Abstract
Mycoplasma genitalium, a human pathogen associated with sexually transmitted diseases, is unique in that it has smallest genome of any known free-living organism. The goal of this study was to investigate if and how M. genitalium uses a minimal genome to generate genetic variations. We analysed the sequence variability of the third gene (MG192 or mgpC) of the M. genitalium MgPa adhesion operon, demonstrated that the MG192 gene is highly variable among and within M. genitalium strains in vitro and in vivo, and identified MG192 sequence shifts in the course of in vitro passage of the G37 type strain and in sequential specimens from an M. genitalium-infected patient. In order to establish the origin of the MG192 variants, we examined nine genomic loci containing partial copies of the MgPa operon, known as MgPar sequences. Our analysis suggests that the MG192 sequence variation is achieved by recombination between the MG192 expression site and MgPar sequences via gene cross-over and, possibly, also by gene conversion. It appears plausible that M. genitalium has the ability to generate unlimited variants from its minimized genome, which presumably allows the organism to adapt to diverse environments and/or to evade host defences by antigenic variation.
Introduction
Mycoplasma genitalium has been recognized as an important cause of non-gonococcal urethritis in men (Taylor-Robinson, 2002; Jensen, 2006) and is likely to be associated with genital tract inflammatory diseases in women, including cervicitis, endometritis, pelvic inflammatory disease and tubal factor infertility (Moller et al., 1984; Clausen et al., 2001; Cohen et al., 2002; Manhart et al., 2003). In addition, there is evidence from at least two studies suggesting that M. genitalium is associated with increased risk of HIV-1 (Perez et al., 1998; Kapiga et al., 2006). A recent molecular study has established that M. genitalium is sexually transmitted (Hjorth et al., 2006). Like other pathogenic mycoplasmas, M. genitalium is capable of causing chronic infections. It has been documented that M. genitalium infection can persist for 5 months in animal models (Taylor-Robinson et al., 1987) and for more than 2 years in humans (Hjorth et al., 2006) although the mechanisms for persistence remain undetermined.
For many bacterial pathogens, antigenic variation of surface-exposed proteins is thought to be an important strategy that allows the organism to persist by adapting to microenvironmental changes and evading host defences (Seifert and So, 1988; van der Woude and Baumler, 2004). Studies of antigenic variation in mycoplasmas have elucidated a variety of molecular mechanisms for generating high-frequency surface protein phase or size variations (Wise, 1993; Citti and Rosengarten, 1997). Well-documented examples include variation of the lipoprotein family in Mycoplasma hyorhinis (Rosengarten and Wise, 1990), M. pulmonis (Bhugra et al., 1995) and M. bovis (Lysnyansky et al., 1996) and of the haemagglutinin in M. synoviae (Noormohammadi et al., 2000). Most of these antigenic variations result from DNA rearrangements and are commonly associated with homologous recombination or site-specific recombination (Dybvig and Voelker, 1996; van der Woude and Baumler, 2004). M. genitalium has the smallest genome (580 kb) known to date for a self-replicating organism; its 470 genes are organized in a frugal manner with minimal spacer regions between them (Fraser et al., 1995). It has been suggested that genome reduction benefits the organism through greater reproductive efficiency in addition to reduction of energy requirements in limited nutrient environments (Maniloff, 1996). With such a minimal genome, the question is how does M. genitalium generate sufficient genetic and antigenic variation to adapt to diverse environments and to evade host defences to permit persistence.
The adhesin protein MgPa of M. genitalium, corresponding to the adhesin molecule P1 of Mycoplasma pneumoniae, is one of the major surface proteins in this organism and has an important role in the attachment of the organism to host epithelial cells (Hu et al., 1987). Serological studies have shown that MgPa is the immunodominant protein of M. genitalium (Hu et al., 1987; Morrison-Plummer et al., 1987; Wang et al., 1997; Svenstrup et al., 2006). The gene encoding MgPa is organized in an operon containing three genes in the order of MG190 (or mgpA), MG191 (or mgpB) and MG192 (or mgpC), with intervening regions of 6 bp and 1 bp respectively (Inamine et al., 1989; Fraser et al., 1995). It is believed that these three genes are co-transcribed (Inamine et al., 1989; Musatovova et al., 2003). Molecular studies of M. genitalium mutants lacking the MG191 or the MG192 gene have revealed that the encoded proteins are essential for the proper assembly and development of the terminal organelle (Burgos et al., 2006). According to the complete genome sequence of the M. genitalium type strain G37 (Fraser et al., 1995), designated as G37T, the expression site for each of these three genes is present in only one copy but there are nine repetitive elements in the form of truncated copies of the MG191 and MG192 genes dispersed throughout the genome, which are designated as MgPa repeats or MgPar sequences (Fraser et al., 1995; Iverson-Cabral et al., 2006). A few studies have demonstrated that MG191 sequence variation occurs extensively within and among in vitro cultured strains and in vivo specimens obtained from patients and chimpanzees with chronic M. genitalium infection (Peterson et al., 1995; Iverson-Cabral et al., 2006; Jensen, 2006). Some of these variations could be explained by homologous recombination between the MG191 expression site and MgPar sequences present in the type strain G37T. These observations led to the hypothesis that such recombination would generate antigenic variation, allowing M. genitalium to evade the host immune response. However, the molecular mechanisms for these proposed recombination events have not been defined. In fact, nothing is known of the MgPar loci of M. genitalium strains other than those in the published G37T genome sequence (Fraser et al., 1995). Because most MgPars contain not only regions homologous to MG191 but also regions homologous to MG192, it is of particular interest to ask whether MG192 also undergoes genetic and antigenic variation. In order to address this question more information is needed on sequence homology of the MgPars and MG192 as well as the exact location of these homologous regions.
In this article, we describe the number and architecture of MG192 homologous regions in the MgPars, show the degree of sequence variation of MG192 among and within M. genitalium strains in vitro and in vivo, demonstrate MG192 sequence shifts in the type strain G37 during in vitro passage as well as in clinical specimens obtained from an M. genitalium-infected patient, and report remarkable sequence variation of MgPar regions among M. genitalium strains. Furthermore we performed preliminary investigations of the molecular mechanism of recombination between the MG192 expression site and MgPar sequences, and provide evidence that gene cross-over and, possibly, also gene conversion mechanisms are involved in recombination.
Results
The homology between the MG192 gene and MgPar sequences in the genome of the M. genitalium type strain G37T
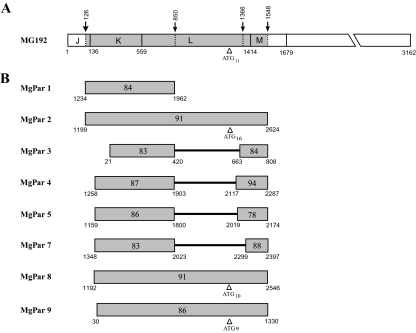
First, we did pairwise comparisons of the G37T MG192 gene with each of the nine MgPars reported by Iverson-Cabral et al. (2006) and found three distinct regions in the MG192 gene: the first and third regions spanning from nucleotide (nt) 1–125 and 1549–3162, respectively, are non-repetitive or conserved regions which have no homology to any of the nine MgPar sequences; the second region (nt 126–1548) is variable and has significant homology (78–94% identity) to all nine MgPar sequences except for MgPar 6 (Fig. 1). Based on the restriction maps described by Dallo and Baseman (1991) and Peterson et al. (1995), the MG192 variable region can be divided into three divisions: JKL (nt 126–850), L (nt 851–1365) and LM (nt 1366–1548). MgPar 1 contains a 729 bp fragment that is homologous only to the JKL division. MgPars 3, 4, 5 and 7 each contains two regions that are homologous to the JKL and LM divisions and which are separated by a segment with homology to MG191 (see Table S1). MgPars 2, 8 and 9 have segments which are homologous to all three divisions. An AGT tandem repeat motif is present in MG192 homologous regions of MgPars 2, 8 and 9 and in the L division of MG192, with the repeat number varying from 9 to 16. While there has been an alternative assignment (TAG) for the repeat unit of this repeat motif (van Belkum et al., 1998; Iverson-Cabral et al., 2006) we think that it is more appropriate to assign the repeat unit as AGT as it is the codon for serine based on the MG192 open reading frame (ORF).
