Abstract
Changes in oxygen levels cause widespread changes in gene expression in organisms ranging from bacteria to humans. In Saccharomyces cerevisiae, this response is mediated in part by Hap1, originally identified as a heme-dependent transcriptional activator that functions during aerobic growth. We show here that Hap1 also plays a significant and direct role under hypoxic conditions, not as an activator, but as a repressor. The repressive activity of Hap1 controls several genes, including three ERG genes required for ergosterol biosynthesis. Chromatin immunoprecipitation experiments showed that Hap1 binds to the ERG gene promoters, while additional experiments showed that the corepressor Tup1/Ssn6 is recruited by Hap1 and is also required for repression. Furthermore, mutational analysis demonstrated that conserved Hap1 binding sites in the ERG5 5′ regulatory region are required for repression. The switch of Hap1 from acting as a hypoxic repressor to an aerobic activator is determined by heme, which is synthesized only in the presence of oxygen. The ability of Hap1 to function as a ligand-dependent repressor and activator is a property shared with mammalian nuclear hormone receptors and likely allows greater transcriptional control by Hap1 in response to changing oxygen levels.
Oxygen is essential for several important biological processes, including aerobic respiration and sterol synthesis. Oxygen can also be harmful, through the production of reactive oxygen species (26). As part of the response to changes in the level of this vital yet potentially toxic molecule, cells produce extensive changes in transcription (5, 32, 34, 53, 61). In many cases, cells sense and respond to changes in oxygen levels by oxygen-dependent pathways. For example, in human cells, the stability and activity of hypoxia-inducible factor 1α, required for the transcription of hypoxic-specific genes, are regulated by the hydroxylation of proline residues, a modification that is catalyzed by a family of oxygen-dependent prolyl hydroxylases. In the absence of oxygen, hydroxylation does not occur, hypoxia-inducible factor 1α protein is stabilized, and hypoxic-specific genes are induced (27). In Saccharomyces cerevisiae, oxygen is sensed, at least in part, by the level of heme, whose synthesis is dependent upon oxygen (23). Heme is an essential molecule for signaling oxygen levels in many organisms (42).
One of the most extensively studied roles for heme with respect to controlling oxygen-mediated transcription in S. cerevisiae is its ability to bind to and activate the transcriptional activator protein Hap1. Hap1 is required to activate the transcription of many aerobic genes (genes expressed only in the presence of oxygen), including those required for respiration and controlling oxidative damage (5, 31, 54, 65). Hap1 also activates the transcription of ROX1, which encodes a repressor of many hypoxic genes (genes expressed only in low-oxygen conditions) (5, 32, 54). Thus, Hap1 directly activates aerobic genes and indirectly represses hypoxic genes. Under hypoxic conditions, when heme is not synthesized, Hap1 does not function as an activator and the aerobic genes, including ROX1, are not transcribed. Current evidence suggests that heme binding mediates the ability of Hap1 to activate transcription by controlling the interaction of Hap1 with particular Hsp proteins (24, 35, 65).
Along with its established role as a transcriptional activator, some previous studies have suggested that Hap1 can also repress transcription. One recent study showed that Hap1 directly represses its own transcription in a heme-independent fashion (25). In addition, older experiments suggest that a small number of genes are repressed by Hap1 in the absence of heme or oxygen, although the results did not determine whether Hap1 repression is direct (11, 12, 17, 45, 58, 59). These studies suggest that the prevailing view of Hap1 as an aerobic-specific and heme-dependent activator may be too narrow.
To gain a comprehensive understanding of the functions of Hap1 during both aerobic and hypoxic growth, we performed microarray studies and combined the results with existing in vivo Hap1 DNA binding data (22) to identify direct targets of Hap1 regulation. These analyses led to the conclusion that Hap1 is active under hypoxic conditions as a direct repressor of several genes, including ERG2, ERG5, and ERG11, required for the biosynthesis of ergosterol, the major sterol in S. cerevisiae. In addition, chromatin immunoprecipitation (ChIP) experiments demonstrated that Hap1 binds to the promoters of these genes under both aerobic and hypoxic conditions and that the corepressor Tup1/Ssn6 is recruited by Hap1 in a hypoxic-specific fashion. Furthermore, mutational analyses of conserved promoter elements led to the identification of Hap1 binding sites in the ERG5 promoter that are required for repression. Finally, our data show that heme converts Hap1 from a hypoxic transcriptional repressor to an aerobic transcriptional activator and provide evidence that this mechanism allows for differential regulation of target genes at intermediate oxygen levels. These results, taken together with previous studies, expand our understanding of Hap1, demonstrating key functions for this regulator during both aerobic and hypoxic growth and providing insight into the biological significance of each role.
MATERIALS AND METHODS
Strains.
All S. cerevisiae strains (Table 1) are isogenic with a GAL2+ derivative of S288C (62). The strains were constructed by standard methods, either by crosses or by transformation (4). The mutant hap1 allele of S288C (18) was repaired in two transformation steps. First, the Ty1 insertion at the 3′ end of the open reading frame (ORF) was replaced with the URA3 gene. Second, URA3 was replaced with a wild-type HAP1 sequence from the Σ1278b strain background (49) while selecting for loss of URA3 on 5-floroorotic acid. The presence of the wild-type HAP1 allele was verified by PCR of the HAP1 locus, mRNA expression of a Hap1 target gene (CYC1), DNA sequencing, and Southern blot analysis (see Fig. S1 at http://genetics.med.harvard.edu/∼winston/Winston%20Lab%20Links.html; data not shown). The hap1Δ::kanMX, hem1Δ::kanMX, hsp82Δ::kanMX, ssa2Δ::LEU2, and ssa4Δ::HIS3 alleles were created by replacing the respective ORF with the kanMX, LEU2, or HIS3 marker (6). The hem1Δ::kanMX strain was grown on 200 μg/ml δ-aminolevulinate (δ-ala), except where indicated. The kanMX::GAL1pr-SSA1 and HIS3::GAL1pr-HSC82 alleles were constructed by placing the kanMX or HIS3 marker and the GAL1 promoter upstream of the SSA1 or HSC82 ORF (38). The myc-HAP1 and TUP1-myc alleles were created by inserting three copies of the myc epitope tag at the N-terminal or C-terminal end, respectively, while retaining the endogenous promoter (48). The myc-HAP1 allele is fully functional, as aerobic and hypoxic expression of both ERG5 and CYC1 is not affected (data not shown) (see Fig. 5B). The TUP1-myc allele is functional, as ERG5 repression is not affected (data not shown). The ERG5 promoter mutations were created as follows, with positions indicated relative to the ERG5 ATG. The two Hap1 binding site mutations (erg5-1 and erg5-2) were constructed by in vivo site-directed mutagenesis (50), replacing 12 bp from −426 to −415 or −571 to −560, respectively, with 5′-TACGGATCCACA-3′ (a BamHI site flanked by random sequences) or 5′-AGTAGAATTCTC-3′ (an EcoRI site flanked by random sequences), respectively. The 100-bp mutations (erg5-3, −144 to −243; erg5-4, −244 to −343; erg5-5, −344 to −443; and erg5-6, −444 to −543) were constructed by replacing the designated sequence with DNA encoding three copies of the myc epitope tag, a sequence that is 192 bp in length (48). The 50-bp mutations (erg5-9, −244 to −293; erg5-8, −269 to −318; and erg5-7, −294 to −343) were constructed by in vivo site-directed mutagenesis, replacing the designated sequence with 5′-ATCAAGATACCTGAACTGTTATGGATCCGCGCCGTAAAGCCATCTGGAGT-3′ (a BamHI site flanked by random sequences). The 20-bp mutations (erg5-14, −244 to −263; erg5-13, −264 to −283; erg5-12, −284 to −303; erg5-11, −304 to −323; and erg5-10, −324 to −343) were constructed by in vivo site-directed mutagenesis, replacing the designated sequence with 5′-GTAACATGGATCCCGTGCTA-3′ (a BamHI site flanked by random sequences). Hsp70 and Hsp90 are essential, so conditional strains were constructed. Hsp90 is encoded by two genes, HSP82 and HSC82; HSP82 was deleted while HSC82 was placed under control of the GAL1 promoter. Hsp70 is encoded by four genes, SSA1-4, although SSA3 is not normally expressed. SSA2 and SSA4 were deleted, and SSA1 was placed under GAL1 control. Strains containing only ssa2Δ ssa4Δ or hsp82Δ were used as controls for the carbon source.
