RNA cleavage is a fundamental host response for controlling viral infections in both plants and animals (26). In higher vertebrates, this process is often regulated by interferons (IFNs), a family of antiviral cytokines discovered 50 years ago (50). One of the principal IFN antiviral pathways involves activation of the ubiquitous cellular endoribonuclease RNase L (formerly 2-5A-dependent RNase) (125). Recently, there has been progress in understanding how RNase L affects a range of different types of viral infections and how viruses counteract RNase L. Understanding how RNase L and viruses interact in vivo could contribute to therapeutic strategies for controlling pathogenic viruses (113).
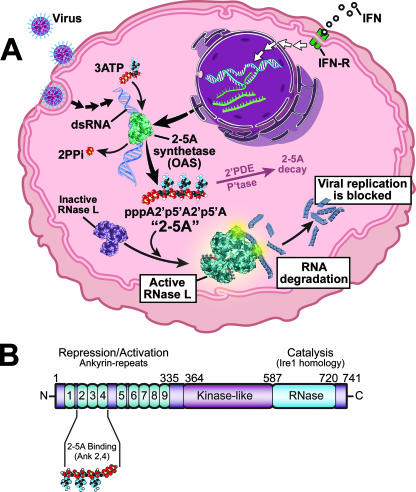
The 2′,5′-oligoadenylate synthetase (OAS)/RNase L system is an innate immunity pathway that responds to a pathogen-associated molecular pattern to induce degradation of viral and cellular RNAs and thereby block viral infections (Fig. 1A). The pathogen-associated molecular pattern is double-stranded RNA (dsRNA), a type of nonself-RNA produced during infections by both RNA and DNA viruses. Viral dsRNAs include replicative intermediates of single-stranded RNA (ssRNA) viruses, viral dsRNA genomes, annealed viral RNAs of opposite polarities, and stem structures in otherwise single-stranded viral RNAs. dsRNA activates the pathogen recognition receptor 2-5A synthetase, or OAS, resulting in production of 2-5A [px5′A(2′p5′A)n; x = 1 to 3; n ≥ 2] from ATP (Fig. 1) (45, 46, 55). IFN signaling induces transcription of the OAS genes through IFN-stimulated response elements in the promoters (94). Therefore, cells exposed to IFN as a result of ongoing viral infections have elevated levels of OAS that contribute to the IFN-induced antiviral state. The trimeric and tetrameric species [(2′-5′)p3A3 and (2′-5′)p3A4] are the principal forms of 2-5A produced in IFN-treated, virus-infected cells (57). Needless to say, 2-5A species are very unusual nucleic acids because of their adjacent 2′ to 5′ phosphodiester bonds. In humans, OAS is a family of 8 to 10 different isoforms encoded by three functional genes (OAS1 to OAS3) and a single OASL gene encoding a related protein with two C-terminal ubiquitin-like domains that does not synthesize 2-5A (42, 52, 75, 88). In mice, there are eight oas1 genes, in addition to oas2, oas3, and two oasl genes (54). OAS1 species (p40/p46) have one catalytic domain and form tetramers, OAS2 species (p69/p71) have two catalytic domains and form dimers, while OAS3 (p100) has three catalytic domains and is a monomer (48, 88). The different forms of OAS occupy different subcellular locations, have different dsRNA optima for activation, and synthesize 2-5A oligomers of different lengths (17, 47, 73). A crystal structure of porcine OAS1 has led to structural and functional insight into the OAS proteins (41).
FIG. 1.
(A) The OAS/RNase L system, an innate immunity pathway that acts against viral infections. PPi, pyrophosphate; 2′PDE, 2′-phosphodiesterase; P'tase, phosphatase; IFN-R, IFN receptor. (B) Domain structure of human RNase L. (Copyright, The Cleveland Clinic Center for Medical Art & Photography; reproduced with permission.)
The only well-established function of 2-5A is activation of RNase L (125). RNase L was detected by cross-linking to radiolabeled 2-5A in extracts of several different mouse organs. In contrast, no 2-5A binding proteins were detected in organs of RNase L−/− mice (126). Those findings suggested that 2-5A is a unique ligand for RNase L. Human RNase L is a 741-amino-acid polypeptide containing, from the N to the C termini, nine ankyrin repeats, several protein kinase-like motifs, and the RNase domain (Fig. 1B). 2-5A binds to ankyrin repeats 2 and 4 (112), causing catalytically inactive RNase L monomers to form activated dimers with potent RNase activity (19, 28). Specifically, RNase L cleaves within single-stranded regions of RNA, principally on the 3′ sides of UpAp and UpUp dinucleotides, leaving 3′-phosphoryl and 5′-hydroxyl groups at the termini of the RNA cleavage products (31, 119). 2-5A is degraded within minutes by 2′-phosphodiesterase and 5′-phosphatase activities within cells and in sera (51, 59, 101, 107). Therefore, 2-5A is an early transient-response molecule or alarmone that signals antiviral innate immunity through RNase L activation. RNase L function is dampened by the RNase L inhibitor (RLI), an ATP binding cassette protein also known as ABCE1 (7). While there is some evidence that RNase L prefers viral to cellular RNA (65), cellular RNAs, including rRNA in intact ribosomes, are also cleaved by RNase L (106, 118).
Lessons learned from studying interactions between the OAS/RNase L system and many different types of RNA and DNA viruses are considered in the following sections, with an emphasis on more-recent in vivo studies.
RNA VIRUSES
Picornaviridae.
Viruses in the Picornaviridae family, including pathogens such as poliovirus, coxsackievirus, and hepatitis A virus, have relatively small (7.2 to 8.4 kb) monopartite positive-stranded RNA genomes that replicate through partially double-stranded RNA intermediates (87). Infections of IFN-treated cells with encephalomyocarditis virus (EMCV), of the Cardiovirus genus, activate OAS, causing accumulation of 2-5A (57, 106, 117). The RNA activators of OAS in EMCV-infected cells are probably the viral replicative intermediates. This was evidenced, for instance, by the isolation from extracts of IFN-treated, EMCV-infected HeLa cells of a complex of OAS1 in an activated state bound to EMCV RNA of both (plus and minus) strands (34). Ectopic expression of either OAS1 or RNase L suppressed replication of mengovirus (another picornavirus) (18) and EMCV (128), respectively. In contrast, IFN was relatively ineffective against EMCV in cells expressing a dominant negative RNase L (43). Furthermore, control and alpha interferon (IFN-α)-treated mice lacking RNase L succumbed to EMCV infections more rapidly and at a higher rate than identically treated and infected wild-type (WT) mice (126). Nevertheless, EMCV is not defenseless against the OAS/RNase L system. EMCV infection of cells that have not been exposed to IFN eliminates RNase L activity through a process that has yet to be elucidated (15, 105). Exposure of cells to IFN prior to EMCV infection, however, prevented the loss of RNase L activity. These studies indicate that the OAS/RNase L system contributes to the anti-EMCV activity of IFN. Nevertheless, mice that either lack RNase L by itself or that are triply deficient for RNase L, RNA-dependent protein kinase (PKR), and Mx1 genes are still able to mount a significant residual IFN response against EMCV (127). These findings illustrate the complexity of and redundancies in the IFN system.
Coxsackieviruses, of the Enterovirus genus, are common picornaviruses linked to a range of different human pathologies, including myocarditis, meningitis, diabetes, and colds (84). RNase L-deficient mice are exquisitely sensitive to coxsackievirus B4 (CV-B4). At 23 days postinfection with 100 PFU by the intraperitoneal route, only 7% of RNase L−/− mice survived; in contrast, 62% of infected WT mice survived (30). In isolated mouse pancreatic islet cells, RNase L was also required for an efficient IFN-α response against CV-B4. However, CV-B4 infection of endogenous pancreatic islets could not be detected in infected mice lacking RNase L, PKR, and Mx1, suggesting that alternative pathways were operating.
Poliovirus (PV), an enterovirus, causes paralytic disease or poliomyelitis in about 1 of 200 susceptible individuals (84). Remarkably, an RNA structure present in the open reading frame for PV proteinase, 3CPro, is a potent inhibitor of RNase L (39). As a result, PV mRNA is resistant to cleavage by RNase L. The inhibitory PV RNA, a highly structured region of 303 nucleotides, is conserved among group C enteroviruses PV-1, PV-2, and PV-3 and coxsackievirus All (CAV-11), CAV-13, CAV-17, CAV-20, PV-21, and PV-24 but is absent in other human enteroviruses. PV yields in HeLa cells were unaffected by expression of either WT or dominant negative mutant (R667A) RNase L. RNase L activity, as measured by rRNA cleavage products, occurred only late in infection in cells containing WT RNase L but not in cells expressing the mutant RNase L. Surprisingly, PV engineered to lack the inhibitory RNA structure grew with a level of efficiency equal to that of the parental PV in HeLa cells expressing WT RNase L. However, the presence of WT RNase L resulted in increased PV plaque size, leading to the suggestion that the apoptotic function of RNase L during PV infections (12, 13, 126) facilitates release of the virus and increases cell-to-cell spread (39). Perhaps a role of the PV RNA inhibitor of RNase L in pathogenesis will be evident once it is possible to test its role in vivo.
