Abstract
Insulator elements can be classified as enhancer-blocking or barrier insulators depending on whether they interfere with enhancer-promoter interactions or act as barriers against the spreading of heterochromatin. The former class may exert its function at least in part by attaching the chromatin fiber to a nuclear substrate such as the nuclear matrix, resulting in the formation of chromatin loops. The latter class functions by recruiting histone modifying enzymes, although some barrier insulators have also been shown to create chromatin loops. These loops may correspond to functional nuclear domains containing clusters of co-expressed genes. Thus, insulators may determine specific patterns of nuclear organization that are important in establishing specific programs of gene expression during cell differentiation and development.
Keywords: insulators, chromatin, loops, gene expression, nuclear organization
1. Introduction
Insulator elements are DNA sequences characterized by two experimentally determined properties that allow their classification into two different subclasses [1]. Enhancer-blocking insulators prevent an enhancer from communicating with a promoter when positioned between the two. This phenomenon occurs without inactivating either the enhancer or the promoter, as both can still communicate with other regulatory sequences. Barrier insulators shield genes from position-effect variegation (PEV) [2] that results from proximity to heterochromatin. If the heterochromatin adjacent to a gene does not have a barrier insulator (also referred to as boundary element), it will spread into the gene in some cells, silencing it, while in other cells it will not. This leads to a mosaic expression pattern. A barrier insulator inserted between heterochromatin and a gene will stop the spread of heterochromatin, allowing the gene to be expressed in all cells. In most cases, these two modes of insulator activity can be uncoupled, although some insulators may have both properties. In this review, we will discuss our current understanding of the mechanisms by which each of these types of insulators affect chromatin structure and gene expression. We will distinguish between the two types by using the “enhancer-blocking” or “barrier” nomenclature proposed by Gaszner and Felsenfeld [1] and we will use the generic term “insulator” when we refer to both types simultaneously.
2. Barrier insulators affect chromatin structure
Barrier insulators alter the structure of chromatin by affecting the covalent modification of histones. Studies in both yeast and vertebrate cells have shed light on barrier insulator function. The HMR mating type locus in S. cerevisiae is heterochromatic and features two silencing elements called E and I. These sequences are bound by proteins that recruit a complex of Sir2p, Sir3p and Sir4p. This Sir protein complex spreads bi-directional from the silencers, interacting with nucleosomes [3]. In particular, Sir2p has a histone deacetylase activity that is required for spreading [4]. The majority of the silenced region lies between E and I, but the borders are located outside these elements. The right border has a well-characterized barrier insulator, of which the principal component is a tRNAThr gene [5]. Transcription of this gene by RNA Polymerase III is required for barrier activity; also important are the acetyltransferases Sas2p and Gcn5p. In fact, artificially tethering these acetyltransferases to chromatin is sufficient to induce barrier activity [6]. These observations lead to a model in which the boundary of the silenced region is determined by a dynamic equilibrium between histone deacetylation activity originating from the heterochromatin and histone acetylation activity centered around the barrier element.
The telomeric regions in yeast also possess flanking sequences that block heterochromatin propagation; these are known as subtelomeric anti-silencing regions, or STARs [7]. These regions contain binding sites for the transcription factors Reb1p and Tbf1p. The activation domains of these transcription factors, as well as of a number of others, are sufficient to confer insulator activity on a sequence when tethered to it. However, direct transcriptional activation of a promoter by the proteins is not required [8]. This effect could be due to the recruitment of activating enzymes such as histone acetylases by these transcription factors, in concordance with the model described above. An alternative model suggests that somehow these proteins create a physical obstruction that spreading heterochromatin cannot pass, perhaps by blocking access to histones. In support of this second model, the transcription factor CTF-1 is known to bind histone H3 directly. This protein possesses insulator activity, for which the only necessary domain is the histone-binding domain. This suggests that the barrier function is simply due to the protein occupying the histone and preventing silencing proteins from interacting with it, rather than to any enzymatic activity of CTF-1 [9].
In vertebrates, the 5′HS4 element at the β-globin locus possesses both enhancer-blocking (see below) and barrier activity. These activities are separable, with CTCF binding sites required for enhancer-blocking but not for barrier function [10]. However, binding sites for the transcription factors USF1 and USF2 are required for barrier activity. These proteins have been shown to interact with the H3K4-specific methyltransferase SET7/9 and the H3-specific histone acetyltransferase PCAF. The modifications conferred by these enzymes are associated with active chromatin. Knockdown of USF1 causes a decrease in H3K4 methylation and H3 acetylation at the 5′HS4 element, as well as an increase in H3K9 methylation, which is associated with inactive chromatin [11]. These results suggest an expansion of the model proposed for barrier function in yeast in which the equilibrium includes not merely acetylation and deacetylation of histones but other types of activating and deactivating chromatin modifications as well. In this case, both acetylation and methylation of histones are specifically found to be involved.
There are also indications that nucleosome positioning can play a role in blocking the spread of heterochromatin. Mutations in RSC2, which is a member of the RSC chromatin remodeling complex, impair the function of the yeast HMR right boundary [12]. Additionally, barrier activity can be induced by tethering Snf6p, a component of the Swi/Snf chromatin remodeling complex, to an arbitrary sequence element [13]. These observations make sense in light of what we know about the propagation of heterochromatin: if spreading of the silencing complex involves proteins on one nucleosome deacetylating and recruiting proteins to bind the next nucleosome, then an interruption in the nucleosome sequence could easily interfere with the spreading process. While this mechanism involves chromatin-activating enzyme function and therefore fits the model that barrier insulators block heterochromatin by recruiting competing enzyme activities, it is also an example of the second general model, namely, that barrier insulators function by creating a physical obstruction that propagating heterochromatin proteins cannot cross.