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype |
|---|---|
| FY2607 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ hap1 |
| FY2608 | MATaarg4-12 ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ hap1 |
| FY2609 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 |
| FY2610 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 |
| FY2611 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ hap1Δ0::kanMX |
| FY2612 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ myc-HAP1 |
| FY2613 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ myc-HAP1 |
| FY2618 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 erg5-3 |
| FY2619 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 erg5-4 |
| FY2620 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 erg5-5 |
| FY2621 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 erg5-6 |
| FY2622 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 hsp82Δ0::KanMX |
| FY2623 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ myc-HAP1 erg5-2 |
| FY2624 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ myc-HAP1 erg5-2 |
| FY2625 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ myc-HAP1 erg5-1 |
| FY2626 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ myc-HAP1 erg5-1 |
| FY2627 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ myc-HAP1 erg5-1,2 |
| FY2628 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ myc-HAP1 erg5-1,2 |
| FY2631 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ myc-HAP1 ssa2Δ0::LEU2 ssa4Δ0::HIS3 |
| FY2635 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 hsp82Δ0::KanMX HIS3::GAL1pr-HSC82 |
| FY2636 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ myc-HAP1 ssa2Δ0::LEU2 ssa4Δ0::HIS3 KanMX::GAL1pr-SSA1 |
| FY2637 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 hem1Δ0::KanMX |
| FY2638 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ hem1Δ0::KanMX hap1Δ0::KanMX |
| FY2639 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ myc-HAP1 erg5-10 |
| FY2640 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ myc-HAP1 erg5-11 |
| FY2641 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ myc-HAP1 erg5-12 |
| FY2642 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ myc-HAP1 erg5-13 |
| FY2643 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ myc-HAP1 erg5-14 |
| FY2644 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ myc-HAP1 erg5-4 |
| FY2645 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ myc-HAP1 erg5-7 |
| FY2646 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ myc-HAP1 erg5-8 |
| FY2647 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ myc-HAP1 erg5-9 |
| FY2673 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 tup1Δ0::KanMX |
| FY2674 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HAP1 TUP1-myc |
| FY2675 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ hap1Δ0::KanMX TUP1-myc |
| O807 | MATα ura3Δ0 his3Δ0::HISG |
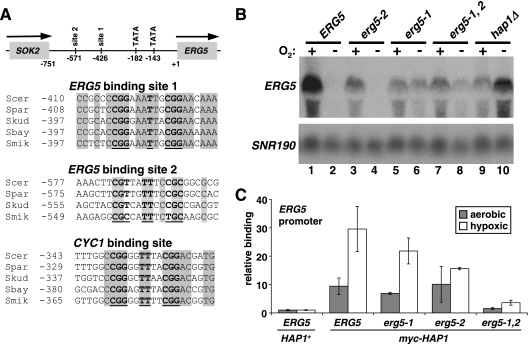
FIG. 5.
Two Hap1 binding sites contribute to ERG5 regulation. (A) At the top is a diagram of the structure of the ERG5 promoter, with numbers denoting locations relative to the ERG5 ATG. The two consensus TATA sequences and putative Hap1 binding sites are shown. Shown below is the alignment of the consensus Hap1 binding sites within the ERG5 promoter of the related yeast species: S. cerevisiae (Scer), S. paradoxus (Spar), S. kudravzevii (Skud), S. bayanus (Sbay), and S. mikitae (Smik). For site 2, there is no consensus site in S. bayanus. Also shown is the alignment for UAS1 from the CYC1 promoter. The numbers denote the first nucleotide of each of the sequences relative to ATG. Nucleotides shown in bold are consensus Hap1 positions, while the nucleotides in shaded boxes denote conserved positions. Site 1 is the reverse complement. (B) ERG5 mRNA levels were measured by Northern blotting in strains containing a wild-type ERG5 promoter (ERG5, FY2612), ERG5 Hap1 site 2 mutant (erg5-2, FY2624), ERG5 Hap1 site 1 mutant (erg5-1, FY2626), ERG5 with both sites mutant (erg5-1,2, FY2628), and hap1Δ (FY2611). Strains that are not hap1Δ contain the myc-HAP1 allele. Cells were grown hypoxically (−) or aerobically (+) for 4 h. Shown is one of the Northern blots from multiple experiments. The averages of the quantitations from multiple Northern analyses are as follows: lane 1, 9.4; lane 2, 1.0; lane 3, 6.1; lane 4, 1.6; lane 5, 6.4; lane 6, 2.9; lane 7, 3.6; lane 8, 2.8; lane 9, 3.8; lane 10, 13.2. (C) Hap1 binding to the ERG5 promoter (−323 to −189) was monitored by ChIP in the following strains: untagged HAP1 (ERG5, HAP1+, FY2610), myc-HAP1 (FY2612 or FY2613), myc-HAP1 erg5-1 (FY2625), myc-HAP1 erg5-2 (FY2623), and myc-HAP1 erg5-1,2 (FY2627). Shown are the means and standard errors for three independent experiments. Note that the data for untagged HAP1 and myc-HAP1 are also shown in Fig. 3.
Media and growth conditions.
Cells were grown at 30°C in YPD (1% yeast extract and 2% peptone supplemented with 2% glucose) except where indicated. YP galactose medium contained 1% yeast extract and 2% peptone supplemented with 2% galactose. For all experiments, mid-log-phase cells (approximately 1 × 107 to 2 × 107 cells/ml) were used for analysis. For hypoxic growth, 25- to 250-ml cultures growing in 250-ml or 500-ml flasks were continuously sparged with ultra-high-purity nitrogen at approximately 3 liters/min. A critical aspect of observing hypoxic effects after the growth of cultures in nitrogen was chilling the cultures by surrounding the flasks with ice for 5 min per 50 ml of culture, with continued exposure to nitrogen prior to the preparation of RNA. Aerobic cultures were chilled in an identical manner. A previous study (9) did not observe hypoxic repression of ERG genes, and the different results may be due to this difference in treatment of the cells. For hypoxic growth and certain other experiments described in the text, the media were supplemented with 20 μg/ml ergosterol (Sigma), 0.5% (vol/vol) Tween 80 (Sigma) as a source of unsaturated fatty acids, and 0.5% (vol/vol) ethanol. For supplementing growth media with heme, a 6.5 mg/ml heme (hemin; Sigma/BioChemika) solution in 0.1 M NaOH was prepared, incubated at 37°C for 1 h, and adjusted to pH 7.0 with HCl. The solution was added to the media within 3 days of preparation, 40 min before exposing the cells to hypoxic conditions. δ-Aminolevulinate (δ-ala; Sigma) was dissolved in water at 20 mg/ml and added to the media at the indicated concentration. Strains containing mutations in the HSP or SSA genes were maintained on YP galactose medium, shifted to YP galactose or YPD for 4 h, and then grown under aerobic or hypoxic conditions for an additional 4 h.
Expression microarray analysis.
Cells were grown under aerobic or hypoxic (supplemented with ergosterol, Tween 80, and ethanol) conditions for 8 h. RNA was isolated using a hot-phenol method (4) and reverse transcribed. The resulting cDNA was labeled with Cy3-dUTP or Cy5-dUTP, using a BioPrime kit (Invitrogen), and hybridized to yeast Y6.4 whole ORF microarrays (Toronto Microarray Centre, University Health Network). Fluorescence was quantitated on a GenePix 4000B scanner with GenePix 5.0 software (Axon Instrumentation). Expression values were subjected to total intensity and LOWESS normalization, using MIDAS software (TIGR) (46). Three biological replicates were performed for wild-type (strain FY2609) and hap1Δ (strain FY2611) cells, and the normalized data are presented in Table S1 in the supplemental material. The mRNA levels that were significantly changed between aerobic and hypoxic growth of HAP1 wild-type cells were identified by a SAM (statistical analysis of microarrays) package, using a false discovery rate cutoff of 9.81% (see Table S1 in the supplemental material) (57). In this way, 878 aerobic and 759 hypoxic genes were identified, and subsequent analyses focused on the genes that changed more than twofold (197 and 217 genes, respectively) (see Table S1 in the supplemental material). Hap1-regulated genes were defined as those that changed more than twofold in hap1Δ compared to the wild type. Hap1 binding to yeast gene promoters was determined previously by ChIP-chip analysis (22); for the current study, a P value of <0.01 was used as a threshold for statistically significant binding. Forty-two of the 6,307 genes (0.7%) represented on the microarray are involved in ergosterol metabolism; 5 out of 10 (50%) of the Hap1 genes repressed in hypoxic conditions were in this category, a significant enrichment (P < 10−162, as determined by the chi-square test for independence).
Northern hybridization analysis.