Fig. 1.
Sequence homology between MG192 and MgPars in M. genitalium G37T. A. Schematic representation of the structure of the full-length MG192 gene based on the complete genome sequence of M. genitalium G37T (GenBank Accession No. NC_000908). The nucleotide positions are in relation to the predicted translational start site of the MG192 gene. Divisions J through M indicated by solid vertical lines represent the restriction fragments described previously (Dallo and Baseman, 1991; Peterson et al., 1995). The region highlighted in grey (nt 126–1548) represents the variable region with homology to eight of the nine MgPar sequences shown in (B). This variable region can be further divided into three segments JKL, L and LM (indicated by dotted vertical lines and arrows), which are homologous to different parts of MgPars. Triangle indicates the location of an 11-copy AGT tandem repeat motif (AGT11). B. Schematic representation of the nucleotide sequence alignment of eight MgPars from the M. genitalium G37T genome. Numbers bordering each shaded area refer to the nucleotide positions in the full-length MgPars as described by Iverson-Cabral et al. (2006). Numbers inside the shaded areas indicate the percentage of identity to the corresponding regions in MG192 shown in (A). Triangles indicate the location of the AGT tandem repeat motif with the number of repeat units shown in subscript. Solid black lines in MgPars 3, 4, 5 and 7 represent regions with homology to MG191.
Sequence variation of MG192 among American Type Culture Collection (ATCC) strains of M. genitalium and their derivatives
We examined the MG192 variable region in M. genitalium American Type Culture Collection (ATCC) strains and their derivatives (Table 1). A fragment of approximately 1.7 kb covering the entire MG192 variable region was amplified by PCR and sequenced directly and after subcloning. For each strain, we identified a mixture of two or three MG192 sequences that differed from each other only in the number of the AGT tandem repeats (Table 2). Aside from the AGT repeat number variation, the MG192 sequences in all ATCC strains except for TW48-5G and TW10-5G were identical to that of the published G37T genome sequence (Fig. 2). The MG192 sequence of TW48-5G contained a total of 28 nucleotide substitutions and one triplet deletion in the regions flanking the AGT tandem repeats compared with the G37T genome sequence. The MG192 sequence of TW10-5G (designated as TW10-5G.ATCC in Fig. 2) had 67 nucleotide substitutions, three triplet insertions and one triplet deletion upstream of the AGT repeats. These nucleotide substitutions, insertions and deletions did not cause frameshifts or stop codons in the predicted ORF but did code for different amino acid sequences.
Table 1.
M. genitalium specimens used in this study.
| Specimen | Description | Reference |
|---|---|---|
| ATCC strains | ||
| G37-D | ATCC33530D, G37 genomic DNA | Tully et al. (1981) |
| G37-P1 | ATCC33530 (G37), frozen culture | Tully et al. (1981) |
| TW10-5G.ATCC | ATCC49123, frozen culture | Baseman et al. (1988) |
| M30 | ATCC49895, frozen culture | Baseman et al. (1988) |
| TW10–6G | ATCC49896, frozen culture | Baseman et al. (1988) |
| R32G [R32] | ATCC49897, frozen culture | Baseman et al. (1988) |
| TW48-5G | ATCC49898, frozen culture | Baseman et al. (1988) |
| UMTB-10G | ATCC49899, frozen culture | Tully et al. (1995) |
| ATCC-related strains | ||
| G37-P17 | G37 (ATCC33530) passage No. 17 | This study |
| G37-P35 | G37 (ATCC33530) passage No. 35 | This study |
| TW10-5G.DK | TW10-5G strain obtained from J.G. Tully prior to deposit into ATCC | Jensen (2006) |
| TW10-5G.SA | TW10-5G strain maintained in the University of Texas Health Sciences Center at San Antonio, Texas | Musatovova et al. (2006) |
| Patient specimens | ||
| 199.0 | Urine specimen from a New Orleans patient (No. 199) at his first visit on 28 January 2003 | Ma and Martin (2004) |
| 199.1 | Follow-up urine specimen from the same patient above on 7 February 2003 | Ma and Martin (2004) |
Table 2.
Variable number of AGT tandem repeats in the M. genitalium MG192 gene.
| Specimena | No. of AGT tandem repeatsb | No. of total clones analysed |
|---|---|---|
| ATCC strains | ||
| G37-D | 1121, 103 and 121 | 25 |
| G37-P1 | 118, 104, 122 and 91 | 15 |
| TW10-5G.ATCC | 1111 and 101 | 12 |
| M30 | 104 and 91 | 5 |
| TW10-6G | 84 and 91 | 5 |
| R32G [R32] | 124, 111 and 101 | 6 |
| TW48-5G | 118, 101 and 91 | 10 |
| UMTB-10G | 112, 102 and 81 | 5 |
| ATCC-related strains | ||
| G37-P17 | 103, 111, 91 and 81 | 6 |
| G37-P35 | 1025, 95, 111, 81 and 71 | 33 |
| TW10-5G.DK | 1111 and 101 | 12 |
| TW10-5G.SA | 11 | –c |
| Patient specimens | ||
| 199.0 | 610 | 10 |
| 199.1 | 618 | 18 |
Same as Table 1.
Numbers in subscript indicate the number of plasmid clones analysed for respective repeats, which are listed in the order from high to low frequency.
Based on the report of Musatovova et al. (2006). Not sequenced in this study.
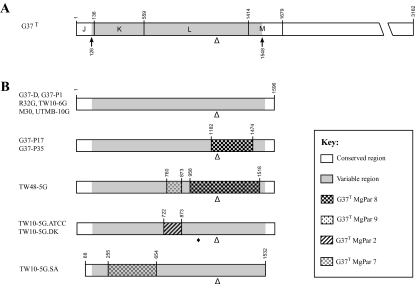
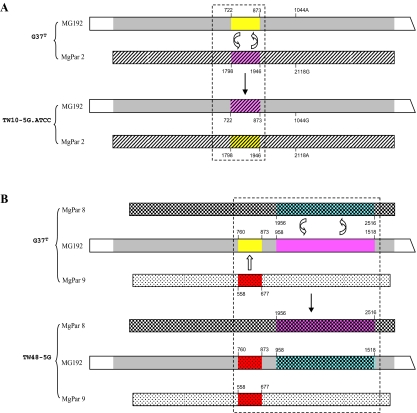
Fig. 2.
MG192 sequence variation among M. genitalium ATCC strains and their derivatives cultured in vitro. A. Diagram of the full-length MG192 gene of G37T, as shown in Fig. 1. B. Schematic drawing of the MG192 variable regions from M. genitalium strains cultured in vitro (see Table 1). Regions showing the longest stretch of sequence identity with the MgPars of G37T are indicated by identical patterns, as shown in the key. Numbers bordering each shaded area correspond to the nucleotide positions in the full-length MG192 gene of G37T. Triangle indicates the location of the AGT tandem repeats, with detailed information on repeat numbers shown in Table 2. Diamond indicates a single-nucleotide substitution from A to G. The sequences of the MG192 genes for G37-P35, TW48-5G and TW10-5G.ATCC have been submitted to GenBank under Accession No. EF117280, EF117281 and EF117282 respectively.
Surprisingly, the MG192 sequence of TW10-5G.ATCC differed from the recently published sequence for the TW10-5G strain maintained in the University of Texas Health Sciences Center at San Antonio (GenBank Accession No. AY679761, designated here as TW10-5G.SA) (Musatovova et al., 2006). We also examined the MG192 sequence of the TW10-5G strain that was provided to one of us (J.S.J.) by J.G. Tully prior to the time that the strain was deposited into the ATCC collection. This strain (designated here as TW10-5G.DK) had been passaged 11 times in Dr Tully's laboratory and subsequently propagated for five passages before being studied as described here. By analysis of the PCR products amplified in three independent experiments using different primers, TW10-5G.ATCC and TW10-5G.DK consistently showed the same MG192 sequence (Fig. 2).