3. Enhancer-blocking insulators mediate the formation of chromatin loops
There is mounting evidence that enhancer-blocking insulators might compartmentalize the chromatin into structural loops, with insulator proteins at the base of the loops, either clustered together or bound to some structural component of the nucleus (Figure 1). These tethering structures can be other insulator elements and/or nuclear components such as the nuclear lamina, the nucleolus or nuclear pores [14, 15]. The activity of various enhancer-blocking insulators has been shown to require this type of tethering, suggesting a common mechanistic theme amongst these elements. Much of the evidence supporting this conclusion comes from the study of insulators in Drosophila. The scs and scs’ insulator elements flank the hsp70A locus in Drosophila and were the first sequences described with insulator properties [16]. This activity is conferred by two different proteins, Zw5 (Zeste-white 5), which interacts with scs, and BEAF-32 (Boundary Element Associated Factor), which interacts with scs’ [17, 18]. Zw5 and BEAF-32 interact in vitro and in vivo, and chromatin capture conformation experiments (3C) have revealed that scs and scs’ are found in close spatial proximity within Drosophila embryonic nuclei [19]. This information suggests that protein-protein interactions between Zw5 and BEAF-32 bring the scs and scs’ insulator elements together and, as a consequence, the connecting chromatin forms a loop.
Figure 1. Insulator elements organize the chromatin fiber in the nucleus by establishing separate compartments of higher-order chromatin structure.
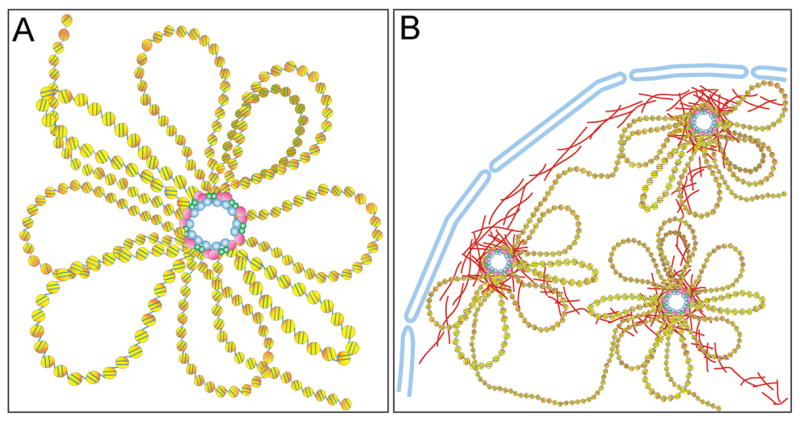
(A) Domains of open chromatin (yellow nucleosomes) are flanked by insulators (pink, blue and green spheres) that interact together to form a loop. (B) Diagram showing part of a nucleus with compartmentalized chromatin, anchored in part to the nuclear periphery by interactions of the insulators with the nuclear lamina (red lines).
Looping due to insulator protein interactions has also been shown for the Drosophila Su(Hw) (Suppressor of Hairy-wing) insulator, which was originally identified in the gypsy retrotransposon. The Su(Hw) protein localizes to hundreds of genomic sites in addition to gypsy as visualized on polytene chromosomes [20]. Recent studies have used bioinformatics to predict the location of these binding sites in the genome and found that they display insulator activity [21, 22]. Looping between Su(Hw) insulators has been visualized in imaginal disc cells by fluorescence in situ hybridization (FISH) after nuclear extraction with 2M NaCl [23]. This study not only showed loop formation between two Su(Hw) insulator elements, but also demonstrated that the addition of a third insulator element between the two original insulators resulted in the formation of two smaller loops. Both the CP190 (Centrosomal Protein 190) and Mod(mdg4)2.2 (Modifier of mdg4 2.2) proteins, also found in the Su(Hw) insulator complex, contain BTB/POZ domains that are thought to be involved in the formation of homodimers or heterodimers [24–26]. These interactions are proposed to facilitate the clustering of insulator elements resulting in the formation of insulator bodies. Insulator bodies are sites where multiple individual insulators coalesce causing the intervening chromatin fiber to form chromatin loops (Figure 1A). Additionally, a fourth component of the Su(Hw) insulator complex, Drosophila dTopors (Topoisomerase I-interacting RS protein), interacts with lamin [27]. This interaction tethers the insulator bodies to the nuclear lamina/matrix and is necessary for insulator function. Therefore, it seems that the Su(Hw) insulator requires both self-interaction between insulator proteins and tethering to the nuclear matrix to maintain its insulator function.
Tethering of insulator elements to a nuclear substrate has also been shown in vertebrate cells. The CTCF protein binds to almost all known vertebrate enhancer-blocking insulators [15]. It has been shown that CTCF interacts with the nucleolar phosphoprotein nucleophosmin in HeLa cells and localizes to the nucleolar surface. Nucleophosmin was found at CTCF insulator elements, suggesting a role for nucleolar localization in insulator function [28]. CTCF has also been found to be present in the nuclear matrix fraction, suggesting that interaction of CTCF with the nuclear matrix, in addition to the nucleolus, might be another mechanism to attach CTCF insulators to a nuclear substrate in vertebrates [29].