Northern hybridization analysis was performed as previously described (4). The following probes were amplified by PCR for random 32P labeling (positions are relative to that of ATG): CYC1 (+6 to + 279), ERG2 (+189 to + 546), ERG5 (+47 to + 1379), ERG11 (+15 to + 731), HIS3 (−27 to + 376), HSP82 (−93 to + 1269; detects both HSP82 and HSC82), SNR190 (entire gene), and SSA1 (+238 to + 831; detects all SSA genes). Note that, based on a comparison to rRNA levels, the SNR190 transcript used as a loading control did not change under the different growth conditions in these experiments. Expression was quantitated on a Molecular Dynamics Storm 860 phosphorimager, using ImageQuant TL software (Amersham Biosciences), and the values are presented in the appropriate figure legends. For ERG5 Northern hybridization analysis, the value for each strain was first normalized to that of the loading control, SNR190, and this ratio was then normalized to the ratio for a wild-type strain after a 4-h shift to hypoxic conditions. The numbers presented are the averages of multiple experiments that were performed at least twice and often multiple times.
ChIP analysis.
ChIP experiments to examine Hap1 binding in vivo were performed as previously described (41), using strains that contained the myc-HAP1 or myc-TUP1 allele. Formaldehyde cross-linking was carried out for 20 min (myc-HAP1 strain) or 90 min (TUP1-myc strain). For immunoprecipitation, we used an anti-myc antibody (clone A14; Santa Cruz Biotechnology). DNA was quantitated by real-time PCR, using a Stratagene MX3000P (2). Binding to the indicated region was calculated relative to binding to an ORF-free region on chromosome I and was normalized to the level of input chromatin. As a control for shearing efficiency, no binding was detected in the two ORFs adjacent to the ERG5 regulatory region (positions are relative to that of ERG5 ATG): ERG5 (+1272 to +1420) and SOK2 (−2174 to −2012) (data not shown). To control for background binding, we normalized myc-Hap1 or Tup1-myc binding to the amount of binding detected by ChIP in a strain expressing untagged (wild-type) HAP1 or TUP1. Each experiment was performed at least three times. The primer sequences are listed in Fig. S6 at http://genetics.med.harvard.edu/∼winston/Winston%20Lab%20Links.html.
Microarray data accession numbers.
The microarray data discussed in this publication have been deposited in the NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database and are accessible through GEO series accession number GSE8613.
RESULTS
Microarray experiments identify new Hap1 targets and suggest that Hap1 is a hypoxic repressor.
Heme-dependent activation by Hap1 during aerobic growth is known to play a widespread role in oxygen-mediated transcription in S. cerevisiae (65). However, previous studies have suggested that Hap1 also has other roles in response to changing oxygen levels (11, 12, 17, 45, 58, 59). In order to determine the global role of Hap1, we performed microarray analyses in the sequenced and systematically characterized S. cerevisiae S288C genetic background. While previous microarray studies have been done to study Hap1-mediated regulation in S. cerevisiae (5, 54), none have been done in the S288C background. Such an analysis in the S288C background would likely benefit from the many types of genome-wide studies that have been performed with S288C strains, including genome-wide analysis of Hap1 binding (22). One drawback of S288C is that it contains a mutant hap1 allele that causes a partial loss of function (18). Therefore, for our studies, we replaced the S288C hap1 mutant gene with either a wild-type HAP1 sequence (hereafter referred to as HAP1) or a complete deletion of HAP1 (hap1Δ; see Materials and Methods). Using these strains, we carried out microarray experiments to compare mRNA levels during aerobic and hypoxic growth for both HAP1 and hap1Δ strains.
Consistent with previous studies, our results show that there are a large number of aerobic-specific and hypoxic-specific mRNAs in HAP1 strains (5, 32-34, 53, 54). Specifically, our experiments identified 197 aerobic genes and 217 hypoxic genes (see Table S1 in the supplemental material; see Materials and Methods for statistical analysis). Approximately 29% of the hypoxic genes, including TIR1 and HEM13, and ∼14% of the aerobic genes, including CYC1 and ROX1, have been previously described (13, 20, 39, 64). Furthermore, our microarray results for hap1Δ mutants show that approximately 58% of hypoxic mRNAs and approximately 45% of aerobic mRNAs are dependent upon Hap1 for their regulation (see Table S1 in the supplemental material). Thus, our results are consistent with those of previous studies that showed that S. cerevisiae regulates transcription of hundreds of genes in response to different oxygen levels and that Hap1 plays a prominent role in this regulation.
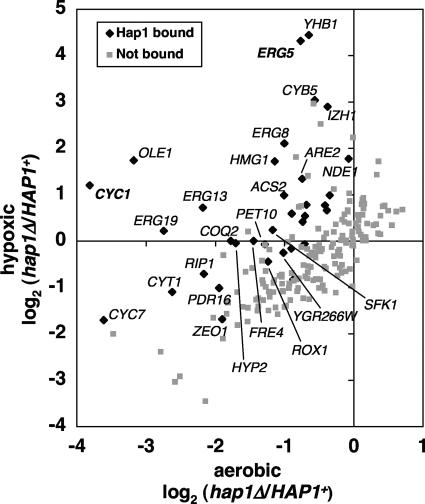
To gain insight into the number of genes that are directly controlled by Hap1, we compared our microarray data to previously published results that analyzed genome-wide binding by Hap1. Although these binding studies (22) were performed under aerobic conditions, this comparison suggests that all of the Hap1-dependent genes expressed in hypoxic conditions are indirectly regulated by Hap1, consistent with the previously proposed model that this mode of Hap1 control occurs via regulation of the repressor Rox1 (28). In contrast, among the 89 Hap1-dependent aerobic genes that we identified, 25 (28%) are putative direct targets of Hap1 (Fig. 1; see the gene list in Fig. S2 at http://genetics.med.harvard.edu/∼winston/Winston%20Lab%20Links.html) (22). These genes fell into two overlapping sets with respect to their regulation by Hap1: aerobically activated and hypoxically repressed. First, in agreement with its known role as an aerobic activator, Hap1 was required for the aerobic expression of 19 (76%) of these 25 putative direct targets. Six of these 19 genes, including ROX1, CYC1, and CYC7, were previously known to be regulated by Hap1 (42), while most of the genes represent previously unknown Hap1 targets. The latter group contains genes involved in respiration (for example, COQ2 and RIP1) and lipid metabolism (ERG13, ERG19, SFK1, and PDR16), among other functions. We did not identify all known Hap1 target genes by this analysis, perhaps due to differences in strain backgrounds or hypoxic conditions. For example, QCR2 was previously identified as an aerobic gene activated by Hap1 (14). Our analysis suggests that QCR2 is indeed activated by Hap1 but that it is not aerobic nor significantly bound by Hap1 (although the level of binding is quite close to the threshold) (see Table S1 in the supplemental material).
FIG. 1.
Hap1 regulates aerobic genes during both aerobic and hypoxic growth conditions. Expression microarray experiments revealed 197 genes that were significantly downregulated in wild-type cells upon a shift to hypoxic growth. The hap1Δ/HAP1 expression ratio of these 197 genes is shown for aerobic and hypoxic cells. The promoters of 34 of these genes (black diamonds) were significantly bound by Hap1 under aerobic conditions. Points labeled with gene names changed more than twofold in hap1Δ compared to HAP1. Statistical analyses are described in the Materials and Methods section.
In addition to its role as an aerobic activator, Hap1 was required for the repression of 10 genes during hypoxic growth, a condition in which Hap1 has been generally viewed as inactive (42, 65). Half of the Hap1-repressed genes that were detected by our microarrays (HMG1, CYB5, ERG5, ERG8, and ARE2) are involved in the metabolism of ergosterol, many more than would be expected by chance (P < 10−162; see Materials and Methods). These data suggest that Hap1 plays a significant but to date largely unexamined role as a hypoxic repressor. This role is the focus of the remaining experiments in this study.
Hap1 is a hypoxic repressor of ERG genes.
We wanted to test by independent methods whether Hap1 functions as a direct transcriptional repressor under hypoxic conditions. Since many of the genes that were identified as repressed by Hap1 during hypoxic growth are involved in ergosterol metabolism, we focused on three ergosterol biosynthetic genes (ERG2, ERG5, and ERG11). One of these genes, ERG5, was in the group of 10 aerobic genes identified by our microarray analysis as direct targets of Hap1 repression during hypoxic growth. ERG2 and ERG11 were initially identified by our microarrays as genes subject to Hap1 repression during hypoxic growth (see Table S1 in the supplemental material), and our subsequent analysis, described below, has shown that they are indeed aerobic genes. Furthermore, genome-wide analysis suggests that Hap1 binds to their promoters (22). In addition, ERG11 was previously suspected to be Hap1 repressed (12, 17, 59). All three genes contain consensus Hap1 binding sites that are conserved among the sequenced sensu stricto yeast strains (ERG5 is described below; for ERG2 and ERG11, see Fig. S3 at http://genetics.med.harvard.edu/∼winston/Winston%20Lab%20Links.html).
