Abstract
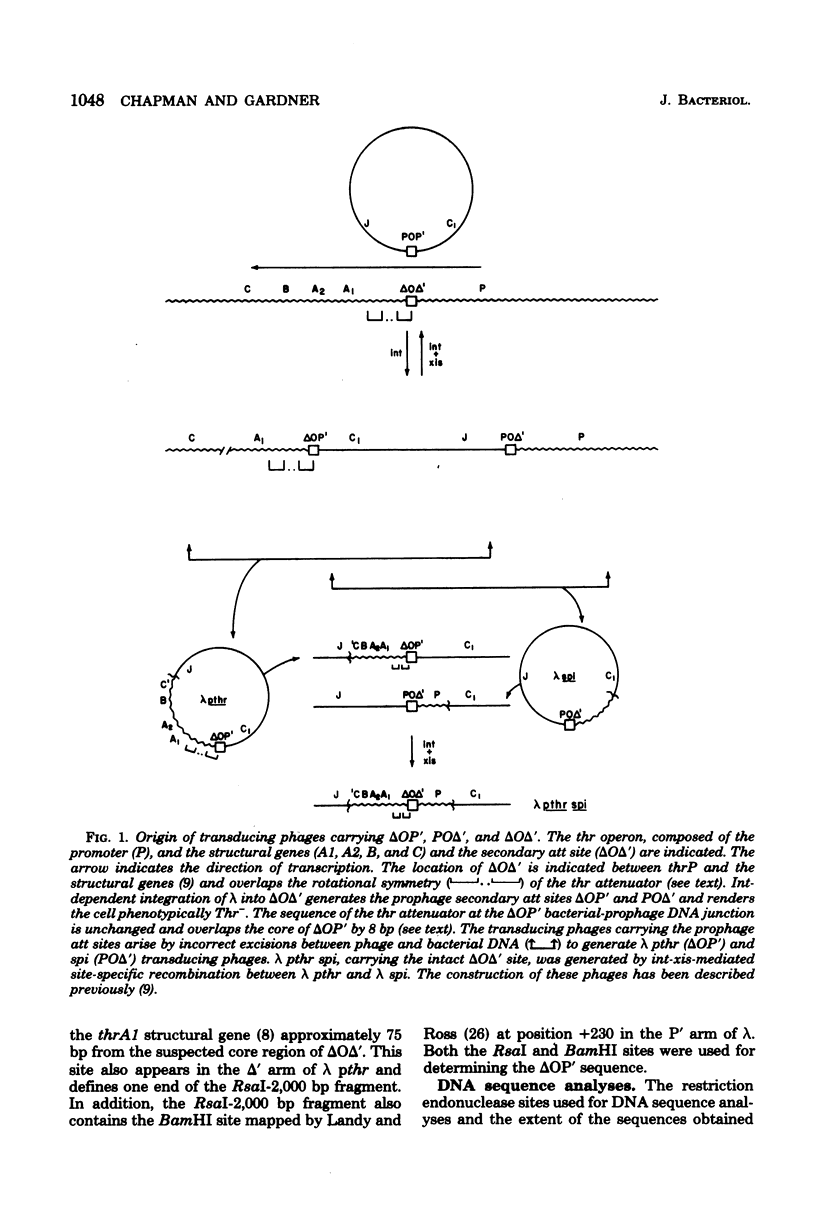
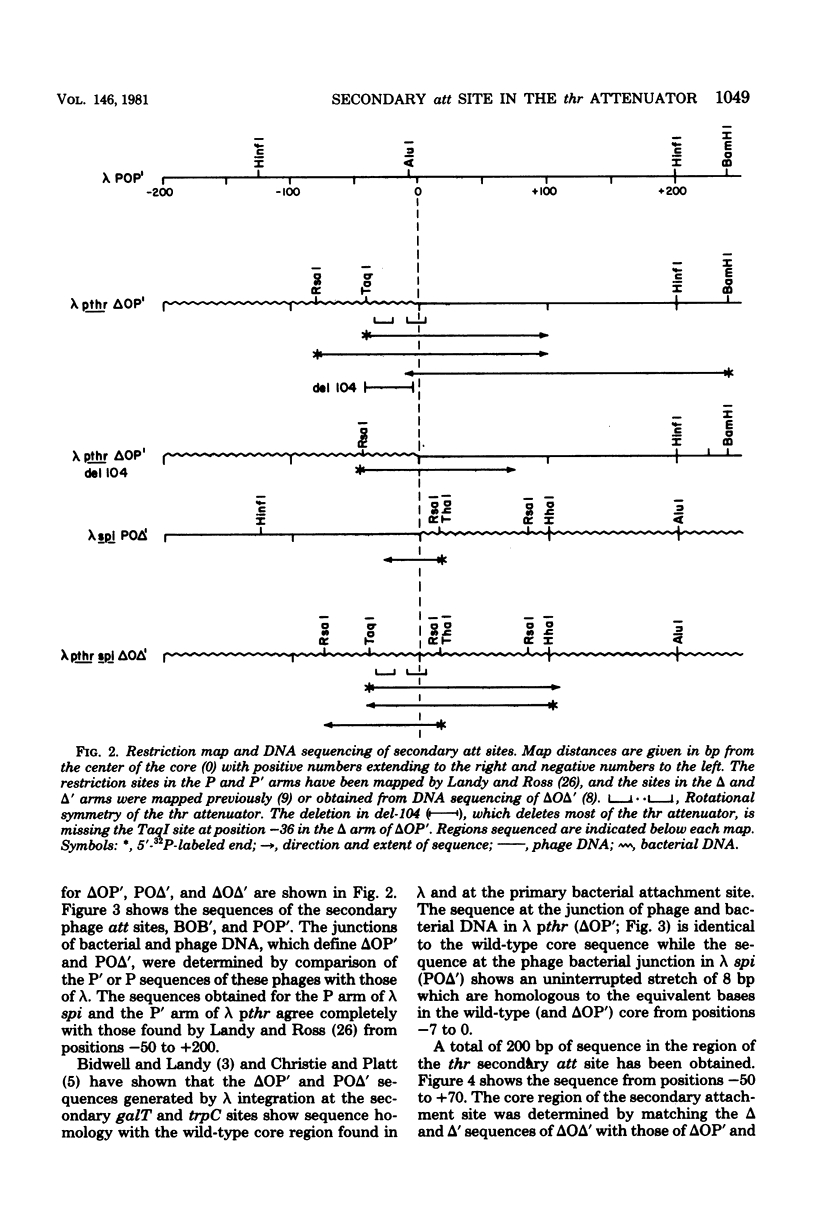
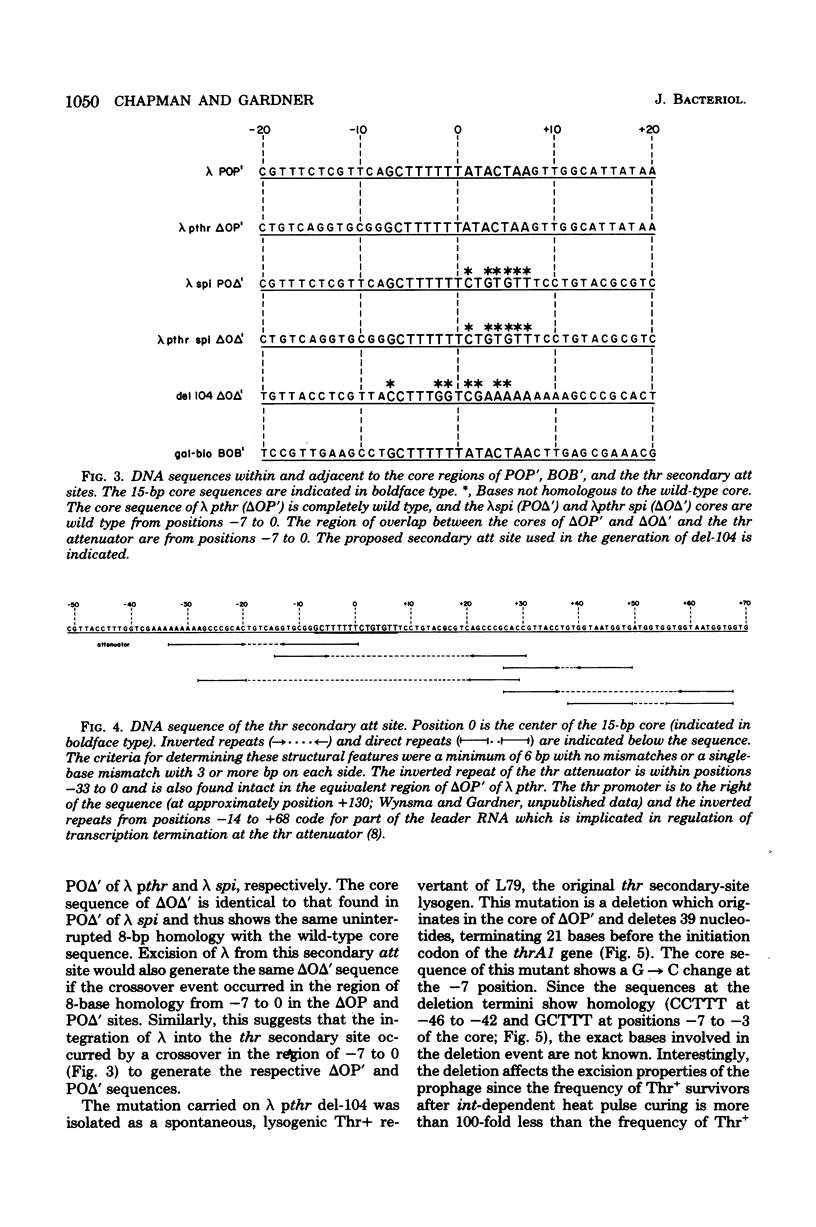
We have determined the nucleotide sequence of a secondary gamma attachment (att) site which overlaps the Escherichia coli threonine (thr) operon attenuator. The secondary att site shows uninterrupted homology (8 out of 15) with the 15 base-pair "common core" sequence found in gamma and at the primary bacterial attachment site. These 8 base paired also overlap the thr operon attenuator. Comparison of the secondary att site with the flanking prophage sites shows that the crossover site for gamma integration lies within the -7 to 0 region of the core. Sequences on both sides of the core show no obvious homology with analogous sequences of the gamma or primary bacterial att sites. The core sequences of the left prophage att site is completely homologous to the wild-type core and also shows the same 8-base pair overlap with the thr operon attenuator. The position of the thr operon attenuator, relative to the left prophage att site, indicates that ribonucleic acid transcripts, initiated at a gamma promoter, are terminated efficiently at the thr attenuator. It is also possible that this prophage att site is able to undergo int dependent site-specific recombination which with another nearby secondary att site. Evidence is also presented which suggests that a base or sequence to the left of position -6 in the core is necessary fo excisive recombination.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abraham J., Mascarenhas D., Fischer R., Benedik M., Campbell A., Echols H. DNA sequence of regulatory region for integration gene of bacteriophage lambda. Proc Natl Acad Sci U S A. 1980 May;77(5):2477–2481. doi: 10.1073/pnas.77.5.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhya S., Cleary P., Campbell A. A deletion analysis of prophage lambda and adjacent genetic regions. Proc Natl Acad Sci U S A. 1968 Nov;61(3):956–962. doi: 10.1073/pnas.61.