Detection of MG192 sequence variation in the M. genitalium type strain G37 during in vitro passage
We passed the G37 ATCC strain serially 35 times in vitro and examined the entire MG192 variable region at three passage levels (designated here as G37-P1, G37-P17 and G37-P35) as well as in the genomic DNA of G37 directly obtained from ATCC (designated here as G37-D). In each passage, sequencing of individual plasmid clones of the PCR products identified a mixture of three to five MG192 sequences that differed from each other only in the number of the AGT tandem repeats (Table 2). Aside from the AGT repeat number variation, the MG192 sequence in G37-D and G37-P1 was identical to that of the published G37T genome sequence, whereas the MG192 sequence in G37-P17 and G37-P35 was identical between them but different from that of G37T (Fig. 2). Compared with the G37T MG192 sequence, G37-P17 and G37-P35 exhibited sequence variation resulting from 16 nucleotide substitutions and one triplet insertion in the regions flanking the AGT repeat motif (Fig. 3A). None of these nucleotide substitutions or insertions introduced frameshifts or stop codons in the predicted ORFs while the deduced amino acid sequence of these variants differed from that of G37T MG192.
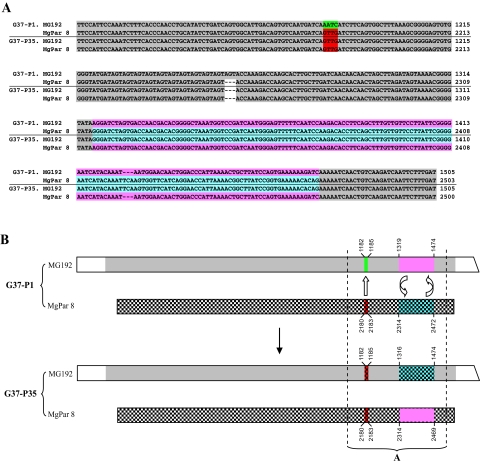
Fig. 3.
Homologous recombination between MG192 and MgPar 8 in the M. genitalium type strain G37 during in vitro passage. A. Alignment of a portion of the MG192 variable region and MgPar 8 in G37-P1 and G37-P35 showing the sequence exchange in the recombination site. Identical sequences are highlighted in the same colour. The number of plasmid clones analysed for each locus is listed in Table 3. All clones analysed for each locus showed homogenous sequence except for the AGT short tandem repeat number variation, with only the most common repeat number shown in this alignment. The sequences of the MG192 and MgPar 8 identified in G37-P35 have been submitted to GenBank under Accession No. EF117280 and EF7289 respectively. B. A model of homologous recombination between MG192 and MgPar 8 in the G37 type strain. Nucleotide positions shown are relative to the full-length MG192 or MgPar 8 of G37-P1, both of which are identical to those of G37T. Regions containing the longest stretch of identical sequences are indicated by the same shading and colour. The regions between dotted vertical lines (corresponding to the sequences shown in A) indicate where sequence exchanges took place, with reciprocal exchange (gene cross-over) shown by two curved arrows and non-reciprocal exchange (gene conversion) shown by one straight arrow.
Extensive variation and rapid shift of the MG192 sequences within a single M. genitalium strain from an infected patient
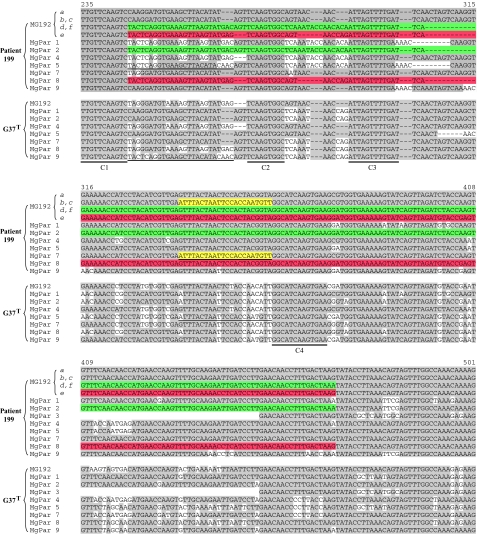
To test the hypothesis that MG192 variation occurs in vivo, we examined two sequential specimens (199.0 and 199.1) obtained 10 days apart from an M. genitalium-infected man (Table 1). These two specimens were previously studied for the variable numbers of tandem repeats (VNTR) in the putative lipoprotein gene, MG309, and single-nucleotide polymorphisms in both the rRNA operon (Ma and Martin, 2004) and the MG191 conserved AB region (Hjorth et al., 2006). All of these loci are present in a single copy in the genome (Fraser et al., 1995). It has been well documented that molecular typing based on these loci, particularly the MG309 VNTR and the MG191 conserved AB region, provides excellent discriminatory power for unrelated M. genitalium strains (Ma and Martin, 2004; Hjorth et al., 2006). The two sequential specimens showed identical genotypes at all three loci, thus demonstrating that this patient was infected with a single M. genitalium strain. Direct sequencing of the MG192 PCR products from both specimens showed a mixture of two or more different sequences. In order to obtain the individual sequences, we studied plasmid clones of the PCR products from two independent assays for each specimen. Three MG192 variant sequences were identified in each specimen (Fig. 4). Remarkably, none of these variant sequences were shared between the two specimens. In silico translation analysis showed that all these six variant sequences remained in the correct reading frame and are predicted to encode divergent amino acid sequences. Comparison of the entire variable region sequence (nt 126–1548) of the G37T MG192 gene to the patient MG192 sequences showed a 14–15% difference at the nucleotide level and a 15–17% difference at the deduced amino acid level.
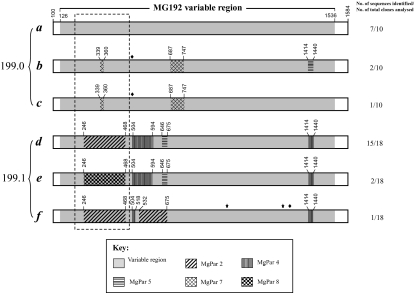
Fig. 4.
representation of MG192 sequence variation and rapid shift identified in two sequential samples (No. 199.0 and 199.1) obtained 10 days apart from an M. genitalium-infected patient. A total of six MG192 variants (a to f) were identified from these two specimens, with three variants present in each specimen. The sequence of the variant a, which is the most predominant variant from the first specimen, is considered the prototype. Various patterns in other five variants represent regions that are different from the variant a and instead have homology to MgPars identified in this patient, as indicated by the key. The diamond indicates single-nucleotide substitutions, which can be traced to a specific MgPar sequence, whereas the star indicates single-nucleotide substitutions, which are not present in any other MG192 variants or MgPars. Numbers above each region refer to the nucleotide positions in the full-length MG192 of the variant a. Detailed nucleotide sequences for the regions included in the box with dashed lines are given in Fig. 6. The number of plasmid clones analysed for MgPars in the patient specimens is listed in Table 3. All clones analysed for each MgPar showed the same sequence except for the triplet repeat number variation in MgPars 2 and 8. The sequences of MG192 variants a to f and MgPars 1 through 9 identified from this patient's strain have been submitted to GenBank under Accession No. EF117283 to EF117288 and EF117293 to EF117301 respectively.
Sequence variation of the MgPar repeats
To date, there are no published data on MgPar sequences from any strains other than the ATCC G37 type strain. We determined the partial or entire sequences of three MgPars (2, 8 and 9) in G37-P1 and all nine MgPars in G37-P35, TW10-5G.ATCC, TW48-5G, and the two sequential patient specimens (No. 199.0 and 199.1). The results of MgPar sequence variation are summarized in Table 3.
Table 3.