4. Shared mechanisms between the two types of insulators
From the previous discussion it appears that barrier insulators and enhancer-blocking insulators act via different mechanistic pathways, with barrier elements recruiting chromatin remodeling enzymes that antagonize the spreading of heterochromatin and enhancer-blocking insulators mediating the formation of chromatin loops. Nevertheless, tethering to fixed structures within the nucleus to form chromatin loops also seems to result in barrier function. It is possible that the formation of a loop tethered to a fixed nuclear substrate interferes with the transmission of signals emanating either from an enhancer or heterochromatin. The first indication of this possibility came from studies by Ishii et al. [30], who created an assay to screen for boundary proteins in S. cerevisiae and surprisingly identified various nuclear transport proteins. They determined that artificially-induced tethering of an arbitrary sequence to the nuclear pore via Nup2p is sufficient to give that sequence barrier activity. This could provide a physical block to heterochromatin in various ways. For example, immobilizing the chromatin could block the spreading of heterochromatin, as propagation of silencing proteins is associated with changes in supercoiling [31]. It is also possible that attachment of the insulator sequence to the nuclear pore complex results in such a large protein assembly that spreading heterochromatin proteins on one side are unable to access nucleosomes on the other side. Alternatively, localization of the insulator sequence to the nuclear pore may place it in a nuclear compartment that favors chromatin activation. Dilworth et al. [32] observed that nup2 mutants exhibit changes in the transcriptional profile, with many genes near the telomeres becoming active and many genes in the interiors of chromosomes becoming repressed. They interpret this to mean that Nup2p plays a role in maintaining endogenous chromatin domains, and suggest that, because Nup2p interacts with the nuclear pore complex transiently rather than stably, it may possess barrier activity due to an ability to transfer insulator sequences between silencing and activating nuclear compartments.
Interestingly, a cassette containing several Su(Hw) binding sites, together with transgenes expressing the Su(Hw) and Mod(mdg4)2.2 proteins, was shown to possess heterochromatin barrier activity in yeast [6]. It is unclear whether chromatin-activating enzymes would be recruited to this insulator in yeast. As this insertion contained Su(Hw) binding sites at only one locus, presumably this activity was independent of Su(Hw) insulator clustering. However, the possibility of loop formation mediated by interaction with some other substrate, such as the nuclear pores, cannot be excluded. Additionally, the gypsy insulator lacks enhancer blocking activity in yeast [33], suggesting that the two properties involve different mechanisms, and that clustering may be important for the enhancer-blocking function. It is also possible that the distinction is due to the two activities having differential requirements for other Drosophila proteins. It has been suggested that S. pombe also uses a looping mechanism to create functional insulator elements. The transcription factor TFIIIC is recruited to the regions flanking the mating loci in the absence of RNA Polymerase III (Pol III), and is necessary to prevent the spread of heterochromatin. Additional TFIIIC binding sites, independent of Pol III recruitment, were identified throughout the genome, and TFIIIC complexes where shown to localize in 5–10 foci at the nuclear periphery. These TFIIIC binding sites, called chromosome-organizing clamps (COCs), cluster at the nuclear periphery in a TFIIIC-dependent manner and are thought to form looped chromatin domains very similar to those described for the Su(Hw) insulator [34].
5. Mechanisms of insulator function by loop formation
The interaction of enhancer-blocking and barrier insulators with other nuclear structures to create chromatin loops is a common theme found in all eukaryotes. It is possible that these interactions lead to the formation of large complexes that create a physical barrier to regulatory elements. Though this by no means excludes other mechanisms to explain insulator activity, it does suggest a universal process used by these elements to create chromatin domains shielded from chromatin states and regulatory elements in the other parts of the genome. Several different possibilities can be postulated to explain how enhancer-promoter communication or heterochromatin spreading is affected by the formation of chromatin loops by insulators. One idea is that insulator-mediated loop formation may direct particular regions of the chromatin into proximity with specific nuclear compartments, such as transcription factories. Transcription factories are regions of the nucleus where RNA polymerases and their attendant transcription complexes cluster together, so that instead of the polymerase tracking along an immobile DNA strand, the DNA strand feeds through an immobile polymerase [35]. The factories are hypothesized to self-assemble and to exist only during the act of transcription. Presumably, transcription of a particular promoter would be facilitated by proximity to a transcription factory, either because of an elevated concentration of polymerase and transcription factors in that microenvironment, or because polymerase function is enhanced by participation in a factory. Thus, insulator elements could aid transcriptional activation of a locus by forming a loop that projects toward a transcription factory. One aspect that remains unclear is the relationship between chromatin loops mediated by insulators and chromatin loops formed by transcription factory assembly. It has been postulated that the mechanisms will eventually be seen to converge – that insulator activity is dependent on transcriptional activity [36, 37]. In support of this theory, some insulator elements, such as scs and scs’, have been shown to be transcribed [38], and DNA sequences made to function as transcriptional activators can also have insulator function [39].
Insulator-established chromatin loops could support intra-loop enhancer action, they could prevent inter-loop enhancer action, or they could do both. Enhancers are able to act on genomically distant promoters by coming in close physical proximity within the nucleus in a different form of chromatin looping. This type of interaction has been shown by 3C analysis between the murine β-globin locus and enhancer elements in the locus control region (LCR) found 40–60 kb away [40]. An interaction was only observed in cells where the β-globin locus is transcriptionally active, suggesting that this interaction plays a role in gene activation. A complementary study using a form of RNA fluorescence in situ hybridization (FISH) called RNA-TRAP to label chromatin near the transcriptionally active β-globin locus also demonstrated that enhancer regions of the LCR and the active β-globin locus come in close contact in vivo [41]. These observations indicate that physical interaction is necessary for an enhancer to activate a promoter and therefore suggest chromatin domains formed by insulator-mediated looping could regulate these interactions. An enhancer might have a higher probability of coming in contact with a promoter located within the same insulator-established chromatin loop than one located in a neighboring loop. In this way promoters located in the same loop as an enhancer could sequester that enhancer from interactions with the rest of the genome.