Theiler's virus is a murine picornavirus that persists in the central nervous systems of susceptible strains of mice (8). The L* viral protein, encoded by an alternative reading frame, is implicated in viral persistence and localizes to the mitochondrial membrane, where it antagonizes the type I IFN response by interfering with the OAS/RNase L system (F. Sorgeloos and T. Michiels, personal communication).
Reoviridae.
Reoviruses, members of the Orthoreovirus genus, have 10 dsRNA genomic segments that can reassort upon coinfection (100). Reovirus infections of HeLa cells resulted in IFN-dependent RNase L activation, as measured by specific rRNA cleavages (82). Interestingly, a recent study showed that RNase L contributes to host shutoff of protein synthesis during reovirus infections (110). There was a decreased level of shutoff of host translation by reovirus strains in mouse embryonic fibroblasts (MEFs) lacking either PKR or RNase L, whereas no shutoff was observed in MEFs lacking both PKR and RNase L. Presumably, viral genomic dsRNA activates PKR and OAS, resulting in phosphorylation of eiF2α and production of 2-5A, respectively. As a result, rather than inhibiting viral replication, some strains of reovirus actually produce higher viral yields in WT MEFs than in PKR−/− or RNase L−/− MEFs (110). The S4 gene segment of reovirus encodes σ3 protein, which has a structural role in the outer capsid but is also a dsRNA binding protein (22, 83). Therefore, σ3 is able to inhibit OAS and PKR activation in the context of a recombinant vaccinia virus (VV) lacking the E3L gene (5) (E3L encodes 25- and 20-kDa dsRNA binding proteins [116]).
Togaviridae.
Members of the Alphavirus genus, including Sindbis virus and Semliki Forest virus, are small, lipid-enveloped, monopartite positive-stranded RNA viruses (35). In RNase L-deficient MEFs, Sindbis virus showed continuous synthesis of minus-strand templates (instead of shutting off at 4 h postinfection) and formed persistent infections (98). It was concluded that RNase L has a role in the cessation of alphavirus minus-strand synthesis. However, mice lacking RNase L, PKR, and Mx1 (triply deficient) (127) develop only subclinical infections with Sindbis virus, unlike mice defective in the type I IFN receptor (IFNAR1−/− mice), which rapidly succumb to infections (95). Nevertheless, higher titers of the virus were observed in the draining lymph nodes of the triply deficient mice than in the identically infected WT mice. These findings suggest that while RNase L and PKR partially suppress Sindbis virus replication in vivo, alternative IFN antiviral pathways protect mice from fatal Sindbis virus infections.
Paramyxoviridae.
Respiratory syncytial virus (RSV), a Pneumovirus in the Paramyxoviridae family of monopartite negative-stranded RNA viruses, is a major cause of lower respiratory tract infections in very young children, immunocompromised patients, and institutionalized elderly patients (20). Inhibition of OAS1 (p40) and OAS2 (p69) expression in cells with an antisense oligonucleotide inhibited the antiviral effect of IFN-γ against RSV (6). In addition, expression of RNase L inhibitor (RLI) suppressed the IFN-γ effect against RSV. The potential of RNase L activator drugs to block RSV infections was demonstrated by targeting RSV genomic RNA with 2-5A linked to an antisense oligonucleotide against the RSV repetitive-gene start site (60). The 2-5A antisense drug administered by the intranasal route to RSV-infected African green monkeys reduced viral yields by 4-log10 units.
Orthomyxoviridae.
Influenza A viruses contain multipartite genomes of eight single-stranded negative-sense RNAs and include major human pathogens responsible for devastating respiratory disease, notably the infamous 1918 Spanish influenza pandemic (120). The NS1 protein of influenza A virus (NS1A) is a multifunctional virulence factor that counteracts innate immunity through binding both proteins and dsRNA (80). A point mutation in the N terminus of NS1A, R38A, ablates dsRNA binding activity. A recombinant influenza virus, strain A/Udorn/72 with the NS1A R38A mutation, is highly sensitivity to inhibition by IFN-β, in contrast to the WT virus, which is resistant to IFN-β. The enhanced IFN susceptibility of the mutant virus was mediated predominantly through RNase L, because its depletion by small interfering RNA (siRNA) or its absence in RNase L−/− MEFs largely relieved the IFN-β-mediated inhibition. The conclusion was that NS1A sequesters dsRNA from OAS, thereby preventing synthesis of 2-5A and activation of RNase L.
Flaviviridae.
Members of the Flavivirus genus of the Flavivirdae family are monopartite, positive-stranded RNA enveloped viruses, including human pathogens propagated through mosquitoes that cause encephalitic syndromes (West Nile virus [WNV] and Japanese encephalitis virus) or severe hemorrhagic disease (yellow fever virus and dengue virus) (66). In mice, a single gene on chromosome 5, the flavivirus resistance gene (flv), reduces flavivirus yields by up to 4-log10 units and is specific for this group of viruses (97). Astonishingly, flv was identified as the OAS1b (or L1) gene, one of eight OAS1 genes in the mouse genome, by the group of M. Brinton (86), and the finding was confirmed soon thereafter by P. Despres and coworkers (76). Combining results from these two studies, a C-to-T transition mutation in exon 4 of oas1b, which truncates 30% of the OAS1b protein from the carboxy terminus, occured among 11 flavivirus-susceptible strains, whereas 9 resistant mouse strains encoded full-length OAS1b. Expression of OAS1b cDNA in C3H/He cells at low (but not high) levels modestly inhibited WNV replication and delayed the cytopathic effect (86). Also, induction of OAS1b but not the truncated mutant OAS1b (under control of a Tet-Off system) reduced WNV yields by 1- to 2.5-log10 units in MEFs at early stages in the viral life cycle, correlating with reduced levels of positive-strand viral RNA (53).
These results are intriguing and potentially immensely important to understanding the host defense to flaviviruses. How OAS1b functions to inhibit flaviviruses remains to be elucidated. The complete form of OAS1b lacks several amino acids believed to be required for 2-5A synthesis. Therefore, OAS1b likely has an alternative mode of action compared with those of other OAS species. For instance, it has been suggested that the OAS1b protein might recognize and bind to a conserved RNA structure unique to flaviviruses (86). Alternatively, it has been proposed, based on sequence homologies, that OAS proteins are nucleases (92). While this concept remains hypothetical, if some OASs are nucleases, OAS1b could potentially degrade WNV RNAs. Human polymorphisms in the RNASEL and OAS genes have been examined in isolates from patients hospitalized with WNV infections (123). A synonymous single-nucleotide polymorphism in exon 2 of OASL that could possibly enhance mRNA splicing had a higher frequency in WNV-infected patients (P < 0.004), leading to the suggestion that the RNA transcript might undergo increased splicing resulting in a dominant negative isozyme.
There is also a well-substantiated anti-WNV effect of RNase L itself. In RNase L−/− MEFs, WNV yields were increased 5- to 10-fold compared with those of WT MEFs (99). A similar effect was obtained by expressing a dominant negative RNase (99) or siRNA knockdown of RNase L (53). The absence of RNase L in mice resulted in increased susceptibility to peripheral (footpad) inoculations with WNV (96). Deficiencies in both PKR and RNase L resulted in an enhanced susceptibility to WNV compared to that of the RNase L−/− mice. These results demonstrate significant contributions of both pathways to host resistance to WNV (loss of the type I IFN receptor produced an even greater susceptibility to WNV). PKR and RNase L contributed to the anti-WNV effect of type I IFN in primary macrophages and cortical neurons but not in peripheral neurons of the cervical ganglia. However, the absence of both PKR and RNase L failed to suppress the IFN-β inhibition of dengue virus in MEF cultures (23). Despite cell type variability, the anti-WNV effect of RNase L in mice appears to be nonredundant with effects mediated by the oas1b/flv gene. Therefore, the OAS/RNase L system has at least two mechanisms for suppressing replication of flaviviruses; one is the classical pathway mediated through RNA cleavage by RNase L, and the other is a yet-to-be-defined alternative mechanism mediated by the OAS1b protein.