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidwell K., Landy A. Structural features of lambda site-specific recombination at a secondary att site in galT. Cell. 1979 Feb;16(2):397–406. doi: 10.1016/0092-8674(79)90015-1. [DOI] [PubMed] [Google Scholar]
- Christie G. E., Platt T. A secondary attachment site for bacteriophage lambda in trpC of E. coli. Cell. 1979 Feb;16(2):407–413. doi: 10.1016/0092-8674(79)90016-3. [DOI] [PubMed] [Google Scholar]
- Csordás-Tóth E., Boros I., Venetianer P. Nucleotide sequence of a secondary attachment site for bacteriophage lambda on the Escherichia coli chromosome. Nucleic Acids Res. 1979 Nov 10;7(5):1335–1341. doi: 10.1093/nar/7.5.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. W., Parkinson J. S. Deletion mutants of bacteriophage lambda. 3. Physical structure of att-phi. J Mol Biol. 1971 Mar 14;56(2):403–423. doi: 10.1016/0022-2836(71)90473-6. [DOI] [PubMed] [Google Scholar]
- Gardner J. F. Regulation of the threonine operon: tandem threonine and isoleucine codons in the control region and translational control of transcription termination. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1706–1710. doi: 10.1073/pnas.76.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner J. F., Reznikoff W. S. Identification and restriction endonuclease mapping of the threonine operon regulatory region. J Mol Biol. 1978 Dec 5;126(2):241–258. doi: 10.1016/0022-2836(78)90361-3. [DOI] [PubMed] [Google Scholar]
- Gardner J. F., Smith O. H., Fredricks W. W., McKinney M. A. Secondary-site attachment of coliphage lambda near the thr operon. J Mol Biol. 1974 Dec 25;90(4):613–631. doi: 10.1016/0022-2836(74)90528-2. [DOI] [PubMed] [Google Scholar]