It is also possible that insulator loops interfere with enhancer-promoter communication by blocking the propagation of a signal along the chromatin between an enhancer and a promoter. Chip is a Drosophila protein that interacts with a variety of homeodomain factors and is thought to facilitate enhancer-promoter communication. It has been proposed that Chip functions by aiding in the spread of homeodomain proteins from an enhancer to its target promoter bringing the enhancer and promoter together [24, 42]. Chip was originally identified in a screen for enhancers of insulation by the gypsy retrotransposon [43]. Therefore, it is possible that one way insulators may block enhancer-promoter communication is by interrupting the propagation of activating signals, such as Chip-mediated spreading, from enhancer to promoter [15]. As a consequence, an enhancer located in one loop would only be able to send its activating signal to a promoter within the same loop. This type of mechanism could also explain the effect of loop formation on barrier activity against heterochromatin spreading.
An additional piece of information that gives insights into the mechanisms of insulator function comes from studies that show insulators are not completely impassable barriers to various signals. The placement of two Su(Hw) insulators in tandem between an enhancer and a promoter seems to neutralize boundary activity and possibly augment enhancer-promoter communication [44, 45]. This form of insulator bypass has been explained by the formation of a small loop between an enhancer and a promoter that actually brings the two in close physical proximity and therefore facilitates enhancer regulation of the promoter. The details of this mechanism, however, are not completely understood, partially due to conflicting data in the field. For example, three Su(Hw) insulators placed between an enhancer and a promoter could result in an insulator cluster forming two small chromatin loops. This would still neutralize boundary activity. Alternatively, two insulators could form a neutralizing pair leaving the third to interact with outside insulators restoring boundary activity. Both of these results have been obtained by different groups, suggesting multiple insulator mechanisms may exist [45–47]. An interesting question arising from these observations is whether or not insulator elements with different binding proteins can interact to form chromatin loops. Studies have suggested that the Su(Hw) insulator and binding sites for GAGA factor can form heterologous interactions that result in insulator bypass while other combinations of insulator elements cannot [46, 48, 49]. Again this suggests there may not be one universal mechanism for insulator function or that sequences defined experimentally as insulators may actually play different roles in the cell.
The ability of insulator pairs to result in insulator bypass argues against the tracking model of insulator activity, which would predict two insulators placed between an enhancer and a promoter would have the same or enhanced boundary activity. Also, it suggests insulators may affect enhancer-promoter communication by altering the physical distance between the two elements. According to this model, being in separate chromatin loops is not sufficient to block enhancer access to a promoter; the loops must also be physically displaced from one another. However, it has also been suggested that the loops formed during insulator bypass are not able to interact with outside insulators and therefore do not establish chromatin domains [50]. Though the exact mechanism of insulator bypass is not understood, it suggests that at least some insulators function through chromatin looping and emphasizes the importance of a physical interaction between an enhancer and a promoter.
6. The role of insulators in establishing functional domains of gene expression
Whatever the mechanism of insulator-mediated formation of chromatin loops, an important question is whether these structural loops correspond to functional domains of co-expressed genes. If this is the case, genes within each loop should have similar expression patterns, supporting a role for insulators in genome organization. Polytene chromosomes in Drosophila larval salivary glands have provided an excellent system in which to visualize chromatin domains. These chromosomes are characterized by regions of alternating high and low density chromatin, which are called bands and interbands due to their respective bright and dim appearance when stained with a DNA dye such as DAPI. Most transcription occurs in the relatively decondensed interbands whereas the bands contain inactive genes. The bands and interbands may thus correspond to functional domains of gene expression equivalent to the insulator-induced loops described above. Are insulators involved in forming the band/interband pattern? A number of different insulator proteins have been shown to be present at the boundaries between bands and interbands in polytene chromosomes, in agreement with a putative role for insulators in establishing or maintaining the band/interband domains. Several protein components of the gypsy insulator, Su(Hw), Mod(mdg4)2.2 and CP190, localize to a large number of sites on polytene chromosomes located at the boundaries between bands and interbands [26]. The insulator protein BEAF-32, which binds scs’, is also located at band/interband boundaries and at the borders of many developmental puffs, temporarily decondensed areas where high levels of transcription occur [18]. Inhibition of BEAF-32 binding causes polytene chromosomes to take on an expanded, fragile appearance without well-defined bands, suggesting that the scs’ insulator element is required to maintain the band/interband organization and proper chromosome structure [51].
The role of insulators in maintaining the band/interband domains has also been observed by elimination of particular DNA sequences rather than insulator proteins. The facet-strawberry (faswb) deletion in the interband between bands 3C6 and 3C7 causes the two bands to fuse, with the interband disappearing [52]. The faswb phenotype includes a rough, variably glossy eye, thought to be due to PEV affecting the nearby Notch promoter. Analysis of the deleted sequence indicates that it possesses insulator activity [53]. This result suggests that the faswb insulator is important for the maintenance of the interband between the 3C6 and 3C7 bands. If we think about each of these band/interbands as chromatin domains, deletion of the faswb insulator causes two domains to become one, suggesting that the role of the insulator is to form or maintain these domains.
Genomic mapping of insulator elements at the level of DNA sequence also suggests a correlation between insulator localization and gene arrangement. Clusters of co-expressed genes have now been identified in the yeast, fly, mouse, and human genomes [54–57], and there is growing evidence that these clusters may be flanked by insulators. In Drosophila predicted Su(Hw) binding sites are over-represented in regions of the genome that do not encode proteins and between gene dense regions, suggesting they may in fact play a role in organizing the genome into transcriptional domains [22]. Binding sites for the CTCF insulator proteins have also been mapped in the human genome [58, 59]. The distribution of these sites strongly correlates with the distribution of genes. Contrary to the distribution of a general transcription factor, which maps in close proximity to the 5′ start sites of genes, CTCF maps an average of 48 kb from gene promoters. Interestingly, CTCF sites are depleted with respect to the average in some chromosome regions that include clusters of transcriptionally co-expressed gene families (Kim et al., 2007). In addition, divergent gene pairs separated by CTCF binding sites show a reduced correlation in gene expression patterns (Xie et al., 2007). These two observations support a role for CTCF insulators in the formation of functional chromatin domains.