Hepatitis C virus (HCV), of the Hepacivirus genus of the Flavividae family, is a virus that has infected about four million adults in the U.S. and is a major cause of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (2). Combination therapy with IFN and ribavirin can produce sustained responses against HCV in some patients (78). However, genotype 1 HCVs, most common in the United States, are relatively IFN resistant in comparison to genotypes 2 and 3 (72). In cell-free systems, HCV mRNA can activate OAS to produce 2-5A, causing the RNA to be degraded into fragments of 200 to 500 bases by RNase L. Perhaps the most cogent evidence that RNase L poses a threat to HCV is that IFN sensitivity correlates with the susceptibility of HCV mRNA to cleavage by RNase L (38). Data suggest that RNase L activity during HCV infections causes selection of viral strains with decreased frequencies of UU and UA dinucleotides. Accordingly, there are fewer RNase L cleavage sites in IFN-resistant genotypes (1a and 1b) than in IFN-sensitive genotypes (2a, 2b, 3a, and 3b). Furthermore, silent mutations accumulate at these sites during IFN therapy of HCV1b-infected individuals. A relatively small number of UU and UA dinucleotides mediate the overall susceptibility of HCV RNA to RNase L (40). Recently, UA and UU dinucleotide frequencies were compared among 162 HCV RNA sequences and were found to be among the least-abundant dinucleotides in HCV open reading frames (115). Expression of the HCV1b polyprotein from a VV vector induced apoptosis in a manner that was RNase L dependent but PKR independent (33). These results suggest that HCV-mediated activation of RNase L could lead to apoptosis and elimination of HCV-infected cells.
Retroviridae.
Human immunodeficiency virus (HIV), the cause of AIDS, is a lentivirus that has infected an estimated 33 to 46 million people worldwide (Joint United Nations Programme on HIV/AIDS; www.unaids.org). While dsRNA per se is not an intermediate in the life cycles of retroviruses, the transactivation responsive (TAR) region at the 5′ termini of all HIV-1 mRNAs activates OAS (67, 103), whereas Tat binding to TAR prevents OAS activation (102). Ectopic expression of RLI decreased RNase L activity and caused a twofold increase in HIV yields (74). However, interpretation of these experiments is complicated by the involvement of RLI (HP68) in the assembly of HIV-1 capsids (129). Expression of antisense RNA against RNase L mRNA decreased RNase L protein levels, increased HIV-1 yields, and reduced the anti-HIV effect of IFN-α (68). The latter findings suggest that RNase L exerts a modest effect against HIV-1 as part of the IFN antiviral response.
Xenotropic murine leukemia virus-related virus (XMRV) is a gammaretrovirus present in tumor-bearing prostate tissues of men that are homozygous for a variant of RNase L (R462Q) (114). The human RNase L gene (RNASEL) was identified in 2002 as a candidate for the hereditary prostate cancer 1 (HPC1) gene based on a positional cloning/candidate gene method (9, 104). Mutations at several sites in RNASEL, including the Q variant, which reduced nuclease activity by threefold, were observed in isolates from prostate cancer patients (11, 122). Screening isolates from prostate cancer patients with a virus DNA microarray (ViroChip) led to the discovery of XMRV (114). Remarkably, XMRV was found in 8 of 20 QQ patients but in only 1 of 62 RR or RQ patients, thus implicating RNase L in the suppression of XMRV infections of the prostate. XMRV infections were visualized by immunohistochemistry and fluorescence in situ hybridizations in a small proportion of prostate stromal cells. A full-length, infectious viral molecular clone of XMRV was cloned from prostate cDNA (27). XMRV replication in the prostate cancer cell line DU145 was sensitive to inhibition by IFN-β. However, the IFN antiviral effect was greatly reduced in LNCaP cells in which there is epigenetic silencing of the JAK1 gene and a mutation in one allele of RNASEL. In DU145 cells, siRNA knockdown of RNase L levels resulted in a partial reversal of the sensitivity of XMRV to IFN-β treatment. These experiments demonstrated that RNase L is necessary for a complete IFN antiviral response against XMRV. In addition, XMRV integration sites were mapped in prostate DNA. Evidence suggests that XMRV infects and persists in the prostate when there is a deficiency in RNase L activity. While chronic infections and inflammation are suspected cofactors in prostate cancer (21), a role for XMRV in prostate cancer etiology, although suspected, has not been established.
DNA VIRUSES
Poxviridae.
Although most viral studies on the OAS/RNase L system have been performed with RNA viruses, it is clear that some DNA viruses produce dsRNA species (through annealing of RNA strands of opposite polarity) that stimulate OAS to generate 2-5A. However, activation of RNase L does not always occur at the same time as 2-5A accumulation. In fact, the poxvirus vaccinia virus, a large DNA virus that replicates in the cytoplasm, is the most-potent known viral inducer of 2-5A (90). Up to 5 μM of 2-5A was produced in VV-infected, IFN-treated cells (90), about 25-fold more 2-5A than was obtained from EMCV-infected, IFN-treated cells (58). VV induces a complex mixture of phosphorylated and nonphosphorylated 2-5A species as well as related compounds of unknown structure (89). Accumulation of 2-5A occurs despite the presence of the VV-encoded E3L proteins that sequester dsRNA (116). Therefore, one might have expected RNase L to have a potent effect against VV. However, despite high levels of 2-5A early after VV infection, activation of RNase L as measured by rRNA cleavage did not occur until late in infection (90). Furthermore, VV yields were identical in WT MEFs and in MEFs lacking RNase L, PKR, and Mx1, and the susceptibility of mice to fatal infections with VV was unaffected by the absence of these proteins (121). It is apparent, therefore, that at least in mice, VV is unimpeded by either RNase L or PKR. The mechanism by which VV inhibits RNase L in the presence of high levels of 2-5A remains unknown, but results suggest that VV produces, or induces the cell to produce, a factor that blocks RNase L activity. VV E3L suppresses activation transcription factor IFN regulatory factor 3 (IRF3), which is involved in IFN-α and IFN-β gene expression (109, 121). VV lacking E3L replicated to about 10-fold-higher titers in RNase L−/− MEFs than in WT MEFs (121), suggesting that E3L is one inhibitor of the pathway, in agreement with earlier studies (5, 91). Expression of OAS and RNase L from recombinant VV (25) or overexpression of RNase L alone (128) overcomes inhibition and is highly effective in suppressing VV replication.
Herpesviridae.
Herpes simplex virus type 1 (HSV-1) and HSV-2 (85) induced accumulation of 2-5A up to about 50 nM in IFN-treated human conjunctival Chang cells but not until late in infection (16 h) (14). Furthermore, only low levels of RNase L-mediated rRNA cleavage products were observed, despite the fact that RNase L is typically highly activated by similar levels of 2-5A in IFN-treated EMCV-infected cells. RNase L also has little or no effect on host RNA degradation mediated by VHS, the HSV-1 virion host shutoff protein, in MEFs (111). A possible explanation for the relative inactivity of RNase L in HSV-infected cells emerged from analysis of the types of 2-5A species produced. In HSV-infected cells, unlike in EMCV-infected cells, a complex mixture of 2-5A and 2-5A-related material was found, including some compounds that were inhibitory to RNase L. Other cell culture studies suggest that RNase L does participate in the anti-HSV-1 effect of IFN. In primary mouse trigeminal ganglion cultures, an absence of RNase L reduced the effects of mouse IFN-β expressed against the McKrae strain of HSV-1 (1, 10). The IFN-α effect against HSV-1 strain F was also greatly reduced in RNase L−/− PKR−/− MEFs compared with that in WT or PKR single-gene-knockout MEFs (56). In vivo evidence for a role of RNase L in innate immunity to HSV infections has been mixed. Differences in the strains of HSV-1, routes of inoculation, and genetic backgrounds in the mice make direct comparison of these studies difficult. Intracerebral inoculation with HSV-1 (strain 17) killed all WT and RNase L−/− mice within 10 days (61). Therefore, under these conditions with this strain of HSV-1, RNase L failed to protect WT mice from fatal infections. However, application of the HSV-1 McKrae strain to unscarified corneas causes significantly higher levels of herpetic keratitis and results in a higher mortality rate in RNase L−/− mice than in WT congenic control mice (124). Also, during acute ocular infections, an IFN-β transgene reduced HSV-1 McKrae levels in the eyes and trigeminal ganglia of WT but not RNase L-deficient mice (3).
Surprisingly, lack of RNase L resulted in decreased pathology following vaginal infections of mice with HSV-2 strain 333 (29). There was less-severe genital and neurological disease as well as a delay in mortality in HSV-2 infected RNase L−/− mice than in identically infected WT mice. The decreased pathology in the absence of RNase L was related to a restricted inflammatory response, with decreased CD4+ T cell infiltration in the infected tissues. Skin allograft rejection, accompanied by a dramatic reduction in inflammatory infiltrates, was also delayed in RNase L−/− mice (108). Therefore, the proinflammatory effect of RNase L can actually contribute to virus-induced pathology in some circumstances.
Polyomaviridae.
Simian virus 40 is induced in IFN-α treated CV-1 monkey cells at most at only very low levels (3 nM) of the 2-5A species that activate RNase L (44). However, large amounts (up to 2 μM) of related 2′,5′-linked oligoadenylates accumulate late in infection, but these compounds do not activate RNase L. The diversion of OAS to producing inactive 2-5A analogs is a viral strategy to evade the antiviral effect of RNase L. However, the identities of the 2-5A-related compounds formed in IFN-treated cells infected with VV, HSV, or simian virus 40 have never been determined.
Hepadnaviridae.
