD. The values are expressed as the ratio of the A600 value after 24 h of growth in minimal medium containing 1 mM of the test sugar to the value after growth in the presence of 1 mM glucose. A value close to 1 (dashed line) indicates that the growth is comparable to that in glucose. The histogram bars and error bars show the averages ± standard errors of the means of the results from three independent colonies.
Growth in the presence of DC.
The growth of the mutants in LB medium was found to be identical to that of the WT (data not shown). Because of the reported differential sensitivities of OmpU and OmpT with respect to DC, we investigated whether the OmpU mutants showed sensitivity to DC in their rates of growth. Pilot experiments with strains expressing the WT or some of the mutant OmpU porins indicated that increasing DC concentrations induced prolonged lag times, followed by exponential growth at similar rates regardless of the presence of DC. In some cases, the cultures reached stationary phase early. We believe that the presence of DC might induce an adaptive process (such as drug extrusion, as documented in reference 8) which, after a prolonged lag time, eventually allows the cells to recover a growth rate similar to that in the absence of the drug. Therefore, we decided to use the OD of cultures at the 4-h time point as a measure of the effect of DC on cell growth. This value was measured for cultures grown in the presence of three concentrations of DC (0.001, 0.03, and 1.0%) and compared to that of a culture in the absence of DC. Overall, no significant differences were observed between the WT and most of the mutant strains tested, except for the R46A and R86A mutants (Fig. 3B). The R86A mutant showed a significant reduction in growth at all three DC concentrations tested, while the R46A mutant displayed DC sensitivity only at the highest DC concentration (1%). It is noteworthy that both the R86A and R46A mutants are completely unable to grow in the presence of 1% DC, while the WT strain and other mutants still show some growth. These results are indicative of an increased sensitivity to DC in the R86A and R46A mutants.
Biofilm induction by DC.
Bile and bile acids have been shown to have pleiotropic effects on V. cholerae physiology (8, 21, 22, 26). In particular, DC was found to induce biofilm formation at concentrations of 0.01% (22). According to the current model, DC activates the response regulator VpsR at the translational level, either directly or indirectly, via an unknown regulator of VpsR (22). VpsR is a response regulator that controls exopolysaccharide production and biofilm formation (44, 45). Although DC has detergentlike properties and can partition into phospholipid bilayers, the outer membrane is intrinsically more resistant to such an effect due to the presence of lipopolysaccharide (33). Thus, although some diffusion through the outer membrane bilayer per se cannot be excluded, it is possible that DC gains access to the periplasm in part by going through the porins, as its molecular mass of 392 Da is well within the typical exclusion limit of porins of ∼600 Da. If this is the case, we anticipated that we might see differences in the ability of sublethal concentrations of DC to promote biofilm formation in cells expressing WT or mutant OmpU porin.
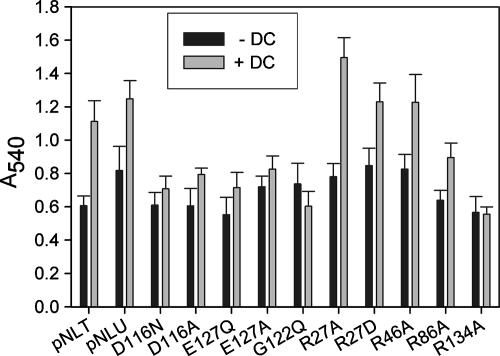
The results shown in Figure 4 show that sublethal concentrations of DC (0.01%) induced biofilm formation in a porinless strain expressing WT ompU from the pNL9 plasmid. Similarly, a strain expressing only WT ompT also produces biofilm in response to DC. The level of induction (∼1.7-fold) is less than has been previously reported for WT O395 cells (22), although the absolute values of crystal violet staining are comparable. In the absence of DC, the A540 readings are similar for all mutant strains and comparable to that for the WT. The mutants with altered residues on the putative L3 loop, i.e., the D116, E127, and G122 mutants, show reductions in the ability to form a biofilm in response to DC. However, the arginine mutants, with the exception of R134A, show inductions of biofilm that are comparable to, if not slightly enhanced over, that of the WT.