7. Regulation of insulator function
If insulators can compartmentalize the genome into functional domains of co-expressed genes, the organization of the chromatin established by insulators could have an important role in establishing broad patterns of gene expression during cell differentiation. If this is the case, cells must have mechanisms to regulate the activity of specific insulators so that, as cells differentiate along different pathways, they can establish different patterns of insulator-mediated nuclear organization (Figure 1B). These mechanisms appear to involve interfering with protein-protein interactions and protein binding to the DNA via competition, protein modification or DNA methylation. The first evidence for regulation of boundary elements came from the analysis of BEAF-32 binding sites in Drosophila. Two regions previously shown to bind BEAF-32 and act as boundary elements were found to also bind another protein, DREF (DNA replication-related element-binding factor), already characterized as an activator of transcription. Analysis of binding revealed BEAF and DREF occupy the sites independently, which lead to the model that these proteins are in competition for DNA binding. This suggests BEAF-32 binding establishes a site of insulation blocking enhancer-promoter communication while DREF binding blocks BEAF-32 and provides a permissive state for enhancer-promoter regulation [60]. A similar mechanism may regulate binding of the CTCF protein to DNA in vertebrates. Regulation of CTCF insulator activity can take place by blocking insulator protein DNA binding at the imprinted control region (ICR) of the mouse Igf2/H19 locus. It is well established that CTCF binds the maternal ICR allele, blocking enhancer activation of the Igf2 gene and resulting in H19 activation. However, in the paternal allele, methylation of the CTCF binding site blocks its binding and Igf2 is activated [61, 62]. This is an example of epigenetic regulation of insulator activity using a mechanism that involves inhibiting protein association with an insulator element.
An alternative mechanism to blocking protein association with chromatin that could result in the regulation of insulator activity involves modulation of insulator looping. Insulator activity has been shown to depend on chromatin looping, and inhibition of protein-protein interactions that mediate the formation of these loops or their interactions with the nuclear lamina/matrix could result in a loss of looping and a loss of insulator activity. Evidence for this method of regulation has been described for CTCF sites throughout the mouse genome. Yu et al. show poly(ADP-ribose) (PAR) co-localizes with 78% of CTCF binding sites genome-wide by ChIP-on-chip analysis of mouse fetal liver cells. Furthermore, they provide convincing evidence that PARlation is necessary for insulator activity at the ICR and notably the rest of the genome, though this modification is not necessary for binding of CTCF to DNA [63]. This suggests CTCF is modified by PARlation in order to facilitate CTCF homo-dimerization, which could be involved in loop formation necessary for insulator activity [64]. Interestingly, though PAR was found at the majority of CTCF binding sites, there were still many loci that contained CTCF but not the PAR modification. Though this could be an artifact from the PAR antibody, this observation could provide insights into the mechanism of activation CTCF insulator function. It is possible that the interaction between CTCF and nucleophosmin at insulator sites may be necessary to localize CTCF binding sites to nucleoli, which are enriched in PAR polymerases [63, 64]). This suggests CTCF insulator activation occurs after DNA binding. It is possible that PAR polymerases are also present in nuclear compartments other than nucleoli or that only a very specific subset of CTCF molecules are PARlated and serve to tether specific DNA sequences to the nucleolus. Alternatively, CTCF-associated sites that do not contain PAR could represent insulator sites that are inactive in the mouse fetal liver cells where this analysis was performed. There is also evidence supporting the idea that CTCF may be present but inactive at particular insulator sites due to the presence of a second protein. For example, CTCF-binding sites at the chicken lysozyme and the human c-myc genes are flanked by thyroid hormone response elements (TREs). In both cases, the presence of thyroid hormone abrogates enhancer blocking, even though CTCF remains bound to the chromatin. While the mechanism of this effect has not been resolved, it seems quite possible that the pathway involves modulation of the looping ability of CTCF.
A similar mechanism of regulation of loop formation has been proposed for the Su(Hw) insulator (Figure 2). In this case two of the proteins that associate with the insulator element, CP190 and Mod(mdg4)2.2, were shown to undergo modification by conjugation to Small Ubiquitin-like Modifier (SUMO). Similar to PARlation of CTCF, SUMO was not found to affect DNA association of CP190 or Mod(mdg4)2.2. However, contrary to PARlation of CTCF, SUMO was found to inhibit the clustering of insulator proteins into insulator bodies and in this way inhibit insulator activity [65]. These findings lead to the model that SUMO conjugation inhibits loop formation and therefore is a form of negative regulation for the Su(Hw) insulator element. Regulation of Su(Hw) insulator activity may also take place at the level of binding of insulator proteins to DNA (Figure 2). Modification of Su(Hw) by the E3 ubiquitin ligase dTopors affects its ability to interact with DNA and results in an increase in insulator activity [27]. A different strategy for regulating Su(Hw) insulator activity has been described more recently. The Su(Hw) insulator requires the RNAi machinery to make RNAs that are part of the insulator complex. These RNAs are required to mediate interactions between individual insulators and form insulator bodies (Figure 2). Mutations that affect components of the RNAi machinery, and presumably affect the formation of these RNAs, result in impaired insulator function. Interestingly, mutations in the RNA helicase Rm62 have the opposite effect on insulator activity, suggesting that this protein may bind insulator RNA and destabilize insulator bodies to regulate insulator function.
Figure 2. Insulator activity can be regulated by ubiquitination and sumoylation of insulator proteins.