IFN-α/β produced in response to poly(I)/poly(C) or to unrelated hepatotropic viruses inhibited hepatitis B virus (HBV) replication noncytopathically in HBV transgenic mice (77). However, HBV transgenic mice that were RNase L+/+ or RNase L−/− showed no difference in HBV replication than control transgenic mice with or without the administration of poly(I)/poly(C) or IFN-γ (36). Therefore, RNase L is not responsible for the anti-HBV effects of dsRNA or IFN-γ observed in these mouse experiments.
HOW RNase L INHIBITS VIRAL INFECTIONS
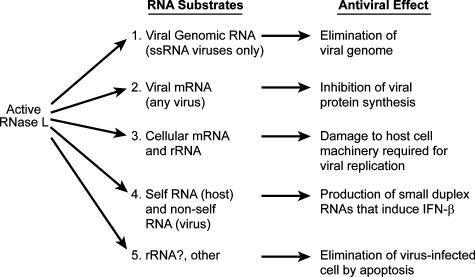
The antiviral effect of RNase L occurs not through a single mechanism but through a combination of effects resulting from cleavages in different RNA substrates and depends on the virus and cell type (Fig. 2 and Table 1). Most but not all RNA viruses are inhibited by RNase L at some level; however, most DNA viruses that have been examined are partially or completely resistant to the OAS/RNase L system. However, antiviral strategies that either enhance RNase L levels or its activity can inhibit DNA viruses from replicating.
FIG. 2.
Antiviral effects of the cleavage of viral and cellular RNA substrates by RNase L.
TABLE 1.
Viral interactions with the OAS/RNase L system
| Virus | Effect of RNase L on viral replication and/or disease | Viral evasion of the OAS/RNase L system | References |
|---|---|---|---|
| RNA viruses | |||
| Encephalomyocarditis virus | Antiviral (mice) | RNase L activity is eliminated in cells not exposed to IFN | 15, 105, 126 |
| Coxsackievirus B4 | Antiviral (mice) | NDa | 30 |
| Poliovirus | No effect on viral yields (cell culture), larger plaques in presence of RNase L | Viral RNA structure inhibits RNase L | 39 |
| Theiler's virus | ND | L* interferes with the OAS/RNase L system | T. Michiels, personal communication |
| Reovirus | Proviral (cell culture); shuts off host translation | DsRNA-binding by σ3 inhibits OAS activation | 5, 110 |
| Sindbis virus | Antiviral with PKR but not protective (mice) | ND | 95 |
| Respiratory syncytial virus | Antiviral, IFN-γ effect (cell culture) | ND | 6 |
| Influenza A virus | No effect (cell culture) | dsRNA binding by NS1 inhibits OAS activation | 80 |
| West Nile virus | Antiviral (mice) | Truncated OAS1b (Flv) renders mice susceptible to West Nile virus and other flaviviruses | 76, 86, 96 |
| Hepatitis C virus | Antiviral (humans) | Selects against RNase L cleavage sites | 38 |
| Human immunodeficiency virus type 1 | Antiviral, IFN-α effect (cell culture) | TAT inhibits OAS activation by TAR | 68, 102 |
| Xenotropic murine leukemia-related virus | Antiviral (humans); causes prostate infections in patients with QQ variant of RNase L | ND | 27, 114 |
| DNA viruses | |||
| Vaccinia virus | No effect (mice) | dsRNA binding by E3L inhibits OAS activation; inactive 2-5A analogs | 5, 89, 90, 121 |
| Herpes simplex virus 1 | Can have or not have an effect (mice) | Inactive 2-5A analogs | 3, 14, 61, 124 |
| Herpes simplex virus 2 | Proinflammatory effect of RNase L contributes to disease (mice) | ND | 29 |
| Simian virus 40 | No effect (cell culture) | Inactive 2-5A analogs | 44 |
| Hepatitis B virus | No effect (mice) | ND | 36 |
ND, not determined (not found in scientific literature).
RNase L SUPPRESSES VIRUS INFECTIONS
(i) Cleavage of viral genomic ssRNA prevents replication.
Even a single cleavage event in a viral ssRNA genome will prevent that genome from replicating unless RNA recombination events regenerate full-length genomic RNA (32). Evidence for genomic RNA strand cleavage by RNase L was obtained for EMCV (65), and the fact that many ssRNA viruses are susceptible to RNase L lends support to the hypothesis (Table 1). A mechanism whereby viral RNA is degraded while the infected cell survives is a preferred scenario for the organism if the infected cells are neurons or other essential cell types such as insulin-producing pancreatic islet cells.
(ii) Cleavage of viral mRNA inhibits viral protein synthesis.
Activated RNase L within cells cleaves both viral and nonviral ssRNA substrates. However, ssRNA molecules linked to double-stranded regions, such as in some viral replicative intermediates, may be preferentially cleaved owing to localized activation of OAS and RNase L (81). In some instances, such a scenario could possibly account for the selective cleavage of viral RNA by RNase L (65). Cleavage of viral mRNA by RNase L, together with the IFN-induced proteins PKR (79) and p56 (37), contributes to IFN-induced inhibition of viral protein synthesis.
(iii) Cleavage of cellular mRNA and rRNA required for viral replication.
Damage to the host cell machinery required for viral replication, in particular the ribosome, contributes to the antiviral effects of RNase L (106, 118). Cleavage of both 28S and 18S rRNA are hallmarks of RNase L activity in virus-infected cells. However, it is worth noting that RNase L-independent cleavage of 28S rRNA occurs in cells infected with mouse hepatitis virus (4). A large number of cellular mRNA species also fall prey to RNase L, as determined by RNA profiling experiments (70). Nevertheless, nonspecific cleavage of RNA is unlikely to account for reovirus-induced shutoff of host protein synthesis because of a lack of correlation of reovirus strain differences in host shutoff with the extent of rRNA cleavages (110). Whether RNase L and PKR can effectively differentiate between viral and cellular mRNA is an open question. However, what is clear is that degradation of cellular RNAs required for viral replication is a very effective strategy for blocking viruses.
(iv) Amplification of IFN-α/β production by RNase L-generated small RNAs.
RNase L cleaves single-stranded regions of RNA, leaving as cleavage products short duplex RNAs with 3′-phosphoryl groups (31, 40, 119). These small RNA cleavage products signal through the RNA helicases RIG-I and MDA5, the adapter IPS-1, and transcription factors IRF-3 and NF-κB to the IFN-β gene (69). As a result, RNase L−/− mice produce significantly less IFN-β in response to viral infections (Sendai virus and EMCV) than identically infected WT mice. In addition, 2-5A induced IFN-β in WT mice but not in RNase L−/− mice, proving that self-RNAs cleaved by RNase L can signal innate immunity. Therefore, by relieving the requirement for sensing nonself (viral)-RNA, RNase L perpetuates and amplifies IFN production during antiviral innate immunity. These effects of RNase L extend beyond the initially infected cells to support a broader antiviral state in the organism.
(v) Elimination of virus-infected cells through apoptosis.
Death of infected cells is the ultimate antiviral pathway, although one that can be costly to the organism. Sustained activation of RNase L or its activation beyond a threshold level causes the cell to spiral into an RNA damage stress response that culminates in apoptosis (12, 13, 126). RNase L causes apoptosis in response to viral and nonviral apoptosis inducers (12, 13, 24, 68, 93, 126, 128). Apoptosis initiated by RNase L is characterized by the release of cytochrome c from mitochondria and requires caspase 3 activity (93). In addition, c-jun NH2-terminal kinases are involved in RNase L-mediated apoptosis (49, 64). RNase L effects on the mRNA stability of mitochondria that could contribute to apoptosis have also been described (16, 62, 63). The ability of RNase L to induce apoptosis suggests a tumor suppressor function of the OAS/RNase L pathway and therapeutic strategies based on RNase L activators (71).
Acknowledgments
I thank David Schumick, The Cleveland Clinic Center for Medical Art & Photography, for artwork.
This work was supported by a grant from the NIH (NCI; grant number CA044059).
Footnotes
Published ahead of print on 5 September 2007.