FIG. 4.
Biofilm induction by DC. Effect of 0.01% DC on biofilm formation as measured by crystal violet staining (A540) of the WT and different mutants. +, present; −, absent. The vertical bars and error bars show the averages ± standard errors of the means of the results obtained from 10 independent colonies.
Antibiotic sensitivity.
An antibiotic disc assay was used to assess the effects of the mutants on the pore function of OmpU. Previous work has shown that the permeation of β-lactam antibiotics and, hence, the sensitivities of the cells to these antibiotics were altered by mutations of the constriction zone (2, 15, 31). Here, we have tested seven β-lactam antibiotics that differ in size, in charge, and in overall architecture. The results are summarized in Table 1.
TABLE 1.
Antibiotic susceptibilities of the OmpU mutants
| Antibiotic | Mol wt | Diam (mm) of zone of inhibition for cells expressing:a
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WT | R27D | R27A | R46A | R86A | D116N | D116A | E127Q | E127A | G122Q | R134A | ||
| Cefaclor | 368 | 32 | 28 | 30 | 30 | 29 | 28 | 28 | 29 | 30 | 28 | 30 |
| Cefadroxil | 364 | 20 | 19 | 20 | 19 | 19 | 19 | 19 | 18 | 20 | 19 | 20 |
| Cefotaxime | 477 | 44 | 40 | 39 | 40 | 40 | 40 | 36 | 40 | 39 | 40 | 42 |
| Cefoxitin | 450 | 23 | 21 | 24 | 22 | 21 | 22 | 23 | 23 | 22 | 22 | 23 |
| Cefamandole | 484 | 31 | 27 | 28 | 29 | 28 | 28 | 27 | 28 | 28 | 27 | 31 |
| Ceftazidime | 546 | 32 | 30 | 29 | 29 | 29 | 30 | 28 | 30 | 30 | 30 | 30 |
| Ceftriaxone | 598 | 38 | 35 | 36 | 37 | 33 | 37 | 32 | 36 | 36 | 38 | 36 |
Measurements were done on three individual colonies and averaged. The antibiotic disc diameter was 6 mm. Bold values represent a greater-than-average resistance to the antibiotic. Standard errors are less than 3%.
In general, no mutation introduced an increase in the sensitivity to the antibiotics; mutations either produced no change or a more-resistant strain. All mutants show about the same, fairly mild, reduction in disc diameter for a given antibiotic. An exception is the D116A mutant, which shows a more drastic increased resistance than the D116N mutant or any other mutant to cefotaxime, ceftazidime, and ceftriaxone in particular (Table 1). As a whole, the effects of the mutations are more pronounced in the case of antibiotics that produce larger zones of inhibition than for those to which the WT cells are already more resistant. For example, WT cells are more resistant to cefadroxil and cefoxitin than to the other drugs tested, and the mutants show very minor differences relative to the WT for these two antibiotics. The case of ceftriaxone is interesting, as this is the antibiotic for which there is the most variation among mutants. Some mutations (R86A and D116A) produced more substantial effects than were seen for the rest of the mutants. Of all the mutants tested, the R134A mutant is the one that shows the least difference relative to the WT for all the antibiotics tested.
Growth on maltodextrins or cyclodextrin.