- Gardner J. F., Smith O. H. Operator-promoter functions in the threonine operon of Escherichia coli. J Bacteriol. 1975 Oct;124(1):161–166. doi: 10.1128/jb.124.1.161-166.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingery R., Echols H. Mutants of bacteriophage lambda unable to integrate into the host chromosome. Proc Natl Acad Sci U S A. 1967 Oct;58(4):1507–1514. doi: 10.1073/pnas.58.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman M. E., Yarmolinsky M. B. Integration-negative mutants of bacteriophage lambda. J Mol Biol. 1968 Feb 14;31(3):487–505. doi: 10.1016/0022-2836(68)90423-3. [DOI] [PubMed] [Google Scholar]
- Guarneros G., Echols H. New mutants of bacteriophage lambda with a specific defect in excision from the host chromosome. J Mol Biol. 1970 Feb 14;47(3):565–574. doi: 10.1016/0022-2836(70)90323-2. [DOI] [PubMed] [Google Scholar]
- Guerrini F. On the asymmetry of lambda integration sites. J Mol Biol. 1969 Dec 28;46(3):523–542. doi: 10.1016/0022-2836(69)90194-6. [DOI] [PubMed] [Google Scholar]
- Hershfield V., Boyer H. W., Chow L., Helinski D. R. Characterization of a mini-ColC1 plasmid. J Bacteriol. 1976 Apr;126(1):447–453. doi: 10.1128/jb.126.1.447-453.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoess R. H., Foeller C., Bidwell K., Landy A. Site-specific recombination functions of bacteriophage lambda: DNA sequence of regulatory regions and overlapping structural genes for Int and Xis. Proc Natl Acad Sci U S A. 1980 May;77(5):2482–2486. doi: 10.1073/pnas.77.5.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoess R. H., Landy A. Structure of the lambda att sites generated by int-dependent deletions. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5437–5441. doi: 10.1073/pnas.75.11.5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. L., Ross W., Landy A. The lambda phage att site: functional limits and interaction with Int protein. Nature. 1980 May 8;285(5760):85–91. doi: 10.1038/285085a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R. A., Walseth T. F. The enzymatic preparation of [alpha-32P]ATP, [alpha-32P]GTP, [32P]cAMP, and [32P]cGMP, and their use in the assay of adenylate and guanylate cyclases and cyclic nucleotide phosphodiesterases. Adv Cyclic Nucleotide Res. 1979;10:135–167. [PubMed] [Google Scholar]