REFERENCES
- 1.Al-khatib, K., B. R. Williams, R. H. Silverman, W. Halford, and D. J. Carr. 2003. The murine double-stranded RNA-dependent protein kinase PKR and the murine 2′,5′-oligoadenylate synthetase-dependent RNase L are required for IFN-beta-mediated resistance against herpes simplex virus type 1 in primary trigeminal ganglion culture. Virology 313:126-135. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, G. L., A. Wasley, E. P. Simard, G. M. McQuillan, W. L. Kuhnert, and M. J. Alter. 2006. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann. Intern. Med. 144:705-714. [DOI] [PubMed] [Google Scholar]
- 3.Austin, B. A., C. James, R. H. Silverman, and D. J. Carr. 2005. Critical role for the oligoadenylate synthetase/RNase L pathway in response to IFN-beta during acute ocular herpes simplex virus type 1 infection. J. Immunol. 175:1100-1106. [DOI] [PubMed] [Google Scholar]
- 4.Banerjee, S., S. An, A. Zhou, R. H. Silverman, and S. Makino. 2000. RNase L-independent specific 28S rRNA cleavage in murine coronavirus-infected cells. J. Virol. 74:8793-8802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beattie, E., K. L. Denzler, J. Tartaglia, M. E. Perkus, E. Paoletti, and B. L. Jacobs. 1995. Reversal of the interferon-sensitive phenotype of a vaccinia virus lacking E3L by expression of the reovirus S4 gene. J. Virol. 69:499-505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behera, A. K., M. Kumar, R. F. Lockey, and S. S. Mohapatra. 2002. 2′-5′ Oligoadenylate synthetase plays a critical role in interferon-γ inhibition of respiratory syncytial virus infection of human epithelial cells. J. Biol. Chem. 277:25601-25608. [DOI] [PubMed] [Google Scholar]
- 7.Bisbal, C., C. Martinand, M. Silhol, B. Lebleu, and T. Salehzada. 1995. Cloning and characterization of a RNAse L inhibitor. A new component of the interferon-regulated 2-5A pathway. J. Biol. Chem. 270:13308-13317. [DOI] [PubMed] [Google Scholar]
- 8.Brahic, M., J. F. Bureau, and T. Michiels. 2005. The genetics of the persistent infection and demyelinating disease caused by Theiler's virus. Annu. Rev. Microbiol. 59:279-298. [DOI] [PubMed] [Google Scholar]
- 9.Carpten, J., N. Nupponen, S. Isaacs, R. Sood, C. Robbins, J. Xu, M. Faruque, T. Moses, C. Ewing, E. Gillanders, P. Hu, P. Bujnovszky, I. Makalowska, A. Baffoe-Bonnie, D. Faith, J. Smith, D. Stephan, K. Wiley, M. Brownstein, D. Gildea, B. Kelly, R. Jenkins, G. Hostetter, M. Matikainen, J. Schleutker, K. Klinger, T. Connors, Y. Xiang, Z. Wang, A. De Marzo, N. Papadopoulos, O. P. Kallioniemi, R. Burk, D. Meyers, H. Gronberg, P. Meltzer, R. Silverman, J. Bailey-Wilson, P. Walsh, W. Isaacs, and J. Trent. 2002. Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat. Genet. 30:181-184. [DOI] [PubMed] [Google Scholar]
- 10.Carr, D. J., K. Al-khatib, C. M. James, and R. Silverman. 2003. Interferon-beta suppresses herpes simplex virus type 1 replication in trigeminal ganglion cells through an RNase L-dependent pathway. J. Neuroimmunol. 141:40-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casey, G., P. J. Neville, S. J. Plummer, Y. Xiang, L. M. Krumroy, E. A. Klein, W. J. Catalona, N. Nupponen, J. D. Carpten, J. M. Trent, R. H. Silverman, and J. S. Witte. 2002. RNASEL Arg462Gln variant is implicated in up to 13% of prostate cancer cases. Nat. Genet. 32:581-583. [DOI] [PubMed] [Google Scholar]
- 12.Castelli, J. C., B. A. Hassel, A. Maran, J. Paranjape, J. A. Hewitt, X. L. Li, Y. T. Hsu, R. H. Silverman, and R. J. Youle. 1998. The role of 2′-5′ oligoadenylate-activated ribonuclease L in apoptosis. Cell Death Differ. 5:313-320. [DOI] [PubMed] [Google Scholar]
- 13.Castelli, J. C., B. A. Hassel, K. A. Wood, X. L. Li, K. Amemiya, M. C. Dalakas, P. F. Torrence, and R. J. Youle. 1997. A study of the interferon antiviral mechanism: apoptosis activation by the 2-5A system. J. Exp. Med. 186:967-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cayley, P. J., J. A. Davies, K. G. McCullagh, and I. M. Kerr. 1984. Activation of the ppp(A2′p)nA system in interferon-treated, herpes simplex virus-infected cells and evidence for novel inhibitors of the ppp(A2′p)nA-dependent RNase. Eur. J. Biochem. 143:165-174. [DOI] [PubMed] [Google Scholar]
- 15.Cayley, P. J., M. Knight, and I. M. Kerr. 1982. Virus-mediated inhibition of the ppp(A2′p)nA system and its prevention by interferon. Biochem. Biophys. Res. Commun. 104:376-382. [DOI] [PubMed] [Google Scholar]
- 16.Chandrasekaran, K., Z. Mehrabian, X. L. Li, and B. Hassel. 2004. RNase-L regulates the stability of mitochondrial DNA-encoded mRNAs in mouse embryo fibroblasts. Biochem. Biophys. Res. Commun. 325:18-23. [DOI] [PubMed] [Google Scholar]
- 17.Chebath, J., P. Benech, A. Hovanessian, J. Galabru, and M. Revel. 1987. Four different forms of interferon-induced 2′,5′-oligo(A) synthetase identified by immunoblotting in human cells. J. Biol. Chem. 262:3852-3857. [PubMed] [Google Scholar]
- 18.Chebath, J., P. Benech, M. Revel, and M. Vigneron. 1987. Constitutive expression of (2′-5′) oligo A synthetase confers resistance to picornavirus infection. Nature 330:587-588. [DOI] [PubMed] [Google Scholar]
- 19.Cole, J. L., S. S. Carroll, and L. C. Kuo. 1996. Stoichiometry of 2′,5′-oligoadenylate-induced dimerization of ribonuclease L. A sedimentation equilibrium study. J. Biol. Chem. 271:3979-3981. [DOI] [PubMed] [Google Scholar]
- 20.Collins, P. L., and J. E. Crowe, Jr. 2007. Respiratory syncytial virus and metapneumovirus, p. 1601-1646. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 21.De Marzo, A. M., E. A. Platz, S. Sutcliffe, J. Xu, H. Gronberg, C. G. Drake, Y. Nakai, W. B. Isaacs, and W. G. Nelson. 2007. Inflammation in prostate carcinogenesis. Nat. Rev. Cancer 7:256-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denzler, K. L., and B. L. Jacobs. 1994. Site-directed mutagenic analysis of reovirus sigma 3 protein binding to dsRNA. Virology 204:190-199. [DOI] [PubMed] [Google Scholar]
- 23.Diamond, M. S., and E. Harris. 2001. Interferon inhibits dengue virus infection by preventing translation of viral RNA through a PKR-independent mechanism. Virology 289:297-311. [DOI] [PubMed] [Google Scholar]
- 24.Díaz-Guerra, M., C. Rivas, and M. Esteban. 1997. Activation of the IFN-inducible enzyme RNase L causes apoptosis of animal cells. Virology 236:354-363. [DOI] [PubMed] [Google Scholar]
- 25.Díaz-Guerra, M., C. Rivas, and M. Esteban. 1997. Inducible expression of the 2-5A synthetase/RNase L system results in inhibition of vaccinia virus replication. Virology 227:220-228. [DOI] [PubMed] [Google Scholar]
- 26.Ding, S. W. 2007. RNAi restricts virus infections. Microbe 2:296-301. [Google Scholar]
- 27.Dong, B., S. Kim, S. Hong, J. Das Gupta, K. Malathi, E. A. Klein, D. Ganem, J. L. Derisi, S. A. Chow, and R. H. Silverman. 2007. An infectious retrovirus susceptible to an IFN antiviral pathway from human prostate tumors. Proc. Natl. Acad. Sci. USA 104:1655-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dong, B., and R. H. Silverman. 1995. 2-5A-dependent RNase molecules dimerize during activation by 2-5A. J. Biol. Chem. 270:4133-4137. [DOI] [PubMed] [Google Scholar]
- 29.Duerst, R. J., and L. A. Morrison. 2006. Herpes simplex virus type 2-mediated disease is reduced in mice lacking RNase L. Virology 360:322-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flodström-Tullberg, M., M. Hultcrantz, A. Stotland, A. Maday, D. Tsai, C. Fine, B. Williams, R. Silverman, and N. Sarvetnick. 2005. RNase L and double-stranded RNA-dependent protein kinase exert complementary roles in islet cell defense during coxsackievirus infection. J. Immunol. 174:1171-1177. [DOI] [PubMed] [Google Scholar]
- 31.Floyd-Smith, G., E. Slattery, and P. Lengyel. 1981. Interferon action: RNA cleavage pattern of a (2′-5′)oligoadenylate-dependent endonuclease. Science 212:1030-1032. [DOI] [PubMed] [Google Scholar]