A. Two active insulators coming together at an insulator body. dTopors is present at the insulator sites, Rm62/Lip is not present, Su(Hw) is ubiquitinated, Mod(mdg4)2.2 and CP190 are not sumoylated and dTopors serves as a bridge to the lamina. B. Two inactive insulators that cannot be part of an insulator body. dTopors is absent and Su(Hw) is not ubiquitinated, whereas Mod(mdg4)2.2 and CP190 are sumoylated. Rm62/Lip is present and bound to RNA. Under these conditions, the two insulator sites cannot interact and form insulator bodies. Absence of dTopors also precludes interactions with the lamina.
Taken together these various strategies for regulating insulator activity at both the level of protein binding to DNA and protein-protein interactions that mediate loop formation are evidence for insulator elements as a dynamic form of chromatin organization. Protein competition and modification are reversible forms of regulation that could be adapted as cells need to change patterns of gene expression throughout the cell cycle and development.
8. Conclusions
Insulators are DNA sequences whose precise role in gene regulation is not well understood. In spite of several years of intense scrutiny, it is not yet clear whether these sequences play a very local role in the regulation of adjacent genes or whether they have a more global function in organizing the chromatin fiber into functional domains that define clusters of co-expressed genes. In the latter case, insulators have the potential to perform important tasks in orchestrating changes in nuclear organization that could regulate gene expression during cell differentiation. It is possible that different cell types posses different insulator-mediated nuclear organization and that cell identity is a function of gene expression patterns that depend on this organization (Figure 1). A corollary of this hypothesis is that undifferentiated stem cells may have a nuclear organization that allows the expression of house keeping genes as well as those required to maintain the pluripotent state. Insulator proteins may then establish and maintain specific arrangements of the chromatin fiber that determine various differentiation outcomes. In vertebrate cells, insulator-induced nuclear organization based on the formation of insulator bodies has been difficult to observe by immunofluorescence microscopy, although structures similar to Drosophila insulator bodies have been found [66]. In Drosophila, differentiated cells posses a nuclear organization that can be visualized by the presence of insulator bodies, but the nature of the specific insulator sequences present at these bodies may vary among cells. Regulatory mechanisms that involve protein modifications may be responsible for allowing binding of insulator proteins to DNA or interactions with other insulator proteins (Figure 2) to determine whether individual insulators participate in the formation of insulator bodies and, therefore, in the formation of chromatin loops that correspond to functional domains of co-expressed genes. Further work is needed to understand if these regulatory mechanisms are used to modulate insulator activity in a locus-specific manner and if this regulation determines cell differentiation by changing patterns of gene expression.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–13. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- 2.Henikoff S. Position-effect variegation after 60 years. Trends Genet. 1990;6:422–6. doi: 10.1016/0168-9525(90)90304-o. [DOI] [PubMed] [Google Scholar]
- 3.Loo S, Rine J. Silencing and heritable domains of gene expression. Annu Rev Cell Dev Biol. 1995;11:519–48. doi: 10.1146/annurev.cb.11.110195.002511. [DOI] [PubMed] [Google Scholar]
- 4.Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci USA. 2000;97:5807–11. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donze D, Adams CR, Rine J, Kamakaka RT. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 1999;13:698–708. doi: 10.1101/gad.13.6.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Donze D, Kamakaka RT. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. Embo J. 2001;20:520–31. doi: 10.1093/emboj/20.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fourel G, Revardel E, Koering CE, Gilson E. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 1999;18:2522–37. doi: 10.1093/emboj/18.9.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fourel G, Boscheron C, Revardel E, Lebrun E, Hu YF, Simmen KC, Muller K, Li R, Mermod N, Gilson E. An activation-independent role of transcription factors in insulator function. EMBO Rep. 2001;2:124–32. doi: 10.1093/embo-reports/kve024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrari S, Simmen KC, Dusserre Y, Muller K, Fourel G, Gilson E, Mermod N. Chromatin domain boundaries delimited by a histone-binding protein in yeast. J Biol Chem. 2004;279:55520–30. doi: 10.1074/jbc.M410346200. [DOI] [PubMed] [Google Scholar]
- 10.Recillas-Targa F, Pikaart MJ, Burgess-Beusse B, Bell AC, Litt MD, West AG, Gaszner M, Felsenfeld G. Position-effect protection and enhancer blocking by the chicken beta- globin insulator are separable activities. Proc Natl Acad Sci USA. 2002;99:6883–8. doi: 10.1073/pnas.102179399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.West AG, Huang S, Gaszner M, Litt MD, Felsenfeld G. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol Cell. 2004;16:453–63. doi: 10.1016/j.molcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Jambunathan N, Martinez AW, Robert EC, Agochukwu NB, Ibos ME, Dugas SL, Donze D. Multiple bromodomain genes are involved in restricting the spread of heterochromatic silencing at the Saccharomyces cerevisiae HMR-tRNA boundary. Genetics. 2005;171:913–22. doi: 10.1534/genetics.105.046938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oki M, Valenzuela L, Chiba T, Ito T, Kamakaka RT. Barrier proteins remodel and modify chromatin to restrict silenced domains. Mol Cell Biol. 2004;24:1956–67. doi: 10.1128/MCB.24.5.1956-1967.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Labrador M, Corces VG. Setting the boundaries of chromatin domains and nuclear organization. Cell. 2002;111:151–4. doi: 10.1016/s0092-8674(02)01004-8. [DOI] [PubMed] [Google Scholar]