Liposome swelling assays have shown that the exclusion limit of OmpU for permeability to sugars is ∼850 Da (7). Larger carbohydrates are believed to utilize the maltoporin homolog OmpS to penetrate inside the cell. It has been previously shown that E. coli strains lacking maltoporin but expressing OmpF or OmpC mutants with enlarged pores can grow on maltodextrins as the sole carbon source (2, 31). In order to test whether the OmpU mutants studied here had the capability of fluxing large carbohydrates, we expressed a representative group of mutated genes from a pBAD24 plasmid in BZB1107, an E. coli strain lacking OmpF, OmpC, and LamB (35). The Western blot shown in Fig. 5A shows that the levels of mutant proteins were comparable to that of the WT. The cells were grown in minimum M63 medium containing 1 mM of glucose, maltotetraose (T4), maltohexaose (H6), or cyclodextrin (CD) as the sole carbon source. The OD600 of the culture was measured after 16 h of incubation at 37°C. The graph in Fig. 5B shows the ratios of the ODs in the presence of the large sugar to those of cultures grown in glucose. A strain expressing WT OmpU grows poorly on these substrates, and the ODs of cultures grown in maltotetraose or maltohexaose remain below or around 20% of that attained with glucose. The growth of the strain expressing WT OmpU is negligible in the presence of CD. The D116A, G122Q, E127A, and R134A mutants show phenotypes comparable to that of the WT with respect to growth in the presence of the two maltodextrins. However, there is a significant improvement in growth in the presence of CD for the D116A, G122Q, and E127A mutants. Most remarkably, the R46A and R86A mutants show growth in the presence of any of the three large sugars that is comparable to that in the presence of glucose. This phenotype is indicative of an enlarged pore size that can accommodate these larger substrates.
DISCUSSION
We have presented here the phenotypic characterization of several mutants of the OmpU porin of V. cholerae that were designed to test the participation of these residues in defining pore properties. In particular, we have assayed functions that are linked to the permeability of the porin to specific compounds, such as β-lactam antibiotics, large sugars, and the bile component DC. DC is a lipophilic compound which is likely to associate with all membranes, although the nature of lipopolysaccharide makes the outer membrane a more effective barrier to these substances than a typical phospholipid bilayer. DC does reach the inner membrane and cytoplasm, as evidenced by a number of processes that are triggered or modulated by bile or this external signal, such as cholera toxin production (21), biofilm formation (22), and the activation of drug efflux systems (8). Although not excluding a membrane-delimited pathway for DC entry, we and others have proposed that DC might also access the cell interior via porins (18, 25, 36), because of the apparent distinct permeability of the two major porins OmpU and OmpT to DC based on (i) growth impairment in the presence of DC which is more pronounced in cells expressing solely OmpT than solely OmpU (37, 38) and (ii) our observation that low concentrations of deoxycholic acid introduce transient interruptions in the flow of ions through OmpT, but not OmpU, as it occludes the pore lumen, presumably during transit (18). Altogether, these results indicate that DC might, in part, gain access to the inner membrane by using the OmpT porin preferentially. They do not exclude the use of OmpU by DC but suggest that permeation through OmpU might occur at higher concentrations. For these reasons, we have also included some assays that depend on DC permeability through porins.
Overall, our results indicate that specific residues play different roles in controlling the passage of various compounds. The permeation of large sugars appears to be essentially controlled by pore size. As shown in Fig. 2B, the two arginines of the barrel wall opposite to the L3 loop, R46 and R86, have elongated side chains that project toward the pore lumen. The truncation of these side chains to that of alanine in the R46A and R86A mutants has the drastic effect of enlarging the effective pore size such as to allow the permeation of maltotetraose, maltohexasose, and CD, as shown in Fig. 5B. Mutations at the L3 loop (D116A and G122Q) have no impact, presumably because of the already small size of their side chains. Electrophysiology experiments have indeed indicated that the R86A and R46A mutants, but not the D116A mutant, have increased conductance relative to that of the WT, as observed for wider pores (B. Lauman and A. H. Delcour, unpublished data). Alanine mutations at E127 and R134 are also ineffective, possibly because these residues are tucked in at the root of the L3 loop. Because of the close proximity of their side chains, they may also have redundant effects such that the shortening of the side chain at a single residue has limited impact.