- 32.Gallei, A., A. Pankraz, H. J. Thiel, and P. Becher. 2004. RNA recombination in vivo in the absence of viral replication. J. Virol. 78:6271-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gómez, C. E., A. M. Vandermeeren, M. A. Garcia, E. Domingo-Gil, and M. Esteban. 2005. Involvement of PKR and RNase L in translational control and induction of apoptosis after hepatitis C polyprotein expression from a vaccinia virus recombinant. Virol. J. 2:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gribaudo, G., D. Lembo, G. Cavallo, S. Landolfo, and P. Lengyel. 1991. Interferon action: binding of viral RNA to the 40-kilodalton 2′-5′-oligoadenylate synthetase in interferon-treated HeLa cells infected with encephalomyocarditis virus. J. Virol. 65:1748-1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Griffin, D. E. 2007. Alphaviruses, p. 1023-1067. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 36.Guidotti, L. G., A. Morris, H. Mendez, R. Koch, R. H. Silverman, B. R. Williams, and F. V. Chisari. 2002. Interferon-regulated pathways that control hepatitis B virus replication in transgenic mice. J. Virol. 76:2617-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo, J., D. J. Hui, W. C. Merrick, and G. C. Sen. 2000. A new pathway of translational regulation mediated by eukaryotic initiation factor 3. EMBO J. 19:6891-6899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han, J. Q., and D. J. Barton. 2002. Activation and evasion of the antiviral 2′-5′ oligoadenylate synthetase/ribonuclease L pathway by hepatitis C virus mRNA. RNA 8:512-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han, J. Q., H. L. Townsend, B. K. Jha, J. M. Paranjape, R. H. Silverman, and D. J. Barton. 2007. A phylogenetically conserved RNA structure in the poliovirus open reading frame inhibits the antiviral endoribonuclease RNase L. J. Virol. 81:5561-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han, J. Q., G. Wroblewski, Z. Xu, R. H. Silverman, and D. J. Barton. 2004. Sensitivity of hepatitis C virus RNA to the antiviral enzyme ribonuclease L is determined by a subset of efficient cleavage sites. J. Interferon Cytokine Res. 24:664-676. [DOI] [PubMed] [Google Scholar]
- 41.Hartmann, R., J. Justesen, S. N. Sarkar, G. C. Sen, and V. C. Yee. 2003. Crystal structure of the 2′-specific and double-stranded RNA-activated interferon-induced antiviral protein 2′-5′-oligoadenylate synthetase. Mol. Cell 12:1173-1185. [DOI] [PubMed] [Google Scholar]
- 42.Hartmann, R., H. S. Olsen, S. Widder, R. Jorgensen, and J. Justesen. 1998. p59OASL, a 2′-5′ oligoadenylate synthetase like protein: a novel human gene related to the 2′-5′ oligoadenylate synthetase family. Nucleic Acids Res. 26:4121-4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hassel, B. A., A. Zhou, C. Sotomayor, A. Maran, and R. H. Silverman. 1993. A dominant negative mutant of 2-5A-dependent RNase suppresses antiproliferative and antiviral effects of interferon. EMBO J. 12:3297-3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hersh, C. L., R. E. Brown, W. K. Roberts, E. A. Swyryd, I. M. Kerr, and G. R. Stark. 1984. Simian virus 40-infected, interferon-treated cells contain 2′,5′-oligoadenylates which do not activate cleavage of RNA. J. Biol. Chem. 259:1731-1737. [PubMed] [Google Scholar]
- 45.Hovanessian, A. G., R. E. Brown, and I. M. Kerr. 1977. Synthesis of low molecular weight inhibitor of protein synthesis with enzyme from interferon-treated cells. Nature 268:537-540. [DOI] [PubMed] [Google Scholar]
- 46.Hovanessian, A. G., and J. Justesen. 2007. The human 2′-5′oligoadenylate synthetase family: unique interferon-inducible enzymes catalyzing 2′-5′ instead of 3′-5′ phosphodiester bond formation. Biochimie 89:779-788. [DOI] [PubMed] [Google Scholar]
- 47.Hovanessian, A. G., J. Svab, I. Marie, N. Robert, S. Chamaret, and A. G. Laurent. 1988. Characterization of 69- and 100-kDa forms of 2-5A-synthetase from interferon-treated human cells. J. Biol. Chem. 263:4945-4949. [PubMed] [Google Scholar]
- 48.Hovnanian, A., D. Rebouillat, M. G. Mattei, E. R. Levy, I. Marie, A. P. Monaco, and A. G. Hovanessian. 1998. The human 2′,5′-oligoadenylate synthetase locus is composed of three distinct genes clustered on chromosome 12q24.2 encoding the 100-, 69-, and 40-kDa forms. Genomics 52:267-277. [DOI] [PubMed] [Google Scholar]
- 49.Iordanov, M. S., J. M. Paranjape, A. Zhou, J. Wong, B. R. G. Williams, E. F. Meurs, R. H. Silverman, and B. E. Magun. 2000. Activation of p38 mitogen-activated protein kinase and c-Jun NH2-terminal kinase by double-stranded RNA and encephalomyocarditis virus: involvement of RNase L, protein kinase R, and alternative pathways. Mol. Cell. Biol. 20:617-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Isaacs, A., and J. Lindenmann. 1957. Virus interference. I. The interferon. Proc. R. Soc. Lond. B Biol. Sci. 147:258-267. [PubMed] [Google Scholar]
- 51.Johnston, M. I., and W. G. Hearl. 1987. Purification and characterization of a 2′-phosphodiesterase from bovine spleen. J. Biol. Chem. 262:8377-8382. [PubMed] [Google Scholar]
- 52.Justesen, J., R. Hartmann, and N. O. Kjeldgaard. 2000. Gene structure and function of the 2′-5′-oligoadenylate synthetase family. Cell. Mol. Life Sci. 57:1593-1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kajaste-Rudnitski, A., T. Mashimo, M. P. Frenkiel, J. L. Guenet, M. Lucas, and P. Despres. 2006. The 2′,5′-oligoadenylate synthetase 1b is a potent inhibitor of West Nile virus replication inside infected cells. J. Biol. Chem. 281:4624-4637. [DOI] [PubMed] [Google Scholar]
- 54.Kakuta, S., S. Shibata, and Y. Iwakura. 2002. Genomic structure of the mouse 2′,5′-oligoadenylate synthetase gene family. J. Interferon Cytokine Res. 22:981-993. [DOI] [PubMed] [Google Scholar]
- 55.Kerr, I. M., and R. E. Brown. 1978. pppA2′p5′A2′p5′A: an inhibitor of protein synthesis synthesized with an enzyme fraction from interferon-treated cells. Proc. Natl. Acad. Sci. USA 75:256-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khabar, K. S., M. Dhalla, Y. Siddiqui, A. Zhou, M. N. Al-Ahdal, S. D. Der, R. H. Silverman, and B. R. Williams. 2000. Effect of deficiency of the double-stranded RNA-dependent protein kinase, PKR, on antiviral resistance in the presence or absence of ribonuclease L: HSV-1 replication is particularly sensitive to deficiency of the major IFN-mediated enzymes. J. Interferon Cytokine Res. 20:653-659. [DOI] [PubMed] [Google Scholar]
- 57.Knight, M., P. J. Cayley, R. H. Silverman, D. H. Wreschner, C. S. Gilbert, R. E. Brown, and I. M. Kerr. 1980. Radioimmune, radiobinding and HPLC analysis of 2-5A and related oligonucleotides from intact cells. Nature 288:189-192. [DOI] [PubMed] [Google Scholar]
- 58.Knight, M., D. H. Wreschner, R. H. Silverman, and I. M. Kerr. 1981. Radioimmune and radiobinding assays for A2′p5′A2′p5′A, pppA2′p5′A, and related oligonucleotides. Methods Enzymol. 79:216-227. [PubMed] [Google Scholar]
- 59.Kubota, K., K. Nakahara, T. Ohtsuka, S. Yoshida, J. Kawaguchi, Y. Fujita, Y. Ozeki, A. Hara, C. Yoshimura, H. Furukawa, H. Haruyama, K. Ichikawa, M. Yamashita, T. Matsuoka, and Y. Iijima. 2004. Identification of 2′-phosphodiesterase, which plays a role in the 2-5A system regulated by interferon. J. Biol. Chem. 279:37832-37841. [DOI] [PubMed] [Google Scholar]
- 60.Leaman, D. W., F. J. Longano, J. R. Okicki, K. F. Soike, P. F. Torrence, R. H. Silverman, and H. Cramer. 2002. Targeted therapy of respiratory syncytial virus in African green monkeys by intranasally administered 2-5A antisense. Virology 292:70-77. [DOI] [PubMed] [Google Scholar]
- 61.Leib, D. A., M. A. Machalek, B. R. Williams, R. H. Silverman, and H. W. Virgin. 2000. Specific phenotypic restoration of an attenuated virus by knockout of a host resistance gene. Proc. Natl. Acad. Sci. USA 97:6097-6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Le Roy, F., C. Bisbal, M. Silhol, C. Martinand, B. Lebleu, and T. Salehzada. 2001. The 2-5A/RNase L/RNase L inhibitor (RLI) pathway regulates mitochondrial mRNAs stability in interferon α-treated H9 cells. J. Biol. Chem. 276:48473-48482. [DOI] [PubMed] [Google Scholar]
- 63.Le Roy, F., M. Silhol, T. Salehzada, and C. Bisbal. 2007. Regulation of mitochondrial mRNA stability by RNase L is translation-dependent and controls IFNα-induced apoptosis. Cell Death Differ. 14:1406-1413. [DOI] [PubMed] [Google Scholar]