- 15.West AG, Gaszner M, Felsenfeld G. Insulators: many functions, many mechanisms. Genes Dev. 2002;16:271–88. doi: 10.1101/gad.954702. [DOI] [PubMed] [Google Scholar]
- 16.Udvardy A, Maine E, Schedl P. The 87A7 chromomere. Identification of novel chromatin structures flanking the heat shock locus that may define the boundaries of higher order domains. J Mol Biol. 1985;185:341–58. doi: 10.1016/0022-2836(85)90408-5. [DOI] [PubMed] [Google Scholar]
- 17.Hart CM, Zhao K, Laemmli UK. The scs’ boundary element: characterization of boundary element- associated factors. Mol Cell Biol. 1997;17:999–1009. doi: 10.1128/mcb.17.2.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao K, Hart CM, Laemmli UK. Visualization of chromosomal domains with boundary element-associated factor BEAF-32. Cell. 1995;81:879–89. doi: 10.1016/0092-8674(95)90008-x. [DOI] [PubMed] [Google Scholar]
- 19.Blanton J, Gaszner M, Schedl P. Protein:protein interactions and the pairing of boundary elements in vivo. Genes Dev. 2003;17:664–75. doi: 10.1101/gad.1052003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerasimova TI, Corces VG. Polycomb and trithorax group proteins mediate the function of a chromatin insulator. Cell. 1998;92:511–21. doi: 10.1016/s0092-8674(00)80944-7. [DOI] [PubMed] [Google Scholar]
- 21.Parnell TJ, Kuhn EJ, Gilmore BL, Helou C, Wold MS, Geyer PK. Identification of genomic sites that bind the Drosophila suppressor of Hairy-wing insulator protein. Mol Cell Biol. 2006;26:5983–93. doi: 10.1128/MCB.00698-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramos E, Ghosh D, Baxter E, Corces VG. Genomic organization of gypsy chromatin insulators in Drosophila melanogaster. Genetics. 2006;172:2337–49. doi: 10.1534/genetics.105.054742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Byrd K, Corces VG. Visualization of chromatin domains created by the gypsy insulator of Drosophila. J Cell Biol. 2003;162:565–74. doi: 10.1083/jcb.200305013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gause M, Morcillo P, Dorsett D. Insulation of enhancer-promoter communication by a gypsy transposon insert in the Drosophila cut gene: cooperation between suppressor of hairy-wing and modifier of mdg4 proteins. Mol Cell Biol. 2001;21:4807–17. doi: 10.1128/MCB.21.14.4807-4817.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghosh D, Gerasimova TI, Corces VG. Interactions between the Su(Hw) and Mod(mdg4) proteins required for gypsy insulator function. EMBO J. 2001;20:2518–27. doi: 10.1093/emboj/20.10.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pai CY, Lei EP, Ghosh D, Corces VG. The Centrosomal Protein CP190 Is a Component of the gypsy Chromatin Insulator. Mol Cell. 2004;16:737–48. doi: 10.1016/j.molcel.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 27.Capelson M, Corces VG. The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol Cell. 2005;20:105–16. doi: 10.1016/j.molcel.2005.08.031. [DOI] [PubMed] [Google Scholar]
- 28.Yusufzai TM, Tagami H, Nakatani Y, Felsenfeld G. CTCF Tethers an Insulator to Subnuclear Sites, Suggesting Shared Insulator Mechanisms across Species. Molecular Cell. 2004;13:291–8. doi: 10.1016/s1097-2765(04)00029-2. [DOI] [PubMed] [Google Scholar]
- 29.Yusufzai TM, Felsenfeld G. The 5′-HS4 chicken {beta}-globin insulator is a CTCF-dependent nuclear matrix-associated element. PNAS. 2004;101:8620–4. doi: 10.1073/pnas.0402938101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishii K, Arib G, Lin C, Van Houwe G, Laemmli UK. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 2002;109:551–62. doi: 10.1016/s0092-8674(02)00756-0. [DOI] [PubMed] [Google Scholar]
- 31.Bi X, Broach JR. DNA in transcriptionally silent chromatin assumes a distinct topology that is sensitive to cell cycle progression. Mol Cell Biol. 1997;17:7077–87. doi: 10.1128/mcb.17.12.7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dilworth DJ, Tackett AJ, Rogers RS, Yi EC, Christmas RH, Smith JJ, Siegel AF, Chait BT, Wozniak RW, Aitchison JD. The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J Cell Biol. 2005;171:955–65. doi: 10.1083/jcb.200509061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J, Shen B, Dorsett D. The Drosophila melanogaster suppressor of Hairy-wing zinc finger protein has minimal effects on gene expression in Saccharomyces cerevisiae. Genetics. 1993;135:343–55. doi: 10.1093/genetics/135.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noma K, Cam HP, Maraia RJ, Grewal SI. A role for TFIIIC transcription factor complex in genome organization. Cell. 2006;125:859–72. doi: 10.1016/j.cell.2006.04.028. [DOI] [PubMed] [Google Scholar]
- 35.Iborra FJ, Pombo A, Jackson DA, Cook PR. Active RNA polymerases are localized within discrete transcription “factories’ in human nuclei. J Cell Sci. 1996;109 ( Pt 6):1427–36. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- 36.Cook PR. Nongenic transcription, gene regulation and action at a distance. J Cell Sci. 2003;116:4483–91. doi: 10.1242/jcs.00819. [DOI] [PubMed] [Google Scholar]
- 37.Marenduzzo D, Faro-Trindade I, Cook PR. What are the molecular ties that maintain genomic loops? Trends Genet. 2007;23:126–33. doi: 10.1016/j.tig.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 38.Glover DM, Leibowitz MH, McLean DA, Parry H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 1995;81:95–105. doi: 10.1016/0092-8674(95)90374-7. [DOI] [PubMed] [Google Scholar]