If the mutations R46A and R86A make the pore wider, based on the experiment testing growth on maltodextrins, why are they relatively ineffective with respect to antibiotic sensitivities? It is legitimate to assume that the susceptibilities to a β-lactam antibiotic for a given set of isogenic strains with different porin mutations are directly correlated with the flow of the antibiotic through porin, everything else being equal. Therefore, it is anticipated that different mutations might affect antibiotic permeation in different ways. In fact, computational and functional studies have shown that the factors controlling antibiotic permeation are multifaceted and include steric, electrostatic, and positional contributions (14, 42). The subtle interplay of these factors is variable for different antibiotics, and thus, the impact of specific mutations varies with the mutation type and the nature of the antibiotic. Danelon et al. have argued that the presence of an internal binding site for a given antibiotic favors its translocation as drug-channel interactions compensate for desolvation energy and entropy changes associated with the passage of a large molecule through the constriction zone (14). Therefore, making the pore wider may not necessarily be a better way to achieve higher permeation and increased sensitivity, as suggested by our results with the R46A and R86A mutants. On the other hand, a decreased interaction at a crucial binding site is likely to decrease sensitivity. The results obtained with D116A indicate that this residue may indeed belong to a binding site, as we see increased resistance to cefotaxime, ceftazidime, and ceftriaxone (Table 1). The homologous residue, D113, in OmpF has also been found, by computational analysis, to be a prime site of drug-channel interactions for cefotaxime and ceftazidime (42). By the same token, our results indicate that other residues besides D116 might play a relatively minor role in forming a binding site for the antibiotics tested. This is somewhat surprising but may be indicative that the passage of the antibiotic molecules through OmpU is less of a tight fit than the passage through OmpF. Indeed, the size of the pore may be somewhat larger than for OmpF, based on the fact that OmpU is permeable to stachyose (molecular weight, 666) while OmpF is not (7). We have also found, in electrophysiology experiments, that ampicillin was unable to induce the current interruptions that are indicative of the transient blocking of the pore that is typically seen in OmpF (32) (G. Duret and A. H. Delcour, unpublished data); therefore, the OmpU channel may be wide enough not to be fully blocked by the antibiotic while it occupies the pore.
Assuming that porins represent a major pathway for DC permeation through the outer membrane, as discussed above, the enlarged pore size afforded by the R46A and R86A mutants might be responsible for the increased growth sensitivities of these two mutants, particularly at higher concentrations, due to increased penetration of DC into the cell. None of the other mutants showed significant differences from the WT with this assay. This is in contrast to the results obtained for biofilm formation. Here, we find that mutations at residues D116, E127, G122, and R134 abolish the induction of biofilm by 0.01% DC. Interestingly, these mutations are all located on the L3 loop or, in the case of R134A, at the root of the L3 loop. A straightforward explanation would be that these mutations reduce the flux of DC through OmpU, but how can these results be reconciled with the lack of effect of the same mutations on cell growth in the presence of DC (Fig. 3)? It is possible that small differences in the DC permeation efficiency of the mutants relative to that of the WT may not have been sufficient to induce a substantial improvement in the growth of these mutants at the 4-h time point, even at 0.03% DC, as indicated by the results shown in Fig. 3. On the other hand, with the biofilm assay requiring 24 h and the possibility that extrusion pumps might have been induced, a small difference in flux through a mutated porin may lead to a steady-state level of periplasmic DC that is lower than for a WT strain. Depending on the sensitivity of the DC detection system involved in biofilm induction (which is unknown at this point), such a reduced level may lead to lack of induction of biofilm formation. We do not think that the lack of effect is due to reduced expression of the mutants, as the Western blot results shown in Fig. 3A show the levels of OmpU mutants in a cell envelope preparation to be comparable to those of the WT.
Interestingly, the levels of biofilm induction are similar for strains expressing OmpU, OmpT, or the β-barrel arginine mutants. This is suggestive of similar steady-state levels of periplasmic DC and, possibly, saturation of the DC response in these strains. This would explain why similar biofilm levels are observed in a strain expressing WT OmpU and in strains where DC permeation might be enhanced, such as in the presence of OmpT (which appears more permeable to deoxycholic acid than OmpU [18]) or of OmpU mutants with enlarged pores. Alternatively, we should not dismiss the possibility that the role of OmpU in DC-induced biofilm formation may be more intricate than that of simply allowing DC flux. Mathur and colleagues have proposed recently that OmpU may serve as a sensor of membrane perturbation, induced by antimicrobial peptides, that activates the σE-dependent cell envelope stress response (30). If such a pathway were involved in DC-induced biofilm production, we might anticipate that the sensory process would be controlled by a set of mutations in OmpU that were different from those related to permeation and cell growth in the presence of DC.