- 64.Li, G., Y. Xiang, K. Sabapathy, and R. H. Silverman. 2004. An apoptotic signaling pathway in the interferon antiviral response mediated by RNase L and c-Jun NH2-terminal kinase. J. Biol. Chem. 279:1123-1131. [DOI] [PubMed] [Google Scholar]
- 65.Li, X. L., J. A. Blackford, and B. A. Hassel. 1998. RNase L mediates the antiviral effect of interferon through a selective reduction in viral RNA during encephalomyocarditis virus infection. J. Virol. 72:2752-2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lindenbach, B. D., H. J. Thiel, and C. M. Rice. 2007. Flaviviridae: the viruses and their replication, p. 1101-1152. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 67.Maitra, R. K., N. A. McMillan, S. Desai, J. McSwiggen, A. G. Hovanessian, G. Sen, B. R. Williams, and R. H. Silverman. 1994. HIV-1 TAR RNA has an intrinsic ability to activate interferon-inducible enzymes. Virology 204:823-827. [DOI] [PubMed] [Google Scholar]
- 68.Maitra, R. K., and R. H. Silverman. 1998. Regulation of human immunodeficiency virus replication by 2′,5′-oligoadenylate-dependent RNase L. J. Virol. 72:1146-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Malathi, K., B. Dong, M. Gale, Jr., and R. H. Silverman. 2007. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature 448:816-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Malathi, K., J. M. Paranjape, E. Bulanova, M. Shim, J. M. Guenther-Johnson, P. W. Faber, T. E. Eling, B. R. Williams, and R. H. Silverman. 2005. A transcriptional signaling pathway in the IFN system mediated by 2′-5′-oligoadenylate activation of RNase L. Proc. Natl. Acad. Sci. USA 102:14533-14538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Malathi, K., J. M. Paranjape, R. Ganapathi, and R. H. Silverman. 2004. HPC1/RNASEL mediates apoptosis of prostate cancer cells treated with 2′,5′-oligoadenylates, topoisomerase I inhibitors, and tumor necrosis factor-related apoptosis-inducing ligand. Cancer Res. 64:9144-9151. [DOI] [PubMed] [Google Scholar]
- 72.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 73.Marié, I., J. Blanco, D. Rebouillat, and A. G. Hovanessian. 1997. 69-kDa and 100-kDa isoforms of interferon-induced (2′-5′)oligoadenylate synthetase exhibit differential catalytic parameters. Eur. J. Biochem. 248:558-566. [DOI] [PubMed] [Google Scholar]
- 74.Martinand, C., C. Montavon, T. Salehzada, M. Silhol, B. Lebleu, and C. Bisbal. 1999. RNase L inhibitor is induced during human immunodeficiency virus type 1 infection and down regulates the 2-5A/RNase L pathway in human T cells. J. Virol. 73:290-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mashimo, T., P. Glaser, M. Lucas, D. Simon-Chazottes, P. E. Ceccaldi, X. Montagutelli, P. Despres, and J. L. Guenet. 2003. Structural and functional genomics and evolutionary relationships in the cluster of genes encoding murine 2′,5′-oligoadenylate synthetases. Genomics 82:537-552. [DOI] [PubMed] [Google Scholar]
- 76.Mashimo, T., M. Lucas, D. Simon-Chazottes, M. P. Frenkiel, X. Montagutelli, P. E. Ceccaldi, V. Deubel, J. L. Guenet, and P. Despres. 2002. A nonsense mutation in the gene encoding 2′-5′-oligoadenylate synthetase/L1 isoform is associated with West Nile virus susceptibility in laboratory mice. Proc. Natl. Acad. Sci. USA 99:11311-11316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McClary, H., R. Koch, F. V. Chisari, and L. G. Guidotti. 2000. Relative sensitivity of hepatitis B virus and other hepatotropic viruses to the antiviral effects of cytokines. J. Virol. 74:2255-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McHutchison, J. G., S. C. Gordon, E. R. Schiff, M. L. Shiffman, W. M. Lee, V. K. Rustgi, Z. D. Goodman, M. H. Ling, S. Cort, and J. K. Albrecht. 1998. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. N. Engl. J. Med. 339:1485-1492. [DOI] [PubMed] [Google Scholar]
- 79.Meurs, E., K. Chong, J. Galabru, N. S. Thomas, I. M. Kerr, B. R. Williams, and A. G. Hovanessian. 1990. Molecular cloning and characterization of the human double-stranded RNA-activated protein kinase induced by interferon. Cell 62:379-390. [DOI] [PubMed] [Google Scholar]
- 80.Min, J. Y., and R. M. Krug. 2006. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: inhibiting the 2′-5′ oligo (A) synthetase/RNase L pathway. Proc. Natl. Acad. Sci. USA 103:7100-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nilsen, T. W., and C. Baglioni. 1979. Mechanism for discrimination between viral and host mRNA in interferon-treated cells. Proc. Natl. Acad. Sci. USA 76:2600-2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nilsen, T. W., P. A. Maroney, and C. Baglioni. 1982. Synthesis of (2′-5′) oligoadenylate and activation of an endoribonuclease in interferon-treated HeLa cells infected with reovirus. J. Virol. 42:1039-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Olland, A. M., J. Jane-Valbuena, L. A. Schiff, M. L. Nibert, and S. C. Harrison. 2001. Structure of the reovirus outer capsid and dsRNA-binding protein σ3 at 1.8 Å resolution. EMBO J. 20:979-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pallansch, M. A., and R. Roos. 2007. Enteroviruses, polioviruses, coxsackieviruses, echoviruses, and newer enteroviruses, p. 839-893. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 85.Pellett, P. E., and B. Roizman. 2007. The family herpesviridae: a brief introduction, p. 2479-2499. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 86.Perelygin, A. A., S. V. Scherbik, I. B. Zhulin, B. M. Stockman, Y. Li, and M. A. Brinton. 2002. Positional cloning of the murine flavivirus resistance gene. Proc. Natl. Acad. Sci. USA 99:9322-9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rancaniello, V. 2007. Picornaviridae: the viruses and their replication, p. 795-838. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 88.Rebouillat, D., and A. G. Hovanessian. 1999. The human 2′,5′-oligoadenylate synthetase family: interferon-induced proteins with unique enzymatic properties. J. Interferon Cytokine Res. 19:295-308. [DOI] [PubMed] [Google Scholar]
- 89.Rice, A. P., S. M. Kerr, W. K. Roberts, R. E. Brown, and I. M. Kerr. 1985. Novel 2′,5′-oligoadenylates synthesized in interferon-treated, vaccinia virus-infected cells. J. Virol. 56:1041-1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rice, A. P., W. K. Roberts, and I. M. Kerr. 1984. 2-5A accumulates to high levels in interferon-treated, vaccinia virus-infected cells in the absence of any inhibition of virus replication. J. Virol. 50:220-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rivas, C., J. Gil, Z. Melkova, M. Esteban, and M. Diaz-Guerra. 1998. Vaccinia virus E3L protein is an inhibitor of the interferon (i.f.n.)-induced 2-5A synthetase enzyme. Virology 243:406-414. [DOI] [PubMed] [Google Scholar]
- 92.Rogozin, I. B., L. Aravind, and E. V. Koonin. 2003. Differential action of natural selection on the N and C-terminal domains of 2′-5′ oligoadenylate synthetases and the potential nuclease function of the C-terminal domain. J. Mol. Biol. 326:1449-1461. [DOI] [PubMed] [Google Scholar]
- 93.Rusch, L., A. Zhou, and R. H. Silverman. 2000. Caspase-dependent apoptosis by 2′,5′-oligoadenylate activation of RNase L is enhanced by IFN-beta. J. Interferon Cytokine Res. 20:1091-1100. [DOI] [PubMed] [Google Scholar]
- 94.Rutherford, M. N., G. E. Hannigan, and B. R. Williams. 1988. Interferon-induced binding of nuclear factors to promoter elements of the 2-5A synthetase gene. EMBO J. 7:751-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ryman, K. D., L. J. White, R. E. Johnston, and W. B. Klimstra. 2002. Effects of PKR/RNase L-dependent and alternative antiviral pathways on alphavirus replication and pathogenesis. Viral Immunol. 15:53-76. [DOI] [PubMed] [Google Scholar]