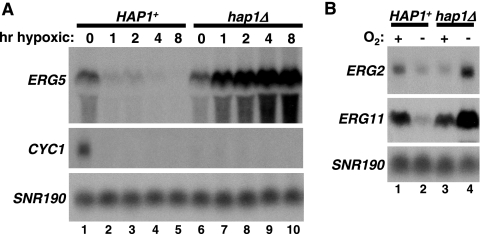
To measure mRNA levels for ERG2, ERG5, and ERG11 during aerobic and hypoxic growth, we performed Northern analysis. Our results (Fig. 2) show that ERG2, ERG5, and ERG11 are all expressed during aerobic growth and repressed during hypoxic growth in HAP1 cells. Importantly, the hypoxic repression of these genes is Hap1 dependent, as they are no longer repressed, but rather are induced, in a hap1Δ mutant. In addition, Hap1 appears to have a modest role in activation of ERG5 during aerobic growth, as hap1Δ caused a reproducible decrease in ERG5 aerobic mRNA levels. However, hap1Δ did not cause a detectable defect in ERG2 or ERG11 activation during aerobic growth. An activating role for Hap1 in the aerobic expression of ERG genes has been previously shown (9, 29, 52, 56). To contrast the role of Hap1 as a hypoxic repressor with its role as an aerobic activator (44), we also measured CYC1 mRNA levels in HAP1 and hap1Δ strains. In HAP1 strains, CYC1 transcription mimics that of the three ERG genes, on during aerobic growth and off during hypoxic growth (Fig. 2A). In striking contrast to the regulation of ERG2, ERG5, and ERG11, however, a hap1Δ mutation abolished aerobic expression of CYC1, while not affecting its lack of expression during hypoxic growth. These results demonstrate that Hap1 plays a significant role in transcriptional repression of a specific set of genes during hypoxic growth.
FIG. 2.
Hap1 is both an aerobic activator of CYC1 and a hypoxic repressor of ERG genes. (A) ERG5 and CYC1 mRNA levels were measured by Northern analysis in a HAP1 strain (FY2609) and a hap1Δ strain (FY2611) grown in hypoxic conditions for the indicated times. Shown is a typical Northern blot with the averages of the results of multiple Northern analyses (as described in Materials and Methods) as follows: lane 1, 5.4; lane 2, 1.7; lane 3, 2.0; lane 4, 1.0; lane 5, 0.9; lane 6, 4.5; lane 7, 8.7; lane 8, 11.8; lane 9, 15.9; lane 10, 19.8. hr, hour. (B) Northern analyses were performed to measure ERG2 and ERG11 mRNA levels in HAP1 (FY2609) and hap1Δ (FY2611) strains grown aerobically (+) and hypoxically (−) for 8 h. The averages of the quantitations from Northern analyses for ERG2 are as follows: lane 1, 1.8; lane 2, 1.0; lane 3, 1.3; lane 4, 4.8. The averages of the quantitations from Northern analyses for ERG11 are lane 1, 2.9; lane 2, 1.0; lane 3, 2.7; lane 4, 7.3.
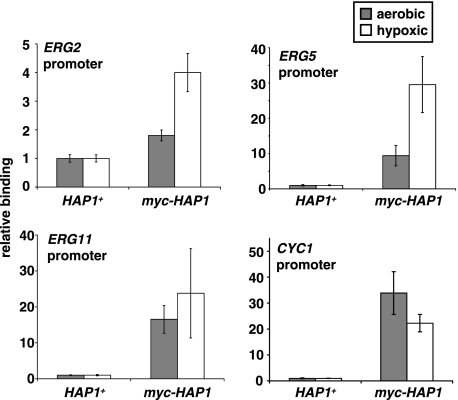
Previous studies that examined Hap1 DNA binding throughout the genome (22) and specifically at the ERG3 gene (9) used aerobic growth conditions. To determine whether Hap1 acts directly to repress transcription of ERG2, ERG5, and ERG11 during hypoxic growth, we performed Hap1 ChIP experiments to compare Hap1 binding in both growth conditions. In this experiment, we included CYC1 for comparison. Our results (Fig. 3) show that Hap1 is specifically associated with the promoters of ERG2, ERG5, and ERG11 under both aerobic and hypoxic conditions and suggest that there is a greater level of binding under hypoxic conditions when Hap1 represses transcription of these genes. The level of binding to ERG2 was significantly less than to ERG5 and ERG11, correlating with the more modest regulation of this gene by Hap1. As expected, Hap1 bound specifically to the CYC1 promoter under aerobic conditions, when Hap1 is known to activate CYC1 (Fig. 3). Under hypoxic conditions, Hap1 was also bound to CYC1, despite the fact that Hap1 does not function at CYC1 under these conditions. Taken together, our microarray, Northern analysis, and ChIP results are most consistent with the conclusion that Hap1 has two distinct and direct regulatory functions in vivo: transcriptional activation during aerobic growth and transcriptional repression during hypoxic growth.
FIG. 3.
Hap1 binds to the promoters of ERG and CYC1 genes under aerobic and hypoxic conditions. Hap1 binding was measured by ChIP (see Materials and Methods), using an α-myc antibody, and by quantitating the following gene sequences by real-time PCR: ERG2 (−491 to −371), ERG5 (−323 to −189), ERG11 (−607 to −490), and CYC1 (−397 to −268). Cells expressing Hap1 fused to a myc epitope tag (myc-HAP1, strain FY2612 or FY2613) or untagged HAP1 (HAP1+, strain FY2609 or FY2610) were grown under aerobic and hypoxic conditions for 4 h. Shown are the means and standard errors of at least three independent experiments.
Tup1 is required for Hap1-mediated repression.
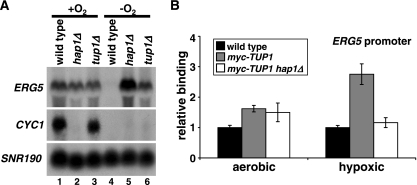
Transcriptional repressors often function through the recruitment of corepressor complexes. One candidate corepressor complex to function with Hap1 in repression is Tup1/Ssn6 (40), previously shown to play roles in repression during hypoxic growth (for examples, see references 1, 8, and 11). We tested whether Tup1/Ssn6 is required for Hap1-mediated repression of ERG5. Our Northern blots show that Tup1, like Hap1, is required for ERG5 repression under hypoxic conditions but has little role in ERG5 aerobic activation (Fig. 4A). In contrast, neither the aerobic activation nor the hypoxic repression of CYC1 is dependent upon Tup1, consistent with a role for Tup1 specifically in Hap1-dependent repression. To test whether the role of Tup1 is direct, we performed ChIP experiments (Fig. 4B). These experiments revealed that Tup1 is physically associated with the ERG5 promoter preferentially under hypoxic conditions when the gene is repressed. Furthermore, the association of Tup1 is Hap1 dependent. These results strongly suggest that Hap1 functions as a direct transcriptional repressor under hypoxic conditions, at least in part by the recruitment of the corepressor Tup1/Ssn6.
FIG. 4.
Hap1 represses ERG5 through recruitment of the Tup1 corepressor. (A) ERG5 and CYC1 expression were monitored by Northern blotting of wild-type (FY2609), hap1Δ (FY2611), and tup1Δ (FY2673) cells grown hypoxically (−O2) or aerobically (+O2) for 4 h. The averages of the quantitations from Northern analyses are as follows: lane 1, 9.4; lane 2, 3.8; lane 3, 6.5; lane 4, 1.0; lane 5, 13.2; lane 6, 7.3. (B) Tup1 binding to the ERG5 promoter (−323 to −189) was monitored by ChIP experiments in aerobic or hypoxic conditions, using an untagged (wild-type, FY2609), TUP1-myc (FY2674), or TUP1-myc hap1Δ (FY2675) strain. Shown are the means and standard errors for three independent experiments.
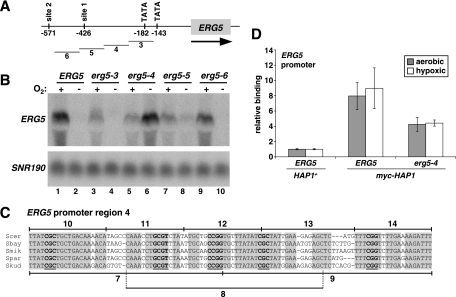
Multiple Hap1 binding sites contribute to ERG5 regulation.