Acknowledgments
This work was supported by NIH grant AI34905 and grant E-1597 from the Robert A. Welch Foundation.
We thank Huy Vu for technical assistance and Guillaume Duret for critical reading of the manuscript.
Footnotes
Published ahead of print on 28 September 2007.
REFERENCES
- 1.Baslé, A., G. Rummel, P. Storici, J. P. Rosenbusch, and T. Schirmer. 2006. Crystal structure of osmoporin OmpC from E. coli at 2.0 Å. J. Mol. Biol. 362:933-942. [DOI] [PubMed] [Google Scholar]
- 2.Benson, S. A., J. L. Occi, and B. A. Sampson. 1988. Mutations that alter the pore function of the OmpF porin of Escherichia coli K12. J. Mol. Biol. 203:961-970. [DOI] [PubMed] [Google Scholar]
- 3.Benz, R. (ed.). 2004. Bacterial and eukaryotic porins: structure, function, mechanism. Wiley-VCH, Weinheim, Germany.
- 4.Benz, R., E. Maier, and T. Chakraborty. 1997. Purification of OmpU from Vibrio cholerae classical strain 569B: evidence for the formation of large cation-selective ion-permeable channels by OmpU. Microbiologia 13:321-330. [PubMed] [Google Scholar]
- 5.Bredin, J., N. Saint, M. Malléa, E. Dé, G. Molle, J. M. Pagès, and V. Simonet. 2002. Alteration of pore properties of Escherichia coli OmpF induced by mutation of key residues in anti-loop 3 region. Biochem. J. 363:521-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bredin, J., V. Simonet, R. Iyer, A. H. Delcour, and J. M. Pagès. 2003. Colicins, spermine and cephalosporins: a competitive interaction with the OmpF eyelet. Biochem. J. 376:245-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakrabarti, S. R., K. Chaudhuri, K. Sen, and J. Das. 1996. Porins of Vibrio cholerae: purification and characterization of OmpU. J. Bacteriol. 178:524-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chatterjee, A., S. Chaudhuri, G. Saha, S. Gupta, and R. Chowdhury. 2004. Effect of bile on the cell surface permeability barrier and efflux system of Vibrio cholerae. J. Bacteriol. 186:6809-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chevalier, J., M. Malléa, and J. M. Pagès. 2000. Comparative aspects of the diffusion of norfloxacin, cefepime and spermine through the F porin channel of Enterobacter cloacae. Biochem. J. 348:223-227. [PMC free article] [PubMed] [Google Scholar]
- 10.Chevalier, J., J. M. Pagès, A. Eyraud, and M. Malléa. 2000. Membrane permeability modifications are involved in antibiotic resistance in Klebsiella pneumoniae. Biochem. Biophys. Res. Commun. 274:496-499. [DOI] [PubMed] [Google Scholar]
- 11.Cowan, S. W., R. M. Garavito, J. N. Jansonius, J. A. Jenkins, R. Karlsson, N. König, E. F. Pai, R. A. Pauptit, P. J. Rizkallah, J. P. Rosenbusch, G. Rummel, and T. Schirmer. 1995. The structure of OmpF porin in a tetragonal crystal form. Structure 3:1041-1050. [DOI] [PubMed] [Google Scholar]
- 12.Cowan, S. W., T. Schirmer, G. Rummel, M. Steiert, R. Ghosh, R. A. Pauptit, J. N. Jansonius, and J. P. Rosenbusch. 1992. Crystal structures explain functional properties of two E. coli porins. Nature 358:727-733. [DOI] [PubMed] [Google Scholar]
- 13.Crawford, J. A., J. B. Kaper, and V. J. DiRita. 1998. Analysis of ToxR-dependent transcription activation of ompU, the gene encoding a major envelope protein in Vibrio cholerae. Mol. Microbiol. 29:235-246. [DOI] [PubMed] [Google Scholar]