- 96.Samuel, M. A., K. Whitby, B. C. Keller, A. Marri, W. Barchet, B. R. Williams, R. H. Silverman, M. Gale, Jr., and M. S. Diamond. 2006. PKR and RNase L contribute to protection against lethal West Nile Virus infection by controlling early viral spread in the periphery and replication in neurons. J. Virol. 80:7009-7019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Sangster, M. Y., N. Urosevic, J. P. Mansfield, J. S. Mackenzie, and G. R. Shellam. 1994. Mapping the Flv locus controlling resistance to flaviviruses on mouse chromosome 5. J. Virol. 68:448-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sawicki, D. L., R. H. Silverman, B. R. Williams, and S. G. Sawicki. 2003. Alphavirus minus-strand synthesis and persistence in mouse embryo fibroblasts derived from mice lacking RNase L and protein kinase R. J. Virol. 77:1801-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Scherbik, S. V., J. M. Paranjape, B. M. Stockman, R. H. Silverman, and M. A. Brinton. 2006. RNase L plays a role in the antiviral response to West Nile virus. J. Virol. 80:2987-2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schiff, L. A., M. L. Nibert, and K. L. Tyler. 2007. Orthoreoviruses and their replication, p. 1853-1915. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 101.Schmidt, A., Y. Chernajovsky, L. Shulman, P. Federman, H. Berissi, and M. Revel. 1979. An interferon-induced phosphodiesterase degrading (2′-5′) oligoisoadenylate and the C-C-A terminus of tRNA. Proc. Natl. Acad. Sci. USA 76:4788-4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Schröder, H. C., D. Ugarkovic, R. Wenger, P. Reuter, T. Okamoto, and W. E. Muller. 1990. Binding of Tat protein to TAR region of human immunodeficiency virus type 1 blocks TAR-mediated activation of (2′-5′) oligoadenylate synthetase. AIDS Res. Hum. Retrovir. 6:659-672. [DOI] [PubMed] [Google Scholar]
- 103.SenGupta, D. N., and R. H. Silverman. 1989. Activation of interferon-regulated, dsRNA-dependent enzymes by human immunodeficiency virus-1 leader RNA. Nucleic Acids Res. 17:969-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Silverman, R. H. 2003. Implications for RNase L in prostate cancer biology. Biochemistry 42:1805-1812. [DOI] [PubMed] [Google Scholar]
- 105.Silverman, R. H., P. J. Cayley, M. Knight, C. S. Gilbert, and I. M. Kerr. 1982. Control of the ppp(a2′p)nA system in HeLa cells. Effects of interferon and virus infection. Eur. J. Biochem. 124:131-138. [DOI] [PubMed] [Google Scholar]
- 106.Silverman, R. H., J. J. Skehel, T. C. James, D. H. Wreschner, and I. M. Kerr. 1983. rRNA cleavage as an index of ppp(A2′p)nA activity in interferon-treated encephalomyocarditis virus-infected cells. J. Virol. 46:1051-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Silverman, R. H., D. H. Wreschner, C. S. Gilbert, and I. M. Kerr. 1981. Synthesis, characterization and properties of ppp(A2′p)nApCp and related high-specific-activity 32P-labelled derivatives of ppp(A2′p)nA. Eur. J. Biochem. 115:79-85. [DOI] [PubMed] [Google Scholar]
- 108.Silverman, R. H., A. Zhou, M. B. Auerbach, D. Kish, A. Gorbachev, and R. L. Fairchild. 2002. Skin allograft rejection is suppressed in mice lacking the antiviral enzyme, 2′,5′-oligoadenylate-dependent RNase L. Viral Immunol. 15:77-83. [DOI] [PubMed] [Google Scholar]
- 109.Smith, E. J., I. Marie, A. Prakash, A. Garcia-Sastre, and D. E. Levy. 2001. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or IκB kinase but is blocked by vaccinia virus E3L protein. J. Biol. Chem. 276:8951-8957. [DOI] [PubMed] [Google Scholar]
- 110.Smith, J. A., S. C. Schmechel, B. R. Williams, R. H. Silverman, and L. A. Schiff. 2005. Involvement of the interferon-regulated antiviral proteins PKR and RNase L in reovirus-induced shutoff of cellular translation. J. Virol. 79:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Smith, T. J., R. H. Silverman, and D. A. Leib. 2003. RNase L activity does not contribute to host RNA degradation induced by herpes simplex virus infection. J. Gen. Virol. 84:925-928. [DOI] [PubMed] [Google Scholar]
- 112.Tanaka, N., M. Nakanishi, Y. Kusakabe, Y. Goto, Y. Kitade, and K. T. Nakamura. 2004. Structural basis for recognition of 2′,5′-linked oligoadenylates by human ribonuclease L. EMBO J. 23:3929-3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Thakur, C. S., B. K. Jha, B. Dong, J. Das Gupta, K. M. Silverman, H. Mao, H. Sawai, A. O. Nakamura, A. K. Banerjee, A. Gudkov, and R. H. Silverman. 2007. Small-molecule activators of RNase L with broad-spectrum antiviral activity. Proc. Natl. Acad. Sci. USA 104:9585-9590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Urisman, A., R. J. Molinaro, N. Fischer, S. J. Plummer, G. Casey, E. A. Klein, K. Malathi, C. Magi-Galluzzi, R. R. Tubbs, D. Ganem, R. H. Silverman, and J. L. Derisi. 2006. Identification of a novel gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathogens 2:e25. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 115.Washenberger, C. L., J. Q. Han, K. J. Kechris, B. K. Jha, R. H. Silverman, and D. J. Barton. 29 June 2007. Hepatitis C virus RNA: dinucleotide frequencies and cleavage by RNase L. Virus Res. doi: 10.1016/j.virusres.2007.05.020. [DOI] [PMC free article] [PubMed]
- 116.Watson, J. C., H. W. Chang, and B. L. Jacobs. 1991. Characterization of a vaccinia virus-encoded double-stranded RNA-binding protein that may be involved in inhibition of the double-stranded RNA-dependent protein kinase. Virology 185:206-216. [DOI] [PubMed] [Google Scholar]
- 117.Williams, B. R., R. R. Golgher, R. E. Brown, C. S. Gilbert, and I. M. Kerr. 1979. Natural occurrence of 2-5A in interferon-treated EMC virus-infected L cells. Nature 282:582-586. [DOI] [PubMed] [Google Scholar]
- 118.Wreschner, D. H., T. C. James, R. H. Silverman, and I. M. Kerr. 1981. Ribosomal RNA cleavage, nuclease activation and 2-5A(ppp(A2′p)nA) in interferon-treated cells. Nucleic Acids Res. 9:1571-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wreschner, D. H., J. W. McCauley, J. J. Skehel, and I. M. Kerr. 1981. Interferon action—sequence specificity of the ppp(A2′p)nA-dependent ribonuclease. Nature 289:414-417. [DOI] [PubMed] [Google Scholar]
- 120.Wright, P. F., G. Neumann, and Y. Kawaoka. 2007. Orthomyxoviruses, p. 1691-1740. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA.
- 121.Xiang, Y., R. C. Condit, S. Vijaysri, B. Jacobs, B. R. Williams, and R. H. Silverman. 2002. Blockade of interferon induction and action by the E3L double-stranded RNA binding proteins of vaccinia virus. J. Virol. 76:5251-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Xiang, Y., Z. Wang, J. Murakami, S. Plummer, E. A. Klein, J. D. Carpten, J. M. Trent, W. B. Isaacs, G. Casey, and R. H. Silverman. 2003. Effects of RNase L mutations associated with prostate cancer on apoptosis induced by 2′,5′-oligoadenylates. Cancer Res. 63:6795-6801. [PubMed] [Google Scholar]
- 123.Yakub, I., K. M. Lillibridge, A. Moran, O. Y. Gonzalez, J. Belmont, R. A. Gibbs, and D. J. Tweardy. 2005. Single nucleotide polymorphisms in genes for 2′-5′-oligoadenylate synthetase and RNase L inpatients hospitalized with West Nile virus infection. J. Infect. Dis. 192:1741-1748. [DOI] [PubMed] [Google Scholar]
- 124.Zheng, X., R. H. Silverman, A. Zhou, T. Goto, B. S. Kwon, H. E. Kaufman, and J. M. Hill. 2001. Increased severity of HSV-1 keratitis and mortality in mice lacking the 2-5A-dependent RNase L gene. Investig. Ophthalmol. Vis. Sci. 42:120-126. [PubMed] [Google Scholar]
- 125.Zhou, A., B. A. Hassel, and R. H. Silverman. 1993. Expression cloning of 2-5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell 72:753-765. [DOI] [PubMed] [Google Scholar]
- 126.Zhou, A., J. Paranjape, T. L. Brown, H. Nie, S. Naik, B. Dong, A. Chang, B. Trapp, R. Fairchild, C. Colmenares, and R. H. Silverman. 1997. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J. 16:6355-6363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhou, A., J. M. Paranjape, S. D. Der, B. R. Williams, and R. H. Silverman. 1999. Interferon action in triply deficient mice reveals the existence of alternative antiviral pathways. Virology 258:435-440. [DOI] [PubMed] [Google Scholar]
- 128.Zhou, A., J. M. Paranjape, B. A. Hassel, H. Nie, S. Shah, B. Galinski, and R. H. Silverman. 1998. Impact of RNase L overexpression on viral and cellular growth and death. J. Interferon Cytokine Res. 18:953-961. [DOI] [PubMed] [Google Scholar]
- 129.Zimmerman, C., K. C. Klein, P. K. Kiser, A. R. Singh, B. L. Firestein, S. C. Riba, and J. R. Lingappa. 2002. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature 415:88-92. [DOI] [PubMed] [Google Scholar]