Studies of DNA binding by Hap1 to known target genes have identified a consensus site for Hap1 binding that contains two tandem CGG triplets separated by 6 bp, although functional variations of this consensus have been identified (43, 65). We identified two putative Hap1 sites within the 750 bp between the ERG5 start codon and the stop codon of the adjacent gene (SOK2). Both sites are conserved among the Saccharomyces sensu stricto yeast species (7), although the proximal site 1 exhibits greater conservation than site 2 (Fig. 5A). To test whether the sites are important for ERG5 regulation, the 12-bp sites were mutated and then analyzed for effects on ERG5 mRNA levels during both aerobic and hypoxic growth. Northern blotting revealed that while each site is required for full activation of ERG5 during aerobic growth, only the site closer to ERG5, site 1, is required for repression of ERG5 during hypoxic growth (Fig. 5B). In this mutant (erg5-1), some derepression of ERG5 is observed during hypoxic growth, although the level is significantly less than that seen in a hap1Δ mutant (Fig. 5B, compare lane 6 to lane 10). The double site mutant (erg5-1,2) (Fig. 5B, lanes 7 and 8) exhibited an ERG5 expression pattern similar to that of the site 1 mutant, further suggesting that only site 1 plays a role in ERG5 repression. These results demonstrate that these two Hap1 binding sites participate in ERG5 regulation. However, they are not sufficient for conferring the full level of Hap1-dependent repression of ERG5 that occurs during hypoxic growth.
To test whether the binding of Hap1 to the ERG5 promoter depends on site 1 and site 2, we performed ChIP experiments using wild-type and site mutant strains. Our results (Fig. 5C) show that each site contributes to Hap1 binding at ERG5. In the individual site 1 or site 2 mutants, there were modest decreases in Hap1 binding, while in the double mutant, Hap1 binding was greatly diminished, although it was still greater than background levels. From these results, we concluded that Hap1 binds to each site in vivo.
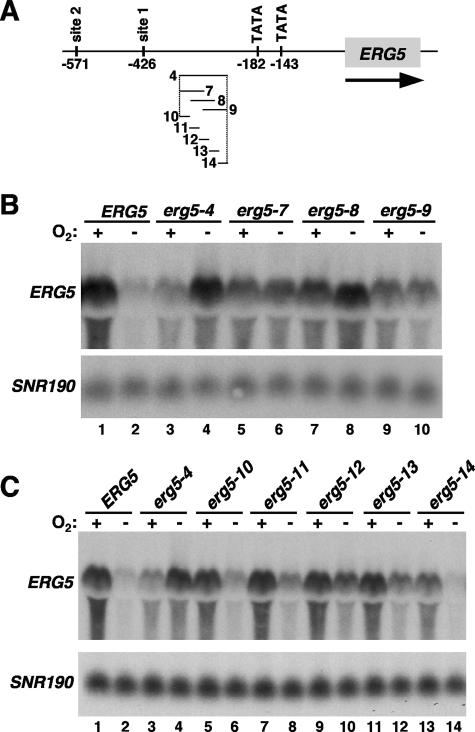
Our analysis of the two Hap1 consensus binding sites suggested that additional ERG5 promoter elements are likely required for ERG5 repression during hypoxic growth. To identify such promoter elements, we systematically mutated 100-bp sequences in the large intergenic region 5′ of ERG5 (Fig. 6A). Northern blots of strains containing these mutations showed that two regions, 4 and 5, are required for full repression. The sequences changed by mutation 5 contain Hap1 binding site 1, and mutation 5 had a modest effect on ERG5 repression, similar to that of mutation 1 (compare erg5-1 in Fig. 5B with erg5-5 in Fig. 6B). In contrast, mutation 4 caused a dramatic effect on ERG5 repression, similar to that caused by a hap1Δ mutation. Interestingly, although this 100 bp does not contain a consensus Hap1 binding site, it does contain five conserved Hap1 half-sites, more than would be expected by chance (P < 10−26) (Fig. 6C). Such half-sites have been shown to be sufficient for Hap1 binding in certain sequence contexts (25, 43). Indeed, mutating region 4 caused a reduction in Hap1 binding at ERG5 (Fig. 6D) that was similar in degree to the mutation of either of the full Hap1 sites (Fig. 5C).
FIG. 6.
Additional ERG5 promoter elements are important for repression. (A) Structure of the ERG5 promoter denoting regions 3 to 6 that were mutated. (B) ERG5 mRNA levels were measured by Northern blotting in strains containing the following ERG5 promoter mutations: ERG5 (FY2609), erg5-3 (FY2618), erg5-4 (FY2619), erg5-5 (FY2620), or erg5-6 (FY2621). Cells were grown aerobically (+) or hypoxically (−) for 4 h. Shown is one of the Northern blots from multiple experiments. The averages of the quantitations are as follows: lane 1, 9.4; lane 2, 1.0; lane 3, 6.2; lane 4, 0.8; lane 5, 4.5; lane 6, 9.2; lane 7, 4.4; lane 8, 2.5; lane 9, 9.7; lane 10, 0.8. (C) Conservation of region 4 in the sequenced sensu stricto yeast species S. cerevisiae (Scer), S. paradoxus (Spar), S. kudravzevii (Skud), S. bayanus (Sbay), and S. mikitae (Smik). Putative Hap1 half-sites in the ERG5 promoter are denoted in bold and are underlined, while the nucleotides in the shaded boxes denote conserved nucleotides. Regions 7 to 14 are the same regions noted and analyzed for Fig. 7. Five conserved half-sites were found in region 4, more than the 0.2 CGG or CGC sites expected by chance (P < 10−26, as determined by the chi-square test for independence). The background rate was determined by counting the number of conserved sites (on either strand) within 5,000 bp of the promoter sequence (specifically within −100 to −600 bp of ATG) of 10 randomly chosen genes. (D) Hap1 binding to the ERG5 promoter (−482 to −367) was monitored by ChIP experiments with the following strains: untagged HAP1 (HAP1+ ERG5, strain FY2610), myc-HAP1 (strain FY2612), and myc-HAP1 erg5-4 (strain FY2644). Shown are the means and standard errors of three independent experiments. The lower level of Hap1 binding to the ERG5 promoter compared to the results shown in Fig. 3 and 5C may be due to strain or primer differences; the primers used for the experiments in Fig. 3 and 5C could not be used here because one of the primers hybridized to the mutated region 4.
To define more precisely the sequences within region 4 that are required for repression, we constructed and analyzed mutations that change shorter sequences. First, three mutations that alter 50 bp each were constructed within the region 4 sequence (mutations 7, 8, and 9) (Fig. 7A and 6C). Our results (Fig. 7B) show that each one of these mutations confers a significant level of derepression, with the strongest derepression observed in the region 8 mutant, which alters three of the five half-sites. Then we constructed a series of five mutations across region 4 that altered only 20 bp each. All of these changes caused derepression, with the strongest effects observed when mutating the middle of region 4 (Fig. 7C), consistent with the pattern of derepression seen with the 50-bp changes. These results demonstrate that besides Hap1 binding site 1, another region of the ERG5 promoter that contains multiple Hap1 half-sites is required for Hap1-mediated repression. The promoters of ERG2, ERG11, and CYC1 also contain multiple conserved half-sites (see Fig. S4 at http://genetics.med.harvard.edu/∼winston/Winston%20Lab%20Links.html). The fact that such sites are present at both Hap1-repressed and Hap1-activated genes suggests that they are not the determining factor in the role of Hap1 at a promoter.
FIG. 7.
Multiple sequences within ERG5 region 4 are required for repression. (A) Structure of the ERG5 promoter denoting regions 4 and 7 to 14 that were mutated. (B) Effect of 50-bp mutations in region 4 on ERG5 expression. Northern blotting was performed for strains containing the following ERG5 promoter alleles: ERG5 (FY2612), erg5-4 (FY2644), erg5-7 (FY2645), erg5-8 (FY2646), and erg5-9 (FY2647). The averages of the quantitations from multiple Northern blotting experiments are as follows: lane 1, 9.4; lane 2, 1.0; lane 3, 3.7; lane 4, 8.1; lane 5, 6.0; lane 6, 4.1; lane 7, 6.2; lane 8, 4.3; lane 9, 3.9; lane 10, 2.6. (C) Effect of 20-bp mutations on ERG5 expression. Northern blotting was performed for strains containing the following ERG5 promoter alleles: ERG5 (FY2612), erg5-4 (FY2644), erg5-10 (FY2639), erg5-11 (FY2640), erg5-12 (FY2641), erg5-13 (FY2642), and erg5-14 (FY2643). Cells were grown aerobically (+) or hypoxically (−) for 4 h. The averages of the quantitations from Northern analyses are as follows: lane 1, 9.4; lane 2, 1.0; lane 3, 3.7; lane 4, 8.1; lane 5, 4.9; lane 6, 1.7; lane 7, 5.6; lane 8, 2.1; lane 9, 5.1; lane 10, 3.3; lane 11, 6.1; lane 12, 2.9; lane 13, 9.2; lane 14, 1.7.
Hsp70 is partially required for ERG5 repression, while Hsp90 is dispensable.