- 14.Danelon, C., E. M. Nestorovich, M. Winterhalter, M. Ceccarelli, and S. M. Bezrukov. 2006. Interaction of zwitterionic penicillins with the OmpF channel facilitates their translocation. Biophys. J. 90:1617-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dé, E., A. Baslé, M. Jaquinod, N. Saint, M. Malléa, G. Molle, and J. M. Pagès. 2001. A new mechanism of antibiotic resistance in Enterobacteriaceae induced by a structural modification of the major porin. Mol. Microbiol. 41:189-198. [DOI] [PubMed] [Google Scholar]
- 16.delaVega, A. L., and A. H. Delcour. 1996. Polyamines decrease Escherichia coli outer membrane permeability. J. Bacteriol. 178:3715-3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Delcour, A. H. 2003. Solute uptake through general porins. Front. Biosci. 8:d1055-d1071. [DOI] [PubMed] [Google Scholar]
- 18.Duret, G., and A. H. Delcour. 2006. Deoxycholic acid blocks Vibrio cholerae OmpT but not OmpU porin. J. Biol. Chem. 281:19899-19905. [DOI] [PubMed] [Google Scholar]
- 19.Duret, G., V. Simonet, and A. H. Delcour. 2007. Modulation of Vibrio cholerae porin function by acidic pH. Channels 1:70-79. [DOI] [PubMed] [Google Scholar]
- 20.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graphics 14:33-38. [DOI] [PubMed] [Google Scholar]
- 21.Hung, D. T., and J. J. Mekalanos. 2005. Bile acids induce cholera toxin expression in Vibrio cholerae in a ToxT-independent manner. Proc. Natl. Acad. Sci. USA 102:3028-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hung, D. T., J. Zhu, D. Sturtevant, and J. J. Mekalanos. 2006. Bile acids stimulate biofilm formation in Vibrio cholerae. Mol. Microbiol. 59:193-201. [DOI] [PubMed] [Google Scholar]
- 23.Iyer, R., Z. Wu, P. M. Woster, and A. H. Delcour. 2000. Molecular basis for the polyamine-OmpF porin interactions: inhibitor and mutant studies. J. Mol. Biol. 297:933-945. [DOI] [PubMed] [Google Scholar]
- 24.Jeanteur, D., T. Schirmer, D. Fourel, V. Simonet, G. Rummel, C. Widmer, J. P. Rosenbusch, F. Pattus, and J. M. Pagès. 1994. Structural and functional alterations of a colicin-resistant mutant of OmpF porin from Escherichia coli. Proc. Natl. Acad. Sci. USA 91:10675-10679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klose, K. E. 2001. Regulation of virulence in Vibrio cholerae. Int. J. Med. Microbiol. 291:81-88. [DOI] [PubMed] [Google Scholar]
- 26.Krukonis, E. S., and V. J. DiRita. 2003. From motility to virulence: sensing and responding to environmental signals in Vibrio cholerae. Curr. Opin. Microbiol. 6:186-190. [DOI] [PubMed] [Google Scholar]
- 27.Li, C. C., J. A. Crawford, V. J. DiRita, and J. B. Kaper. 2000. Molecular cloning and transcriptional regulation of ompT, a ToxR-repressed gene in Vibrio cholerae. Mol. Microbiol. 35:189-203. [DOI] [PubMed] [Google Scholar]
- 28.Liu, N., and A. H. Delcour. 1998. The spontaneous gating activity of OmpC porin is affected by mutations of a putative hydrogen bond network or of a salt bridge between the L3 loop and the barrel. Protein Eng. 11:797-802. [DOI] [PubMed] [Google Scholar]
- 29.Malléa, M., J. Chevalier, C. Bornet, A. Eyraud, A. Davin-Regli, C. Bollet, and J. M. Pagès. 1998. Porin alteration and active efflux: two in vivo drug resistance strategies used by Enterobacter aerogenes. Microbiology 144:3003-3009. [DOI] [PubMed] [Google Scholar]
- 30.Mathur, J., B. M. Davis, and M. K. Waldor. 2007. Antimicrobial peptides activate the Vibrio cholerae sigmaE regulon through an OmpU-dependent signalling pathway. Mol. Microbiol. 63:848-858. [DOI] [PubMed] [Google Scholar]