Comparison of MgPars in the M. gentalium strains examined in this study with those of G37T.
| No. of nucleotides different relative to G37T MgParsa | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| MgPar 1 1234−1962 | MgPar 2b 1199−2624 | MgPar 3 21–808 | MgPar 4 1258−2287 | MgPar 5 1159−2174 | MgPar 6 1–385 | MgPar 7 1348−2397 | MgPar 8b 1192−2546 | MgPar 9b 30–1330 | |
| G37-P1 | – | 0 | – | – | – | – | – | 0 (22) | 0 |
| G37-P35 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 18 (30c) | 0 |
| TW10-5G.ATCC | 0 | 35 (9) | 0 | 0 | 0 | 5 | 0 | 0 | 0 |
| TW48-5G | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 43 (10) | 0 (10) |
| 199.0 | 133 (5) | 258 (30c) | 110 (5) | 149 (5) | 212 (7) | 65 | 142 (7) | 207 (10) | 223 (8) |
| 199.1 | 133 (5) | 258 (30c) | 110 (5) | 149 (6) | 212 (5) | 65 | 142 (7) | 207 (8) | 223 (9) |
Based on the report of Iverson-Cabral et al. (2006). The regions included for comparison are listed under the names of MgPars.
The number of plasmid clones analysed for selected MgPars was given in parenthesis. MgPars not examined in G37-P1 are indicated by ‘–’.
Variation in the AGT tandem repeat number is not taken into account for the ATCC strains and their derivatives.
P < 0.005 for finding a different sequence based on the exact binomial 95% confidence interval.
As expected, the MgPars 2, 8 and 9 in G37-P1 were identical to those of G37T (Iverson-Cabral et al., 2006) except for the deletion or insertion of one AGT tandem repeat unit. In G37-P35, MgPar 8 contained a region for which the sequence varied significantly from that of G37T (Fig. 3A) while the other eight MgPars were identical to those of G37T except for the deletion or insertion of one AGT repeat unit in MgPars 2 and 9. We examined 30 plasmid clones of the MgPar 8 PCR product from G37-P35 and found identical sequences in all clones except for variation in the repeat unit number of the AGT tandem repeats. In TW10-5G.ATCC, MgPars 2 and 6 showed sequence variations due to apparent nucleotide substitutions and/or insertions while all other MgPars were identical to the corresponding MgPars of G37T except for the deletion or insertion of one AGT tandem repeat unit in MgPars 8 and 9 (Fig. 5A and Fig. S1). In TW48-5G, MgPar 5 showed only a single base substitution compared with G37T while MgPar 8 had extensive sequence variation in a 561 bp segment (Fig. 5B and Fig. S2). All other TW48-5G MgPars were identical to the corresponding MgPars of G37T.
Fig. 5.
MG192 and MgPar sequence variation in M. genitalium ATCC strains TW10-5G and TW48-5G. A. Schematic representation of the MG192 variable region and MgPar 2 in TW10-5G.ATCC compared with those of G37T. Numbers below and above each box refer to the nucleotide positions in the full-length MG192 and MgPar 2 of G37T. Regions containing identical sequences were indicated by the same shading and colour. The regions included in the box with dashed lines indicate where sequence exchanges potentially took place (detailed sequence alignment for this regions is given in Fig. S1). A reciprocal exchange of a 152 bp fragment is indicated by two curved arrows. Another reciprocal exchange of a single base occurred at nt 1044 of MG192 with nt 2118 of MgPar 2. The MgPar 2 sequence in TW10-5G.ATCC was obtained by sequencing of nine plasmid clones, all of which contained the same sequence except for the expected AGT repeat number variation (from 15 to 17 repeats). The sequences of the MG192 and MgPar 2 in TW10-5G.ATCC have been submitted to GenBank under Accession No. EF117281 and EF117290 respectively. B. Schematic diagram of the MG192 variable region and MgPars 8 and 9 in TW48-5G compared with those of G37T. Numbers below and above each box refer to the nucleotide positions in the full-length MG192, MgPar 8 or MgPar 9 of G37T. Regions containing identical sequences were indicated by the same shading and colour. The regions included in the box with dashed lines indicate where sequence exchanges took place (detailed sequence alignment for this regions is given in Fig. S2). A non-reciprocal sequence exchange of a 114 bp fragment was indicated by one straight arrow, and a reciprocal sequence exchange of a 561 bp fragment was indicated by two curved arrows. Both MgPars 8 and 9 sequences in TW48-5G were obtained by sequencing of 10 plasmid clones for each MgPar; all clones for each MgPar contained identical sequences except for the expected AGT repeat number variation (9–11 repeats for MgPar 8 and 8–10 repeats for MgPar 9). The sequences of the MG192 and MgPar 8 in TW48-5G have been submitted to GenBank under Accession No. EF117282 and EF117292 respectively.
For the two sequential patient specimens 199.0 and 199.1 obtained 10 days apart, we did PCR subcloning and analysed multiple plasmid clones for all of the MgPars except for MgPar 6, which has no identity to MG192 (Table 3). Surprisingly, given the striking sequence variation found in MG192, all clones showed identical sequences for each MgPar between and within the first and second specimen except for the expected tandem repeat variations in MgPar 8. The MgPar 6 sequence obtained by direct sequencing the PCR product was also identical between these two sequential specimens. Of note is the fact that the sequences of all nine patient MgPars differed from those of G37T by 13–20% of nucleotides. Figure 6 shows an alignment of a portion of these MgPar sequences.
Fig. 6.
Alignment of a portion of the six MG192 variants (as shown in Fig. 4) and eight MgPar sequences identified in an M. genitalium-infected patient 199 and comparison with the MG192 and MgPar sequences of G37T. Nucleotide positions are in relation to the predicted translational start site of the full-length G37T MG192 gene. For MgPar 3 in both G37T and the patient's strain, the region preceding the sequence shown has homology to MG191 but not to MG192 (see Fig. 1). Nucleotides identical to the variant a sequence are highlighted in grey shading. Three possible recombination events are shown, which involve replacement of part of the MG192 sequence in variant a by a segment from MgPar 2 (highlighted in green), MgPar 8 (highlighted in red) and MgPar 7 (highlighted in yellow) respectively. Double underlines C1 to C4 indicate highly conserved regions among all sequences shown. Single underlines indicate identical regions between MgPars in the patient's strain and MgPars in G37T. The sequences of MG192 variants a to f and MgPars 1 through 9 identified from this patient's strain have been submitted to GenBank under Accession No. EF117283 to EF117288 and EF117293 to EF117301 respectively. Alignment of the entire MG192 variable regions and their homologous regions in MgPars shown in this figure is available upon request.
The pattern of MG192 variation in vitro is suggestive of DNA recombination with MgPars
To determine the mechanism by which the MgPars recombine into the MG192 expression site, we mapped the regions of sequence exchange in MG192 variantsidentified in this study. As shown in Fig. 3B, by comparative analysis of the MG192 and MgPar sequences in G37-P1 and G37-P35, we observed that the sequence variations noted in MG 192 and MgPar 8 could be accounted for by a reciprocal exchange of a stretch of 155 nucleotides between the MG192 expression site and MgPar 8 as well as a non-reciprocal replacement of four nucleotides in MG192 by the corresponding region in MgPar 8.