- Kaiser A. D., Masuda T. Evidence for a prophage excision gene in lambda. J Mol Biol. 1970 Feb 14;47(3):557–564. doi: 10.1016/0022-2836(70)90322-0. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y., Nash H. A. The bacteriophage lambda int gene product. A filter assay for genetic recombination, purification of int, and specific binding to DNA. J Biol Chem. 1978 Oct 25;253(20):7149–7157. [PubMed] [Google Scholar]
- Kikuchi Y., Nash H. Integrative recombination of bacteriophage lambda: requirement for supertwisted DNA in vivo and characterization of int. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1099–1109. doi: 10.1101/sqb.1979.043.01.122. [DOI] [PubMed] [Google Scholar]
- Landy A., Hoess R. H., Bidwell K., Ross W. Site-specific recombination in bacteriophage lambda: structural features of recombining sites. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1089–1097. doi: 10.1101/sqb.1979.043.01.121. [DOI] [PubMed] [Google Scholar]
- Landy A., Ross W. Viral integration and excision: structure of the lambda att sites. Science. 1977 Sep 16;197(4309):1147–1160. doi: 10.1126/science.331474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee F., Yanofsky C. Transcription termination at the trp operon attenuators of Escherichia coli and Salmonella typhimurium: RNA secondary structure and regulation of termination. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4365–4369. doi: 10.1073/pnas.74.10.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn S. P., Cohen L. K., Kaplan S., Gardner J. F. RsaI: a new sequence-specific endonuclease activity from Rhodopseudomonas sphaeroides. J Bacteriol. 1980 May;142(2):380–383. doi: 10.1128/jb.142.2.380-383.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash H. A. Integration and excision of bacteriophage lambda. Curr Top Microbiol Immunol. 1977;78:171–199. doi: 10.1007/978-3-642-66800-5_6. [DOI] [PubMed] [Google Scholar]