- 31.Misra, R., and S. A. Benson. 1988. Genetic identification of the pore domain of the OmpC porin of Escherichia coli K-12. J. Bacteriol. 170:3611-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nestorovich, E. M., C. Danelon, M. Winterhalter, and S. M. Bezrukov. 2002. Designed to penetrate: time-resolved interaction of single antibiotic molecules with bacterial pores. Proc. Natl. Acad. Sci. USA 99:9789-9794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikaido, H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phale, P. S., A. Philippsen, C. Widmer, V. P. Phale, J. P. Rosenbusch, and T. Schirmer. 2001. Role of charged residues at the OmpF porin channel constriction probed by mutagenesis and simulation. Biochemistry 40:6319-6325. [DOI] [PubMed] [Google Scholar]
- 35.Prilipov, A., P. S. Phale, P. Van Gelder, J. P. Rosenbusch, and R. Koebnik. 1998. Coupling site-directed mutagenesis with high-level expression: large scale production of mutant porins from E. coli. FEMS Microbiol. Lett. 163:65-72. [DOI] [PubMed] [Google Scholar]
- 36.Provenzano, D., and K. E. Klose. 2000. Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc. Natl. Acad. Sci. USA 97:10220-10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Provenzano, D., C. M. Lauriano, and K. E. Klose. 2001. Characterization of the role of the ToxR-modulated outer membrane porins OmpU and OmpT in Vibrio cholerae virulence. J. Bacteriol. 183:3652-3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Provenzano, D., D. A. Schuhmacher, J. L. Barker, and K. E. Klose. 2000. The virulence regulatory protein ToxR mediates enhanced bile resistance in Vibrio cholerae and other pathogenic Vibrio species. Infect. Immun. 68:1491-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saint, N., K. L. Lou, C. Widmer, M. Luckey, T. Schirmer, and J. P. Rosenbusch. 1996. Structural and functional characterization of OmpF porin mutants selected for larger pore size. II. Functional characterization. J. Biol. Chem. 271:20676-20680. [PubMed] [Google Scholar]
- 40.Simonet, V., M. Malléa, and J. M. Pagès. 2000. Substitutions in the eyelet region disrupt cefepime diffusion through the Escherichia coli OmpF channel. Antimicrob. Agents Chemother. 44:311-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simonet, V. C., A. Baslé, K. E. Klose, and A. H. Delcour. 2003. The Vibrio cholerae porins OmpU and OmpT have distinct channel properties. J. Biol. Chem. 278:17539-17545. [DOI] [PubMed] [Google Scholar]
- 42.Vidal, S., J. Bredin, J. M. Pagès, and J. Barbe. 2005. Beta-lactam screening by specific residues of the OmpF eyelet. J. Med. Chem. 48:1395- 1400. [DOI] [PubMed] [Google Scholar]
- 43.Wibbenmeyer, J. A., D. Provenzano, C. F. Landry, K. E. Klose, and A. H. Delcour. 2002. Vibrio cholerae OmpU and OmpT porins are differentially affected by bile. Infect. Immun. 70:121-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yildiz, F. H., N. A. Dolganov, and G. K. Schoolnik. 2001. VpsR, a member of the response regulators of the two-component regulatory systems, is required for expression of vps biosynthesis genes and EPSETr-associated phenotypes in Vibrio cholerae O1 El Tor. J. Bacteriol. 183:1716-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yildiz, F. H., and G. K. Schoolnik. 1999. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc. Natl. Acad. Sci. USA 96:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]