Based on exact matches between multiple variable loci inside and outside the MgPa operon, it appears that all ATCC M. genitalium strains are very closely related to the G37 type strain and possibly could be the same strain (Kokotovic et al., 1999; Ma and Martin, 2004; Hjorth et al., 2006; Jensen, 2006). If one considers G37 as the progenitor of TW48-5G, the MG192 sequence variations in TW48-5G can be explained by a non-reciprocal replacement of a 114 bp segment between MG192 and MgPar 9 as well as a reciprocal exchange of a 561 bp segment between MG192 and MgPar 8 (Fig. 5B). Similarly if one considers G37 as the progenitor of TW10-5G.ATCC, the MG192 sequence variations identified in TW10-5G.ATCC can be explained by a reciprocal exchange of a 152 bp fragment between the G37T MG192 expression site and the G37T MgPar 2 and a reciprocal exchange of a single nucleotide located 250 bp downstream of the 152 bp fragment (Fig. 5A and Fig. S1). In fact these two areas are the only known differences in sequence between TW10-5G.ATCC and G37T. Similarly the MG192 sequence of TW10-5G.SA could have resulted from a single recombination event involving a 400 bp segment near the 5′ end of the G37T MG192 expression site and the corresponding region of G37T MgPar 7 (Fig. 2). This observation is based on comparison of the published MG192 sequence of TW10-5G.SA (Musatovova et al., 2006) with the published MG192 and MgPar sequences of G37T (Fraser et al., 1995; Iverson-Cabral et al., 2006). Whether this recombination is caused by gene cross-over or gene conversion in this case could not be predicted as the sequences of TW10-5G.SA MgPars have not been published.
The pattern of MG192 variation in vivo is suggestive of DNA recombination with MgPars
To determine if the recombination mechanisms identified in vitro as described above also occur in vivo, we analysed the MG192 variants and MgPar sequences identified in the two patient specimens obtained 10 days apart. Figure 4 illustrates the structure of the full-length variable region of the six MG192 variants and their homology to the MgPar sequences identified in this patient. Almost all sequence variations seen in each of the MG192 variants can be traced to a specific MgPar donor sequence. In one variant (f) there were two single-nucleotide changes which were not present in the corresponding sites of any of the MgPar or MG192 variants. Because the variant sequence f was derived from only one clone, it could not be confirmed if these two nucleotide changes are the result of PCR artefacts. Assuming the MG192 variant sequence a (identified as the most predominant type from specimen 199.0 obtained at the first visit) is the prototype, the other five MG192 variants appear to have been generated by two to four segmental recombination events between the MG192 expression site and a specific MgPar identified in this patient's M. genitalium strain. Given that each MgPar was found to be identical between and within the first- and second-visit specimen, the recombination events all appear to have resulted from duplication of the MgPar sequences, with loss of the segment previously present in the MG192 variable region. Therefore, a non-reciprocal recombination mechanism (gene conversion) may be the mechanism involved in the multiple MG192 recombination events occurring in this patient's M. genitalium strain over a relatively short period of 10 days.
Architecture of the MG192 variable region and the MgPar sequences
Alignment of all MG192 variants and MgPar sequences identified in this study revealed 36 discrete blocks of sequence of 8–43 bp which were completely identical among all MG192 and MgPar sequences examined. These conserved blocks were interspersed between highly variable regions, which differed at both the nucleotide and amino acid levels. Figure 6 shows examples of these constant and variable sequences corresponding to nt 235–501 of the G37T MG192 gene. The conserved sequences could act as anchors for the recombination reactions and/or they could be important in maintaining the structural integrity of the molecule. Detailed comparison of the sequences in the highly variable regions indicates that some elements might be able to translocate from one MgPar to another. For example, a 25 bp element between conserved regions C1 and C2 (Fig. 6), which is unique for MgPar 9 of G37T, is present in MgPar 5 rather than MgPar 9 in the patient's strain. Another example is a 22 bp element located between C3 and C4, which is unique for MgPar 5 in G37T but appears to translocate to MgPar 7 in the patient's strain. These observations may suggest that recombination could also occur between two MgPars.
All nine MgPar sequences identified from the patient specimens showed features similar to the G37T MgPars as previously described (Peterson et al., 1995; Iverson-Cabral et al., 2006; Jensen, 2006), including: the presence of AT-rich sequences, the maintenance of partial ORFs in individual regions with similarity to the MG191 and MG192 sequences, the appearance of stop codons near the beginning and end of the partial ORFs, the occurrence of insertions and deletions in multiples of three nucleotides, and the lack of obvious translational signals. All these features indicate that MgPar sequences are not likely to be directly expressed as functional proteins but rather serve as the donor sequences that recombine into the MG191 and MG192 expression sites. The data presented here strongly support this hypothesis.
Discussion
This study has demonstrated that, like the MG191 gene, the MG192 gene is highly variable within and among M. genitalium strains cultured in vitro and from clinical specimens. The variation occurs in the MG192 region (nt 126–1548) that has homology to eight of the nine MgPar loci. To define the molecular mechanisms for the MG192 variation, we have analysed the sequences of the MG192 variable region and all MgPar loci in serial in vitro passages of the M. genitalium G37 type strain and in sequential specimens from an M. genitalium-infected patient. We have found that the MG192 sequence changes rapidly over time in vitro and in vivo, and provided evidence that the MG192 sequence variation is generated by recombination between the MG192 expression site and MgPar sequences via gene cross-over and, possibly, also by gene conversion. As the MG192 gene is co-transcribed with MG191 (Inamine et al., 1989) and both molecules elicit strong immune responses in animal models and in humans during infection (Hu et al., 1987; Morrison-Plummer et al., 1987; Wang et al., 1997; Svenstrup et al., 2006), the finding of MG192 sequence variation generated by DNA recombination supports the hypothesis that M. genitalium is undergoing antigenic variation in the MgPa operon, which may allow the organism to evade the host immune response and establish persistent infection (Peterson et al., 1995; Iverson-Cabral et al., 2006; Jensen, 2006).
This study is the first to identify sequence variability of the MG192 and MgPar loci among ATCC M. genitalium strains and in clinical specimens, and to demonstrate that MgPar loci serve as the donor sequences that are recombined into the MG192 expression site. The hypothesis of DNA recombination is further supported by the presence in the genome of the basic components required for DNA recombination and repair, including recA (MG339), recU (MG352), Holliday junction DNA helicases ruvA (MG358) and ruvB (MG359), formamidopyrimidine-DNA glycosylase mutM (MG262.1) and a likely DNA damage-inducible protein gene (MG360) (Fraser et al., 1995; Glass et al., 2006). Although the homologues of some recombination-related enzymes (such as RecBCD, RecQ, RecO and RecJ) have been described in other organisms, they have not been identified in the M. genitalium genome or discovered experimentally. It is likely that the function of such enzymes is performed by other M. genitalium enzymes (Fraser et al., 1995). For example, the first gene (MG190) in the M. genitalium MgPa operon, which is present in single copy in the genome, contains a predicted phosphoesterase motif and has homology to the RecJ enzyme of Escherichia coli (Aravind and Koonin, 1998), raising the possibility that MG190 might act as an enzyme involved in homologous recombination (Iverson-Cabral et al., 2006).
Our studies suggest that the potential of the MgPa recombination system for generating genetic variation is nearly unlimited. Except for MgPar 6 which has homology only to the MG191 region EF, all MgPars contain 3–5 discrete minicassettes that are homologous to different regions of the MG191 and MG192 expression sites. Studies of the G37 strain maintained in Seattle have shown that sequence variation in the MG191 B, EF and G regions can occur independently (Iverson-Cabral et al., 2006). Similarly our studies of M. genitalium strains from clinical specimens revealed that recombination in the MG192 JKL and LM regions occurred independently with MgPars (Fig. 4 and L. Ma et al., unpubl. data). If we assume conservatively that each recombination event involves one of the six variable regions in MgPa and its entire corresponding region of one MgPar, the number of potential variants would be 187 500 (see Table S1). However, because chimeric sequences can be generated from small blocks within the donor MgPar sequences, in reality the number of possible combinations would be much greater. In addition, the possibilities exist that recombination can occur between MgPars or between two different variable regions within the MG191 and MG192 expression sites and that other recombination mechanisms such as phase variation (Burgos et al., 2006) are also involved in the generation of MG191 and MG192 diversity. All these findings suggest that M. genitalium has evolved an efficient recombination system to generate a vast number of variants from a minimal genome.