Previous studies have shown that Hap1 is physically associated with the chaperones Hsp70 and Hsp90 and that these associations are important in regulating Hap1-mediated activation of CYC1 (24, 35, 36). To test whether these proteins also regulate Hap1 repression of ERG5, we analyzed ERG5 mRNA levels in strains that can be depleted for these essential chaperones (see Materials and Methods). Briefly, to test the role of Hsp70, we used a mutant strain in which the one remaining Hsp70 gene, SSA1, was under the control of the GAL1 promoter. Our results show that when cells are depleted for Hsp70, there is a modest derepression of ERG5 during hypoxic growth (see Fig. S5A at http://genetics.med.harvard.edu/∼winston/Winston%20Lab%20Links.html; compare lane 4 to lane 8). Contrary to previous results (24), however, depletion of Hsp70 in our strains had no effect on the repression of CYC1 (see Fig. S5A at http://genetics.med.harvard.edu/∼winston/Winston%20Lab%20Links.html; compare lane 4 to lane 8) but instead impaired activation (Fig. S5A at http://genetics.med.harvard.edu/∼winston/Winston%20Lab%20Links.html; compare lane 2 to lane 6). We next tested the role of Hsp90 by deleting the HSP82 gene and placing the HSC82 gene under GAL1 control. In contrast to earlier observations (36), Hsp90 did not appear to play a role in CYC1 activation in our strains (Fig. S5B). Upon Hsp90 depletion, the levels of CYC1 mRNA showed the same mild reduction in both the control and depletion strains, possibly a result of the shift in carbon source. Similarly, there was no effect on ERG5 mRNA levels under any growth condition. Taken together, our results suggest that Hsp70 contributes modestly to the ability of Hap1 to serve as a repressor during hypoxic growth. However, the small effect of Hsp70 depletion on ERG5 derepression in comparison to the large effect of a hap1Δ mutation suggests that additional factors play significant roles in determining the activity of Hap1.
ERG genes and CYC1 are differentially regulated at intermediate heme levels.
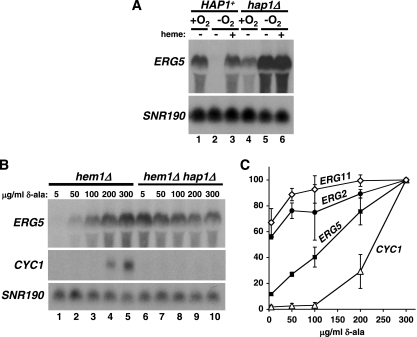
Our studies have shown that ERG5 and other genes are repressed during hypoxic growth by Hap1. As activation of CYC1 and other aerobic genes by Hap1 is heme dependent (19, 65), we tested whether repression of ERG5 by Hap1 is also regulated by heme. To do this, we added heme to cells grown hypoxically, a condition in which heme is normally absent, and measured ERG5 mRNA levels. Our results show that the addition of heme causes derepression of ERG5 transcription (Fig. 8A, compare lanes 2 and 3). In contrast, the addition of heme had no effect on ERG5 expression in hap1Δ cells (Fig. 8A, compare lanes 5 and 6). Thus, Hap1 is a repressor of ERG5 only in the absence of heme.
FIG. 8.
ERG5 and CYC1 are differentially regulated at intermediate heme levels. (A) ERG5 expression was monitored by Northern blotting in HAP1+ (FY2609) and hap1Δ (FY2611) cells grown hypoxically (−O2) or aerobically (+O2) for 8 h in the presence (+) or absence (−) of 50 μg/ml heme. The averages of quantitations are as follows: lane 1, 7.5; lane 2, 1.0; lane 3, 15.5; lane 4, 6.1; lane 5, 25.2; lane 6, 20.6. (B) Northern blotting was performed to measure ERG5 and CYC1 mRNA levels in hem1Δ (FY2637) and hem1Δ hap1Δ (FY2638) mutants. Cells were grown in media containing the indicated concentrations of δ-ala for 4 h. (C) ERG2, ERG5, ERG11, and CYC1 mRNA levels in hem1Δ cells were quantitated as described in the Materials and Methods section. Shown are the means and standard errors for three independent experiments.
The two roles of Hap1, aerobic activation of CYC1 and hypoxic repression of ERG genes, appear to accomplish the same expression pattern of aerobic-specific transcription. To test whether there might be differences of biological consequence between these two modes of regulation by Hap1, we measured ERG2, ERG5, ERG11, and CYC1 mRNA levels at intermediate heme levels. To do this, we used a hem1Δ mutant which cannot synthesize δ-ala, a precursor in the synthesis of heme. In this mutant, heme levels can be adjusted by adding different amounts of δ-ala to the growth medium. Our results (Fig. 8B and C) show that, as expected, when δ-ala levels (and heme levels) were increased, ERG and CYC1 mRNA levels increased. However, there was a clear difference between the two sets of genes, as the three ERG genes were expressed at low levels of δ-ala while CYC1 was not detectably expressed until significantly higher levels of δ-ala were added. As expected, in the absence of Hap1, ERG5 was constitutively derepressed while CYC1 was not detectably transcribed, demonstrating that Hap1 mediated these changes in expression in response to the changes in heme levels (Fig. 8B). These results show that the different modes of Hap1 regulation result in different levels of gene expression at intermediate oxygen levels.
DISCUSSION
In order to understand oxygen-mediated transcription in S. cerevisiae, we set out to identify the direct targets of the transcriptional regulator Hap1. Our microarray experiments, combined with previous in vivo DNA binding studies (22), allowed a comprehensive identification of putative Hap1 direct targets, many of which were previously unidentified. Surprisingly, we found that a significant number of these genes are repressed by Hap1 during hypoxic growth, including many involved in ergosterol metabolism. We focused on three genes in the ergosterol biosynthetic pathway that Hap1 represses (ERG2, ERG5, and ERG11), as well as one gene that Hap1 activates, CYC1, and showed by ChIP experiments that Hap1 binds to their regulatory regions during both hypoxic and aerobic growth. Additional experiments have suggested a model in which Hap1 represses transcription during hypoxic growth by recruiting the corepressor Tup1/Ssn6. Furthermore, our experiments, combined with previous studies, have established that the dual activity of Hap1 is controlled by heme levels, with activation dependent on the presence of heme and repression dependent upon its absence. Finally, our results suggest that the two activities of Hap1 allow it to differentially modulate gene expression at intermediate oxygen levels, a condition often encountered by cells. Taken together, our findings alter the prevailing view of Hap1 as an activator of aerobic genes, providing strong evidence that it plays an equally significant role as a repressor under hypoxic conditions.
One key question raised by these results is how heme controls the switch of Hap1 between activator and repressor. Mammalian nuclear hormone receptors are one previously studied example of factors that can act both positively and negatively. These factors repress transcription in the absence of ligand through the recruitment of corepressor complexes. In the presence of ligand, the corepressor complexes are displaced in favor of coactivators (3, 63). In the case of Hap1, heme may act in a similar manner, dictating the differential association of factors with Hap1. Indeed, we have shown here that Tup1, a subunit of the Tup1/Ssn6 corepressor complex, is required for ERG5 repression and that Hap1 recruits Tup1 to the ERG5 promoter specifically during hypoxic growth. Hap1 presumably functions as an aerobic activator through the recruitment of a coactivator yet to be identified. It will be important to determine how heme regulates Hap1 binding to these and other factors.
Previous studies have yielded ambiguous results regarding the possibility that Hap1 might directly repress transcription during hypoxic growth. A number of studies had suggested that Hap1 can act as a repressor under such conditions (11, 12, 17, 45, 58, 59), and recent evidence has shown that Hap1 directly represses transcription of its own gene in a heme-independent fashion during aerobic growth (25). Prior to our studies, however, there was no conclusive evidence that Hap1 plays a direct role as a repressor during hypoxic growth. With respect to the ability of Hap1 to bind DNA during hypoxic growth, one study found that Hap1 isolated from heme-deficient cells could bind DNA in vitro (17) and a recent study also suggested that Hap1 can bind DNA in heme-deficient cells (25). Other studies, however, suggested that Hap1 has reduced affinity for DNA in the absence of heme (24, 35, 58, 65). Our ChIP and mutational results strongly suggest that Hap1 can bind DNA in vivo in the absence of heme to directly repress the transcription of particular genes. The fact that Hap1 binds in vivo under both aerobic and hypoxic conditions suggests that the conversion of Hap1 between activator and repressor may occur at a step subsequent to DNA binding. There are several examples of transcription factors that bind to their sites under conditions where they do not normally act, including the well-characterized activator Gal4 (15, 47).
A related issue is the nature of the promoter elements at Hap1 target genes that determine whether Hap1 acts as an activator or repressor. Our results have shown that the region of the ERG5 promoter that is most strongly required for Hap1-dependent repression contains five conserved Hap1 half-sites. Such half-sites have been shown to allow Hap1 binding (43) and may be responsible for Hap1 binding to its own promoter, which lacks full Hap1 sites (25). An examination of three genes that are repressed by Hap1 and one gene that is activated by Hap1 (Fig. S4) showed that all four contain several half-sites in a variety of positions, making it unlikely that half-sites are sufficient to confer Hap1-mediated repression. In addition, previous studies suggested that the specific sequence of Hap1 binding sites can determine the level of activation by Hap1 without affecting its DNA binding by an allosteric effect (21, 30). Similarly, the sequence of binding sites might determine repression by Hap1. Additional experiments are required to resolve the role of promoter elements in determining whether Hap1 acts as an activator or as a repressor.