- 39.Sutter NB, Scalzo D, Fiering S, Groudine M, Martin DI. Chromatin insulation by a transcriptional activator. Proc Natl Acad Sci USA. 2003;100:1105–10. doi: 10.1073/pnas.242732999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active beta-globin locus. Mol Cell. 2002;10:1453–65. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 41.Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–6. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 42.Torigoi E, Bennani-Baiti IM, Rosen C, Gonzalez K, Morcillo P, Ptashne M, Dorsett D. Chip interacts with diverse homeodomain proteins and potentiates bicoid activity in vivo. Proc Natl Acad Sci USA. 2000;97:2686–91. doi: 10.1073/pnas.050586397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morcillo P, Rosen C, Dorsett D. Genes regulating the remote wing margin enhancer in the Drosophila cut locus. Genetics. 1996;144:1143–54. doi: 10.1093/genetics/144.3.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cai HN, Shen P. Effects of cis arrangement of chromatin insulators on ennhancer-blocking activity. Science. 2001;291:493–5. doi: 10.1126/science.291.5503.493. [DOI] [PubMed] [Google Scholar]
- 45.Muravyova E, Golovnin A, Gracheva E, Parshikov A, Belenkaya T, Pirrotta V, Georgiev P. Loss of insulator activity by paired Su(Hw) chromatin insulators. Science. 2001;291:495–8. doi: 10.1126/science.291.5503.495. [DOI] [PubMed] [Google Scholar]
- 46.Kuhn EJ, Viering MM, Rhodes KM, Geyer PK. A test of insulator interactions in Drosophila. Embo J. 2003;22:2463–71. doi: 10.1093/emboj/cdg241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Savitskaya E, Melnikova L, Kostuchenko M, Kravchenko E, Pomerantseva E, Boikova T, Chetverina D, Parshikov A, Zobacheva P, Gracheva E, Galkin A, Georgiev P. Study of long-distance functional interactions between Su(Hw) insulators that can regulate enhancer-promoter communication in Drosophila melanogaster. Mol Cell Biol. 2006;26:754–61. doi: 10.1128/MCB.26.3.754-761.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Majumder P, Cai HN. The functional analysis of insulator interactions in the Drosophila embryo. PNAS. 2003;100:5223–8. doi: 10.1073/pnas.0830190100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Melnikova L, Juge F, Gruzdeva N, Mazur A, Cavalli G, Georgiev P. Interaction between the GAGA factor and Mod(mdg4) proteins promotes insulator bypass in Drosophila. Proc Natl Acad Sci USA. 2004;101:14806–11. doi: 10.1073/pnas.0403959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mongelard F, Corces VG. Two insulators are not better than one. Nature Structural Biology. 2001;8:192–4. doi: 10.1038/84905. [DOI] [PubMed] [Google Scholar]
- 51.Gilbert MK, Tan YY, Hart CM. The Drosophila boundary element-associated factors BEAF-32A and BEAF-32B affect chromatin structure. Genetics. 2006;173:1365–75. doi: 10.1534/genetics.106.056002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Welshons WJ, Keppy DO. The recombinational analysis of aberrations and the position of the notch locus on the polytene chromosome of Drosophila. Mol Gen Genet. 1981;181:319–24. doi: 10.1007/BF00425605. [DOI] [PubMed] [Google Scholar]
- 53.Vazquez J, Schedl P. Deletion of an insulator element by the mutation facet-strawberry in Drosophila melanogaster. Genetics. 2000;155:1297–311. doi: 10.1093/genetics/155.3.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cohen BA, Mitra RD, Hughes JD, Church GM. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat Genet. 2000;26:183–6. doi: 10.1038/79896. [DOI] [PubMed] [Google Scholar]
- 55.Lercher MJ, Urrutia AO, Hurst LD. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat Genet. 2002;31:180–3. doi: 10.1038/ng887. [DOI] [PubMed] [Google Scholar]
- 56.Spellman PT, Rubin GM. Evidence for large domains of similarly expressed genes in the Drosophila genome. J Biol. 2002;1:1–8. doi: 10.1186/1475-4924-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caron H, van Schaik B, van der Mee M, Baas F, Riggins G, van Sluis P, Hermus MC, van Asperen R, Boon K, Voute PA, Heisterkamp S, van Kampen A, Versteeg R. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science. 2001;291:1289–92. doi: 10.1126/science.1056794. [DOI] [PubMed] [Google Scholar]
- 58.Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–45. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie X, Mikkelsen TS, Gnirke A, Lindblad-Toh K, Kellis M, Lander ES. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc Natl Acad Sci U S A. 2007;104:7145–50. doi: 10.1073/pnas.0701811104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hart CM, Cuvier O, Laemmli UK. Evidence for an antagonistic relationship between the boundary element- associated factor BEAF and the transcription factor DREF. Chromosoma. 1999;108:375–83. doi: 10.1007/s004120050389. [DOI] [PubMed] [Google Scholar]
- 61.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–5. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 62.Hark AT, Schoenherr CJ, Katz DJ, Ingram RS, Levorse JM, Tilghman SM. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–9. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 63.Yu W, Ginjala V, Pant V, Chernukhin I, Whitehead J, Docquier F, Farrar D, Tavoosidana G, Mukhopadhyay R, Kanduri C, Oshimura M, Feinberg AP, Lobanenkov V, Klenova E, Ohlsson R. Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat Genet. 2004;36:1105–10. doi: 10.1038/ng1426. [DOI] [PubMed] [Google Scholar]
- 64.Klenova E, Ohlsson R. Poly(ADP-ribosyl)ation and epigenetics. Is CTCF PARt of the plot? Cell Cycle. 2005;4:96–101. doi: 10.4161/cc.4.1.1398. [DOI] [PubMed] [Google Scholar]
- 65.Capelson M, Corces VG. SUMO conjugation attenuates the activity of the gypsy chromatin insulator. Embo J. 2006;25:1906–14. doi: 10.1038/sj.emboj.7601068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ishihara K, Oshimura M, Nakao M. CTCF-dependent chromatin insulator is linked to epigenetic remodeling. Mol Cell. 2006;23:733–42. doi: 10.1016/j.molcel.2006.08.008. [DOI] [PubMed] [Google Scholar]