In the context of the above it should be noted that while comparison of the entire MG192 variable region in G37T and our patient specimens showed great variation at both the nucleotide level (14–15% difference) and the deduced amino acid level (15–17% difference) as would be predicted based on the model we are proposing, the ATCC M. genitalium strains were relatively homogenous (Fig. 2). In fact, for four of the ATCC strains the MG192 gene sequences were identical to G37T and in the other two (TW10-5G.ATCC and TW48-5G) the differences could be explained by only one or two recombination events between G37T MG192 and G37T MgPars (Fig. 5). The MgPar sequences from these two strains confirmed that several recombination events indeed may have occurred just as would have been predicted (Fig. 5). In contrast, the MgPar sequences from the patient specimens differed greatly compared with those of G37T (Table 3 and Fig. 6). In our as yet unpublished studies of the MG192 gene and selected MgPar regions of M. genitalium specimens obtained from 12 unrelated patients we have confirmed this extraordinary level of variation. These data provide further evidence that the ATCC M. genitalium strains are extremely closely related to one another if in fact they actually are not all the same strain (Kokotovic et al., 1999; Ma and Martin, 2004; Hjorth et al., 2006; Jensen, 2006).
Recombination of surface protein genes to generate genetic and antigenic variation has been described in many other pathogenic microorganisms. The architecture of the MgPa operon in M. genitalium is very similar to that of the P1 operon in M. pneumoniae (organized as ORF4–P1–ORF6 in which the latter two correspond to MG191 and MG192 respectively), with some portions of the P1 and ORF6 genes repeated seven to nine times throughout the genome (Himmelreich et al., 1996). However, in contrast to the MG191 and MG192 genes, the P1 and ORF6 genes show only limited variation (Ruland et al., 1994; Kenri et al., 1999). While the molecular mechanisms for this variation have not been well defined, preliminary studies suggest that variation in both P1 and ORF6 may be generated by recombination with repetitive elements via a gene conversion mechanism (Ruland et al., 1994; Kenri et al., 1999). In fact, the gene conversion mechanism has been documented as a common mechanism to generate genetic and antigenic variations in surface structures in other mycoplasma species as well as other sexually transmitted pathogens. The MgPa operon of M. genitalium and the P1 operon of M. pneumoniae resemble the genetic architecture of the vlhA gene of M. synoviae (Noormohammadi et al., 2000), the pilin gene of Neisseria gonorrhoeae (Zhang et al., 1992) and the tprk gene of Treponema pallidum (Centurion-Lara et al., 2004), in that all carry a single or no more than two expression sites as well as a repertoire of homologous repeats or pseudogenes that serve as donor sequences and can be recombined into the expression site. However, in contrast to these genes for which the donor sequences are present in tandem arrays, the repetitive elements for the MgPa and P1 operons are dispersed throughout the genome. In all the pathogens mentioned above, studies of the recombination mechanisms have shown a clear bias towards gene conversion, as opposed to gene cross-over (Santoyo and Romero, 2005). This bias has also been demonstrated in studies of the antigenic variation in the vlsE gene of Borrelia burgdorferi (Zhang and Norris, 1998), the vmp gene of Borrelia hermsii (Donelson, 1995) and the msp2 gene of Anaplasma marginale (Brayton et al., 2001).
In the present study, it appeared that gene cross-over, probably together with gene conversion, occurred in vitro in the serial passages of the G37 type strain. The occurrence of the gene cross-over mechanism is strongly supported by the reciprocal exchange of the 150 bp segment between MG192 and MgPar 8 in serial passages of G37-P35 (Fig. 3). The sequence exchange of 4 bp between MG192 and MgPar 8 in the G37-P35 (Fig. 3) likely reflects a non-reciprocal exchange (gene conversion) event as 33 plasmid clones of G37-P35 MG192 and 30 plasmid clones of G37-P35 MgPar 8 were sequenced without a single example of a potential cross-over event at this region (P< 0.005 for finding such a clone from the plasmid library, Table 3). However, because the sequence between the 4 bp and 150 bp segment was identical (aside from the AGT tandem repeat number variation as discussed below), an alternative explanation is that two sequential cross-over events had occurred, with the first cross-over event involving both regions and the second cross-over involving the 4 bp region alone. Nonetheless, definitive proof of such events would require single colony cloning studies.
Analysis of the six MG192 variants and nine MgPars in the sequential patient specimens showed that different MgPar segments had recombined into the MG192 expression site without detectable change in MgPar sequences over time, suggesting gene conversion rather than gene cross-over events. If gene cross-over had occurred, we would have expected the MgPar sequences as determined by PCR subcloning analyses to be highly heterogeneous between and/or within the two sequential specimens. For one of the loci where recombination events appeared to have occurred (nucleotides 248–468 in variants d and f, Fig. 4) 30 MgPar 2 plasmid clones were analysed without a single example of a potential cross-over event, thus supporting gene conversion as the mechanism (P< 0.005, Table 3). As discussed above, gene conversion occurs more frequently than gene cross-over in a number of different bacteria and we hypothesize that the same is the case with M. genitalium. However, given the strong evidence for the occurrence of gene cross-over in the G37 type strain, it is possible that the patient's strain studied here was unique and that gene cross-over also occurs in vivo but could not be distinguished from gene conversion by PCR and sequence analysis of the whole cell population in the specimen as was done here. While single colony cloning would provide the needed definitive evidence for gene conversion as a mechanism of recombination in the M. genitalium MG192 gene, this is not possible given the difficulties with culturing the organism (Hamasuna et al., 2007). Our hypothesis could be supported through study of more patient specimens and statistical analyses of large samples of plasmid clones as done in the example above.
Despite the strong evidence for the role of homologous recombination in generation of MG192 variation presented in this study, other mechanisms might also be involved. The presence of stretches of conserved sequences within the MG192 variable region (Fig. 6) suggests a possible involvement of a site-specific recombination mechanism. In addition, genetic variation of MG192 also involves gain or loss of AGT tandem repeats (Table 2). In all ATCC M. genitalium strains and their derivatives examined in this study, we observed a mixture of two or more AGT repeat variants which differed by only one repeat unit, suggesting that this repeat sequence is undergoing rapid change over time. This observation is further supported by the finding of a significant change in the ratio of populations containing different repeat numbers during serial passage of the G37 type strain in vitro (Table 2). Repeat number variation could be generated by recombination between MG192 and MgPars that contain different numbers of these tandem repeats (Fig. 1), but it also could be due to slipped-strand mispairing (Levinson and Gutman, 1987), which may occur alone or together with recombination. The AGT repeat unit encodes serine and changes in the number of repeat units result in heterogeneity in the size of the polyserine tract. The functional significance of the polyserine tract in M. genitalium is unknown. Whether it functions as a flexible spacer region to optimize protein interactions, as it has been hypothesized for modular molecules in other organisms (Hall et al., 1989; Howard et al., 2004), currently cannot be determined.
The occurrence of MG192 sequence variation in vitro demonstrates that these events can arise spontaneously, as has been observed in other organisms (Borst, 1991; Criss et al., 2005). In N. gonorrhoeae, the frequency of gene conversion in vitro has been determined to be 3.3 × 10−2 per generation (Serkin and Seifert, 1998). In this study we observed change in the G37 MG192 sequence as early as passage 17 but no further change through passage 35. The change may have occurred at a much earlier passage level in response to growth conditions in the laboratory. Perhaps the resulting change in the translated protein conferred some replicative advantage to the organism. The sequence difference found in the TW10-5G strain maintained in different laboratories (TW10-5G.DK and TW10-5G.SA) could have been due to different in vitro growth conditions in the two laboratories where they were maintained but this is only speculation. It would be of interest to determine if predictable changes in MG191 and/or MG192 can be induced by differing in vitro environments including cultivation with different mammalian cell types. The occurrence of frequent spontaneous variation may be necessary to produce escape variants for selection by the host immune system (Borst, 1991). The striking MG192 variation we found here in patient 199's sequential specimens after an interval of only 10 days may represent an example of this phenomenon. In addition, as both MG192 and MG191 proteins appear to be required for cellular adhesion and terminal organelle development (Burgos et al., 2006), variation in the primary amino acid sequence of MG191 and MG192 may change the structure of the MgPa adhesin in response to in vivo environmental changes and/or the availability of certain human cell types.