In both humans and yeast, normal sterol levels are critical for proper membrane function. Sterol biosynthesis is tightly controlled, and it is well known that in humans aberrant cholesterol levels can lead to many serious diseases (16, 51). In S. cerevisiae, two transcription factors, Upc2 and Ecm22, have previously been demonstrated to be required for the activation of transcription of at least some ERG genes in response to low sterol levels (10, 60). Our work has extended our understanding of the regulation of ergosterol levels in S. cerevisiae by elucidating the central role played by Hap1 in modulating the level of ERG gene expression in response to oxygen levels. The finding that Hap1 does not fully repress ERG transcription at intermediate heme levels suggests that cells are able to synthesize ergosterol as long as there is some oxygen available. This ability may be critical for growth, given that oxygen excludes the import of exogenous ergosterol (37, 55). In contrast, the Hap1-activated gene required for aerobic respiration, CYC1, may not be expressed at intermediate heme levels, because cells can generate sufficient ATP in low-oxygen conditions through fermentation. Thus, the fact that the repression and activation functions of Hap1 respond differentially to intermediate heme levels allows S. cerevisiae to differentially regulate two key metabolic pathways in response to changing oxygen levels.
Supplementary Material
Acknowledgments
We thank Dominique Helmlinger, Lisa Laprade, Brendon Monahan, and Li Zhang for helpful comments on the manuscript.
M.J.H. was funded by a fellowship from the National Institutes of Health (F32GM71102). This work was supported by a grant to F.W. from the National Institutes of Health (GM45720).
Footnotes
Published ahead of print on 4 September 2007.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abramova, N. E., B. D. Cohen, O. Sertil, R. Kapoor, K. J. Davies, and C. V. Lowry. 2001. Regulatory mechanisms controlling expression of the DAN/TIR mannoprotein genes during anaerobic remodeling of the cell wall in Saccharomyces cerevisiae. Genetics 157: 1169-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aparicio, O., J. V. Geisberg, E. Sekinger, A. Yang, Z. Moqtaderi, and K. Struhl. 2005. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo, p. 21.3.1-21.3.33. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, Inc., Hoboken, NJ. [DOI] [PubMed]
- 3.Aranda, A., and A. Pascual. 2001. Nuclear hormone receptors and gene expression. Physiol. Rev. 81: 1269-1304. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. E. Struhl. 1991. Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York, NY.
- 5.Becerra, M., L. J. Lombardia-Ferreira, N. C. Hauser, J. D. Hoheisel, B. Tizon, and M. E. Cerdan. 2002. The yeast transcriptome in aerobic and hypoxic conditions: effects of hap1, rox1, rox3 and srb10 deletions. Mol. Microbiol. 43: 545-555. [DOI] [PubMed] [Google Scholar]
- 6.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14: 115-132. [DOI] [PubMed] [Google Scholar]
- 7.Christie, K. R., S. Weng, R. Balakrishnan, M. C. Costanzo, K. Dolinski, S. S. Dwight, S. R. Engel, B. Feierbach, D. G. Fisk, J. E. Hirschman, E. L. Hong, L. Issel-Tarver, R. Nash, A. Sethuraman, B. Starr, C. L. Theesfeld, R. Andrada, G. Binkley, Q. Dong, C. Lane, M. Schroeder, D. Botstein, and J. M. Cherry. 2004. Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 32: D311-D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crisp, R. J., E. M. Adkins, E. Kimmel, and J. Kaplan. 2006. Recruitment of Tup1p and Cti6p regulates heme-deficient expression of Aft1p target genes. EMBO J. 25: 512-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies, B. S., and J. Rine. 2006. A role for sterol levels in oxygen sensing in Saccharomyces cerevisiae. Genetics 174: 191-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davies, B. S., H. S. Wang, and J. Rine. 2005. Dual activators of the sterol biosynthetic pathway of Saccharomyces cerevisiae: similar activation/regulatory domains but different response mechanisms. Mol. Cell. Biol. 25: 7375-7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deckert, J., R. Perini, B. Balasubramanian, and R. S. Zitomer. 1995. Multiple elements and auto-repression regulate Rox1, a repressor of hypoxic genes in Saccharomyces cerevisiae. Genetics 139: 1149-1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Defranoux, N., M. Gaisne, and J. Verdiere. 1994. Functional analysis of the zinc cluster domain of the CYP1 (HAP1) complex regulator in heme-sufficient and heme-deficient yeast cells. Mol. Gen. Genet. 242: 699-707. [DOI] [PubMed] [Google Scholar]
- 13.Donzeau, M., J. P. Bourdineaud, and G. J. Lauquin. 1996. Regulation by low temperatures and anaerobiosis of a yeast gene specifying a putative GPI-anchored plasma membrane protein. Mol. Microbiol. 20: 449-459. [DOI] [PubMed] [Google Scholar]
- 14.Dorsman, J. C., and L. A. Grivell. 1990. Expression of the gene encoding subunit II of yeast QH2: cytochrome c oxidoreductase is regulated by multiple factors. Curr. Genet. 17: 459-464. [DOI] [PubMed] [Google Scholar]
- 15.Dudley, A. M., C. Rougeulle, and F. Winston. 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 13: 2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eberlé, D., B. Hegarty, P. Bossard, P. Ferré, and F. Foufelle. 2004. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie 86: 839-848. [DOI] [PubMed] [Google Scholar]
- 17.Fytlovich, S., M. Gervais, C. Agrimonti, and B. Guiard. 1993. Evidence for an interaction between the CYP1(HAP1) activator and a cellular factor during heme-dependent transcriptional regulation in the yeast Saccharomyces cerevisiae. EMBO J. 12: 1209-1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaisne, M., A. M. Becam, J. Verdiere, and C. J. Herbert. 1999. A ‘natural’ mutation in Saccharomyces cerevisiae strains derived from S288c affects the complex regulatory gene HAP1 (CYP1). Curr. Genet. 36: 195-200. [DOI] [PubMed] [Google Scholar]
- 19.Guarente, L., B. Lalonde, P. Gifford, and E. Alani. 1984. Distinctly regulated tandem upstream activation sites mediate catabolite repression of the CYC1 gene of S. cerevisiae. Cell 36: 503-511. [DOI] [PubMed] [Google Scholar]
- 20.Guarente, L., and T. Mason. 1983. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell 32: 1279-1286. [DOI] [PubMed] [Google Scholar]
- 21.Ha, N., K. Hellauer, and B. Turcotte. 1996. Mutations in target DNA elements of yeast HAP1 modulate its transcriptional activity without affecting DNA binding. Nucleic Acids Res. 24: 1453-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harbison, C. T., D. B. Gordon, T. I. Lee, N. J. Rinaldi, K. D. Macisaac, T. W. Danford, N. M. Hannett, J. B. Tagne, D. B. Reynolds, J. Yoo, E. G. Jennings, J. Zeitlinger, D. K. Pokholok, M. Kellis, P. A. Rolfe, K. T. Takusagawa, E. S. Lander, D. K. Gifford, E. Fraenkel, and R. A. Young. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431: 99-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hon, T., A. Dodd, R. Dirmeier, N. Gorman, P. R. Sinclair, L. Zhang, and R. O. Poyton. 2003. A mechanism of oxygen sensing in yeast. Multiple oxygen-responsive steps in the heme biosynthetic pathway affect Hap1 activity. J. Biol. Chem. 278: 50771-50780. [DOI] [PubMed] [Google Scholar]
- 24.Hon, T., H. C. Lee, A. Hach, J. L. Johnson, E. A. Craig, H. Erdjument-Bromage, P. Tempst, and L. Zhang. 2001. The Hsp70-Ydj1 molecular chaperone represses the activity of the heme activator protein Hap1 in the absence of heme. Mol. Cell. Biol. 21: 7923-7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hon, T., H. C. Lee, Z. Hu, V. R. Iyer, and L. Zhang. 2005. The heme activator protein Hap1 represses transcription by a heme-independent mechanism in Saccharomyces cerevisiae. Genetics 169: 1343-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jamieson, D. J. 1998. Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 14: 1511-1527. [DOI] [PubMed] [Google Scholar]
- 27.Kaelin, W. G. 2005. Proline hydroxylation and gene expression. Annu. Rev. Biochem. 74: 115-128. [DOI] [PubMed] [Google Scholar]