- Ross W., Landy A., Kikuchi Y., Nash H. Interaction of int protein with specific sites on lambda att DNA. Cell. 1979 Oct;18(2):297–307. doi: 10.1016/0092-8674(79)90049-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Hutchinson C. A., 3rd, Harris J. I. A thermostable sequence-specific endonuclease from Thermus aquaticus. Proc Natl Acad Sci U S A. 1977 Feb;74(2):542–546. doi: 10.1073/pnas.74.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K., Weisberg R. A., Gottesman M. E. Prophage lambda at unusual chromosomal locations. I. Location of the secondary attachment sites and the properties of the lysogens. J Mol Biol. 1972 Feb 14;63(3):483–503. doi: 10.1016/0022-2836(72)90443-3. [DOI] [PubMed] [Google Scholar]
- Shimada K., Weisberg R. A., Gottesman M. E. Prophage lambda at unusual chromosomal locations. II. Mutations induced by bacteriophage lambda in Escherichia coli K12. J Mol Biol. 1973 Oct 25;80(2):297–314. doi: 10.1016/0022-2836(73)90174-5. [DOI] [PubMed] [Google Scholar]
- Shimada K., Weisberg R. A., Gottesman M. E. Prophage lambda at unusual chromosomal locations. III. The components of the secondary attachment sites. J Mol Biol. 1975 Apr 25;93(4):415–429. doi: 10.1016/0022-2836(75)90237-5. [DOI] [PubMed] [Google Scholar]
- Shulman M. J., Mizuuchi K., Gottesman M. M. New att mutants of phage lambda. Virology. 1976 Jul 1;72(1):13–22. doi: 10.1016/0042-6822(76)90307-x. [DOI] [PubMed] [Google Scholar]
- Shulman M., Gottesman M. Attachment site mutants of bacteriophage lambda. J Mol Biol. 1973 Dec 25;81(4):461–482. doi: 10.1016/0022-2836(73)90517-2. [DOI] [PubMed] [Google Scholar]
- Zissler J. Integration-negative (int) mutants of phage lambda. Virology. 1967 Jan;31(1):189–189. doi: 10.1016/0042-6822(67)90030-x. [DOI] [PubMed] [Google Scholar]