A pragmatic outcome of the present study is the demonstration that inclusion of the MG192 variable region in a genotyping system as recently suggested by Musatovova et al. (2006) is inappropriate. Here we have shown that the TW10-5G MG192 sequence published by these investigators (TW10-5G.SA) and the sequence of the ATCC strain (TW10-5G.ATCC) differ significantly. We have shown how this may have occurred by demonstrating sequence shifts of the MG192 variable region between the early and late in vitro passages of the G37 strain. Moreover, we have documented dramatic sequence shifts in the MG192 variable region in a human strain over a 10-day period in vivo. Musatovova et al. (2006) used restriction fragment length polymorphism (RFLP) analysis of a fragment which included parts of the MG192 variable region as the basis for their proposed genotyping system. Over this fragment, there are different numbers of restriction sites between TW10-5G.SA and TW10-5G.ATCC, which likely would have resulted in different RFLP patterns and the false impression that they are two different M. genitalium strains. We believe that genotyping should be based on relatively more conserved regions of the M. genitalium genome (Ma and Martin, 2004; Hjorth et al., 2006).
In summary, we have demonstrated sequence variation of the MG192 and MgPar loci among ATCC M. genitalium strains and in clinical specimens, and provided evidence for homologous recombination between the MG192 expression site and MgPars in vitro and in vivo. Based on preliminary analysis of putative donor sites for MG192 variants, we propose a model of recombination involving gene cross-over and, possibly also gene conversion. As the free-living organism with the smallest known genome, M. genitalium appears to have evolved an efficient system to generate a vast number of variants, thus enhancing the organism's survival in differing host environments and allowing it to escape from host defences.
Experimental procedures
Mycoplasma genitalium specimens
We obtained seven M. genitalium strains as freeze-dried culture and/or genomic DNA from ATCC. Detailed information about these specimens is given in Table 1. One vial of each freeze-dried ATCC strain was rehydrated in 0.5 ml of Spiroplasma medium SP-4 and then inoculated into a culture flask containing 10 ml of SP-4. The flask was incubated at 37°C in 5% CO2. When the colour of the SP-4 medium changed from red to orange, the culture was collected and concentrated by centrifugation at 9000 g for 30 min. One aliquot of the cell pellet for each strain was used for DNA analysis. This material was designated as passage 1 for each strain. For the G37 type strain (ATCC33530), one aliquot of the passage 1 cell pellet (G37-P1) was used to inoculate another flask with 25 ml of fresh SP-4 medium to make passage 2. This procedure was repeated until the passage 35 (G37-P35) was obtained. In addition to the ATCC strains, we studied the TW10-5G strain which was provided to one of us (J.S.J.) by J.G. Tully at the passage level 11 prior to deposit into ATCC (designated as TW10-5G.DK, Table 1). The strain had been passaged further five times before being studied for DNA sequences. Two sequential urine specimens were obtained 10 days apart from a man in New Orleans with acute urethritis (Mena et al., 2002). Both samples were positive for M. genitalium by our PCR assay and had been genotyped using multiple genomic loci (Ma and Martin, 2004; Hjorth et al., 2006). Informed consent was obtained from this patient and the study protocol was approved by the LSUHSC Institutional Review Board.
PCR and sequencing of the MG192 gene
Based on an alignment of the published MgPa operon and MgPar sequences of G37T (Fraser et al., 1995; Iverson-Cabral et al., 2006), we designed primers with no homology to any of the MgPar sequences so that only the MG192 gene would be amplified. The sequences of the primers are listed in Table 4. In addition, the previously described primer MG192A (Musatovova et al., 2006) was also used.
Table 4.
Primers used to amplify the M. genitalium MG192 and MgPar sequences.
| Primer name | Sequence (5′→3′) | Target (sequence locationa) |
|---|---|---|
| 5346F | TACCCCAACCAAACCAACTC | MG192 (225850–225869) |
| 227567R | CTTCGCAATTCATTAGGGGAGACA | MG192 (227546–227569) |
| 227529R | GATCTGATCAGTTCTGGAAGGTAAACG | MG192 (227505–227531) |
| MG192Ab | CACTAGCCAATACCTTCCTTGTCAAAGAGG | MG192 (225974–226003) |
| 1F2 | GAATGGCGGTTTTTACCCAG | MgPar 1 (86583–86602) |
| 1R1 | AATTTCACACCAACAATGCC | MgPar 1 (88396–88415) |
| 124F3 | TCAGTTTATTGGTCAAAGAAGTCGC | MgPars 1, 2 and 4 (168272–168296) |
| 2F1 | CAAGGGGTTGTTTCTTTGC | MgPar 2 (166960–166978) |
| 2R1 | GTTCTAGGATCAATTCTTGCAAA | MgPar 2 (168687–168709) |
| 170116R | TACTCACCCGTTCGCCTCTA | MgPar 2 (170095–170114) |
| 3F3 | CCAGCAGTTAAGCACAGTGG | MgPar 3 (174730–174749) |
| 3R4 | CTTTCACTACCTGTTTCACG | MgPar 3 (176120–176139) |
| 4F3 | GGTCAAAGAAGTCGCAAATTTTCTTA | MgPar 4 (214652–214677) |
| 4R1 | ATGCATGCAGTGGCTATGG | MgPar 4 (216074–216092) |
| 5F4 | TGGCGGTTTTTATCAACTAAATAAAAAC | MgPar 5 (230434–230461) |
| 5R2 | GCGGTTTTAATCTTTTGGAG | MgPar 5 (232126–232145) |
| 6F1 | TGCAGCCATTTCTTTGATCC | MgPar 6 (273024–273043) |
| 6R1 | CATCTGGTAACCAAAGACAC | MgPar 6 (273872–273891) |
| 7F2 | TGCTGCGGTGAATTCTAAGG | MgPar 7 (312727–312746) |
| 7F3 | TTAATTAATAAAACCTCGACCC | MgPar 7 (314133–314154) |
| 7R1 | CAGCGCCAAAAAGATTAGGA | MgPar 7 (315817–315836) |
| 7R3 | ATGGATGGTTTGGTCAGCG | MgPar 7 and MG192 (314170–314188) |
| 8F1 | GGTAAGCCAATCCGATATTGAG | MgPar 8 (348804–348825) |
| 8F4 | ATGGTGGTTTTTATCAACTAAATAAAAAAC | MgPar 8 (350254–350283) |
| 8R2 | AATAAAGGTAAAGCCTAGTG | MgPars 5 and 8 (350377–350396) |
| 351964R | CAATCAACTACTTTTCACTTTACACG | MgPar 8 (351940–351965) |
| 9F1 | CTGAAAACTAGCGACTGCATTTAG | MgPar 9 (427858–427881) |
| 9F3 | GGTTTGTTGACATAAACACTCC | MgPar 9 (428248–428269) |
| 9R1 | TTAAGTGCTGCATCCTTCCA | MgPar 9 (430749–430768) |
| 429679R | TTGAATCCTTCACCGGACTATC | MgPar 9 (213693–213714) |
Location of the primer on the M. genitalium G37T genome sequence under GenBank Accession No. NC_000908.
From reference (Musatovova et al., 2006).
M. genitalium ATCC strains and their derivatives
The DNAs from all M. genitalium ATCC strains and their derivatives were amplified using 5346F or MG192A as the forward primer and 227567R as the reverse primer. A single round of PCR amplification was performed with high-fidelity Pfu DNA polymerase (Stratagene) and the following cycling conditions: 95°C for 1 min, 35 cycles of 94°C for 45 s, 50°C for 1 min, and 72°C for 3 min.