- 28.Keng, T. 1992. HAP1 and ROX1 form a regulatory pathway in the repression of HEM13 transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 12: 2616-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kennedy, M. A., R. Barbuch, and M. Bard. 1999. Transcriptional regulation of the squalene synthase gene (ERG9) in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1445: 110-122. [DOI] [PubMed] [Google Scholar]
- 30.King, D. A., L. Zhang, L. Guarente, and R. Marmorstein. 1999. Structure of HAP1-18-DNA implicates direct allosteric effect of protein-DNA interactions on transcriptional activation. Nat. Struct. Biol. 6: 22-27. [DOI] [PubMed] [Google Scholar]
- 31.Kwast, K. E., P. V. Burke, and R. O. Poyton. 1998. Oxygen sensing and the transcriptional regulation of oxygen-responsive genes in yeast. J. Exp. Biol. 201: 1177-1195. [DOI] [PubMed] [Google Scholar]
- 32.Kwast, K. E., L. C. Lai, N. Menda, D. T. James III, S. Aref, and P. V. Burke. 2002. Genomic analyses of anaerobically induced genes in Saccharomyces cerevisiae: functional roles of Rox1 and other factors in mediating the anoxic response. J. Bacteriol. 184: 250-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lai, L. C., A. L. Kosorukoff, P. V. Burke, and K. E. Kwast. 2005. Dynamical remodeling of the transcriptome during short-term anaerobiosis in Saccharomyces cerevisiae: differential response and role of Msn2 and/or Msn4 and other factors in galactose and glucose media. Mol. Cell. Biol. 25: 4075-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai, L. C., A. L. Kosorukoff, P. V. Burke, and K. E. Kwast. 2006. Metabolic-state-dependent remodeling of the transcriptome in response to anoxia and subsequent reoxygenation in Saccharomyces cerevisiae. Eukaryot. Cell 5: 1468-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lan, C., H. C. Lee, S. Tang, and L. Zhang. 2004. A novel mode of chaperone action: heme activation of Hap1 by enhanced association of Hsp90 with the repressed Hsp70-Hap1 complex. J. Biol. Chem. 279: 27607-27612. [DOI] [PubMed] [Google Scholar]
- 36.Lee, H. C., T. Hon, and L. Zhang. 2002. The molecular chaperone Hsp90 mediates heme activation of the yeast transcriptional activator Hap1. J. Biol. Chem. 277: 7430-7437. [DOI] [PubMed] [Google Scholar]
- 37.Lewis, T. A., F. R. Taylor, and L. W. Parks. 1985. Involvement of heme biosynthesis in control of sterol uptake by Saccharomyces cerevisiae. J. Bacteriol. 163: 199-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953-961. [DOI] [PubMed] [Google Scholar]
- 39.Lowry, C. V., and R. S. Zitomer. 1988. ROX1 encodes a heme-induced repression factor regulating ANB1 and CYC7 of Saccharomyces cerevisiae. Mol. Cell. Biol. 8: 4651-4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malavé, T. M., and S. Y. Dent. 2006. Transcriptional repression by Tup1-Ssn6. Biochem. Cell Biol. 84: 437-443. [DOI] [PubMed] [Google Scholar]
- 41.Martens, J. A., P. Y. Wu, and F. Winston. 2005. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 19: 2695-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mense, S. M., and L. Zhang. 2006. Heme: a versatile signaling molecule controlling the activities of diverse regulators ranging from transcription factors to MAP kinases. Cell Res. 16: 681-692. [DOI] [PubMed] [Google Scholar]
- 43.Näit-Kaoudjt, R., R. Williams, B. Guiard, and M. Gervais. 1997. Some DNA targets of the yeast CYP1 transcriptional activator are functionally asymmetric—evidence of two half-sites with different affinities. Eur. J. Biochem. 244: 301-309. [DOI] [PubMed] [Google Scholar]
- 44.Pfeifer, K., K. S. Kim, S. Kogan, and L. Guarente. 1989. Functional dissection and sequence of yeast HAP1 activator. Cell 56: 291-301. [DOI] [PubMed] [Google Scholar]
- 45.Pinkham, J. L., Z. Wang, and J. Alsina. 1997. Heme regulates SOD2 transcription by activation and repression in Saccharomyces cerevisiae. Curr. Genet. 31: 281-291. [DOI] [PubMed] [Google Scholar]
- 46.Quackenbush, J. 2002. Microarray data normalization and transformation. Nat. Genet. 32(Suppl.): 496-501. [DOI] [PubMed] [Google Scholar]
- 47.Ren, B., F. Robert, J. J. Wyrick, O. Aparicio, E. G. Jennings, I. Simon, J. Zeitlinger, J. Schreiber, N. Hannett, E. Kanin, T. L. Volkert, C. J. Wilson, S. P. Bell, and R. A. Young. 2000. Genome-wide location and function of DNA binding proteins. Science 290: 2306-2309. [DOI] [PubMed] [Google Scholar]
- 48.Schneider, B. L., W. Seufert, B. Steiner, Q. H. Yang, and A. B. Futcher. 1995. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast 11: 1265-1274. [DOI] [PubMed] [Google Scholar]
- 49.Siddiqui, A. H., and M. C. Brandriss. 1988. A regulatory region responsible for proline-specific induction of the yeast PUT2 gene is adjacent to its TATA box. Mol. Cell. Biol. 8: 4634-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Storici, F., L. K. Lewis, and M. A. Resnick. 2001. In vivo site-directed mutagenesis using oligonucleotides. Nat. Biotechnol. 19: 773-776. [DOI] [PubMed] [Google Scholar]
- 51.Sturley, S. L. 2000. Conservation of eukaryotic sterol homeostasis: new insights from studies in budding yeast. Biochim. Biophys. Acta 1529: 155-163. [DOI] [PubMed] [Google Scholar]
- 52.Tamura, K., Y. Gu, Q. Wang, T. Yamada, K. Ito, and H. Shimoi. 2004. A hap1 mutation in a laboratory strain of Saccharomyces cerevisiae results in decreased expression of ergosterol-related genes and cellular ergosterol content compared to sake yeast. J. Biosci. Bioeng. 98: 159-166. [DOI] [PubMed] [Google Scholar]
- 53.ter Linde, J. J., H. Liang, R. W. Davis, H. Y. Steensma, J. P. van Dijken, and J. T. Pronk. 1999. Genome-wide transcriptional analysis of aerobic and anaerobic chemostat cultures of Saccharomyces cerevisiae. J. Bacteriol. 181: 7409-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ter Linde, J. J., and H. Y. Steensma. 2002. A microarray-assisted screen for potential Hap1 and Rox1 target genes in Saccharomyces cerevisiae. Yeast 19: 825-840. [DOI] [PubMed] [Google Scholar]
- 55.Trocha, P. J., and D. B. Sprinson. 1976. Location and regulation of early enzymes of sterol biosynthesis in yeast. Arch. Biochem. Biophys. 174: 45-51. [DOI] [PubMed] [Google Scholar]
- 56.Turi, T. G., and J. C. Loper. 1992. Multiple regulatory elements control expression of the gene encoding the Saccharomyces cerevisiae cytochrome P450, lanosterol 14α-demethylase (ERG11). J. Biol. Chem. 267: 2046-2056. [PubMed] [Google Scholar]
- 57.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98: 5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ushinsky, S. C., and T. Keng. 1994. A novel allele of HAP1 causes uninducible expression of HEM13 in Saccharomyces cerevisiae. Genetics 136: 819-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Verdière, J., M. Gaisne, and R. Labbe-Bois. 1991. CYP1 (HAP1) is a determinant effector of alternative expression of heme-dependent transcribed genes in yeast. Mol. Gen. Genet. 228: 300-306. [DOI] [PubMed] [Google Scholar]
- 60.Vik, Å., and J. Rine. 2001. Upc2p and Ecm22p, dual regulators of sterol biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 21: 6395-6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wenger, R. H. 2000. Mammalian oxygen sensing, signalling and gene regulation. J. Exp. Biol. 203: 1253-1263. [DOI] [PubMed] [Google Scholar]
- 62.Winston, F., C. Dollard, and S. L. Ricupero-Hovasse. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11: 53-55. [DOI] [PubMed] [Google Scholar]
- 63.Xu, L., C. K. Glass, and M. G. Rosenfeld. 1999. Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev. 9: 140-147. [DOI] [PubMed] [Google Scholar]
- 64.Zagorec, M., J.-M. Buhler, I. Treich, T. Keng, L. Guarente, and R. Labbe-Bois. 1988. Isolation, sequence, and regulation by oxygen of the yeast HEM13 gene coding for coproporphyrinogen oxidase. J. Biol. Chem. 263: 9718-9724. [PubMed] [Google Scholar]
- 65.Zhang, L., and A. Hach. 1999. Molecular mechanism of heme signaling in yeast: the transcriptional activator Hap1 serves as the key mediator. Cell. Mol. Life Sci. 56: 415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.