Patient specimens
To amplify the DNAs from patient specimens, we carried out a nested PCR strategy using primers 5346F and 227567R for the first round of amplification and primers MG192A plus 227529R for the second round. The first round of amplification was performed with AmpliTaq Gold DNA polymerase (Applied Biosystems) and a touch-down protocol as described previously (Ma et al., 2003). The second round of amplification was performed with high-fidelity Pfu DNA polymerase (Stratagene) and the same conditions described above.
Each specimen was amplified by at least two independent PCR assays. All PCR products were initially directly sequenced and then by sequencing of individual plasmid clones after subcloning. The PCR-Script® Amp Cloning Kit (Stratagene) and the TOPO TA Cloning kit (Invitrogen) were used according to manufacturers' standard protocols. There was no difference in the overall distribution of the MG192 sequences between different PCR runs for the same specimens (Fig. 4). All MG192 sequences obtained contained a constant region of 26–125 bp in the 5′ end and of 49 bp in the 3′ end, which were identical to the corresponding region of the G37T MG192 gene. This finding confirmed that the 5′ and 3′ ends of MG192 are highly conserved and all MG192 sequences we obtained were from the MG192 gene; not from any of the MgPar regions.
PCR and sequencing of the MgPar sequences
The primers used to amplify MgPars were designed from the published genome sequence of the M. genitalium G37 strain (Fraser et al., 1995) and are listed in Table 4. In general, primers for each MgPar were chosen from regions that have no homology to MG192 or other MgPars in order to assure the specificity. There are three primers 124F3, 7R3 and 8R2 that are shared among MgPars 1, 2 and 4, between MgPar 7 and MG192, and between MgPars 5 and 8 respectively. These were used in combination with another primer specific for the MgPar to be evaluated. We amplified three MgPars (2, 8 and 9) in G37-P1, all nine MgPars in G37-P35, TW10-5G.ATCC, TW48-5G, and the two sequential patient specimens (No. 199.0 and 199.1). MgPars 3 and 6 were amplified in their full length by using primer sets 3F3–3R4 and 6F1–6R1 respectively. For all other MgPars, only the regions that were homologous to the MG192 gene were amplified unless otherwise stated below. The primer sets for MgPars 1, 2, 4, 5, 7, 8 and 9 were 1F2–1R1, 124F3–170116R, 4F3–4R1, 5F4–5R2, 7F3–7R1, 8F4–351964R and 9F1–429679R respectively. The full-length MgPar 2 in TW10-5G.ATCC was amplified in two overlapping fragments using primer sets 2F1–2R1 and 124F3–170116R respectively. The full-length MgPar 7 in TW10-5G.ATCC was amplified in two overlapping fragments using primer sets 7F2–7R3 and 7F3–7R1 respectively. The full-length MgPar 8 in G37-P1, G37-P35, TW48-5G and two patient specimens was amplified in two overlapping fragments using primer sets 8F1–8R2 and 8F4–351964R respectively. The full-length MgPar 9 in TW10-5G.ATCC and TW48-5G was amplified by a single round of PCR using the primer set 9F3–9R1. All amplifications were performed with AmpliTaq Gold DNA polymerase (Applied Biosystems) and a touch-down protocol as described previously (Ma et al., 2003). Initially the PCR products of all MgPars were directly sequenced and none of them except MgPars 2, 8 and 9 displayed a mixture of two or more sequences. These sequence mixtures were completely accounted for by variation in the number of AGT repeats; the sequences flanking the repeat region were entirely uniform. However, because the MG192 variable region in the clinical specimens was found to contain sequence mixtures, we further investigated the possibility of existence of sequence mixtures in MgPars by performing subcloning of the PCR products from MgPar 8 in G37-P1 and G37-P35, MgPars 8 and 9 in TW48-5G, MgPars 2 and 8 in TW10-5G, and all MgPars except for MgPar 6 in the two patient specimens. For each PCR product, 5–30 plasmid clones were selected for sequencing. All cloned PCR products showed homogenous sequences within each strain except for the VNTR in MgPar 8 and MgPar 9 as well as the presence of a few scattered single-base substitutions in a few clones, which may represent PCR artefacts.
DNA and predicted protein sequence analysis
DNA sequencing was carried out by use of an ABI PRISM 3100 automated capillary sequencer (Applied Biosystems). Sequence analysis was performed using the CLC Combined Workbench 2.0 (CLC bio, Aarhus C, Denmark) and the MultAlin software available at http://bioinfo.genopole-toulouse.prd.fr/multalin/ (Corpet, 1988). Representative nucleotide sequences obtained in this study have been submitted to the GenBank database under Accession No. EF117280 to EF117301.
Statistical analyses
Calculation of the probability of undetected cross-over events as the explanation of sequence data which uniformly show evidence of a conversion event was performed by calculating the exact binomial 95% confidence interval centred on zero for the proportion of cross-over sequences among a set of n sequences all of which were consistent with conversion events. Then the probability of finding a cross-over event if n + 1 plasmid clones had been sequenced was calculated.
Acknowledgments
This work was supported by the Gulf South Sexually Transmitted Infections/Topical Microbicide Cooperative Research Center grant from NIH-NIAID (1 U19 AI061972). We thank Dr Patricia A. Totten and Dr Stefanie L. Iverson-Cabral from University of Washington (UW) at Seattle for sharing their G37 MG192 and MgPar sequence alignment, Dr Arturo Centurion-Lara from UM for discussion about the recombination mechanisms, and Dr Jun Xie, Kodensha, Osaka, Japan for assistance in electronic artwork.
Supplementary material
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.1365-2958.2007.05911.x (This link will take you to the article abstract).
Please note: Blackwell Publishing is not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Aravind L, Koonin EV. A novel family of predicted phosphoesterases includes Drosophila prune protein and bacterial RecJ exonuclease. Trends Biochem Sci. 1998;23:17–19. doi: 10.1016/s0968-0004(97)01162-6. [DOI] [PubMed] [Google Scholar]
- Baseman JB, Dallo SF, Tully JG, Rose DL. Isolation and characterization of Mycoplasma genitalium strains from the human respiratory tract. J Clin Microbiol. 1988;26:2266–2269. doi: 10.1128/jcm.26.11.2266-2269.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Belkum A, Scherer S, van Alphen L, Verbrugh H. Short-sequence DNA repeats in prokaryotic genomes. Microbiol Mol Biol Rev. 1998;62:275–293. doi: 10.1128/mmbr.62.2.275-293.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhugra B, Voelker LL, Zou N, Yu H, Dybvig K. Mechanism of antigenic variation in Mycoplasma pulmonis: interwoven, site-specific DNA inversions. Mol Microbiol. 1995;18:703–714. doi: 10.1111/j.1365-2958.1995.mmi_18040703.x. [DOI] [PubMed] [Google Scholar]
- Borst P. Molecular genetics of antigenic variation. Immunol Today. 1991;12:A29–A33. doi: 10.1016/S0167-5699(05)80009-X. [DOI] [PubMed] [Google Scholar]
- Brayton KA, Knowles DP, McGuire TC, Palmer GH. Efficient use of a small genome to generate antigenic diversity in tick-borne ehrlichial pathogens. Proc Natl Acad Sci USA. 2001;98:4130–4135. doi: 10.1073/pnas.071056298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos R, Pich OQ, Ferrer-Navarro M, Baseman JB, Querol E, Pinol J. Mycoplasma genitalium P140 and P110 cytadhesins are reciprocally stabilized and required for cell adhesion and terminal organelle development. J Bacteriol. 2006;188:8627–8637. doi: 10.1128/JB.00978-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centurion-Lara A, LaFond RE, Hevner K, Godornes C, Molini BJ, Van Voorhis WC, Lukehart SA. Gene conversion: a mechanism for generation of heterogeneity in the tprK gene of Treponema pallidum during infection. Mol Microbiol. 2004;52:1579–1596. doi: 10.1111/j.1365-2958.2004.04086.x. [DOI] [PubMed] [Google Scholar]
- Citti C, Rosengarten R. Mycoplasma genetic variation and its implication for pathogenesis. Wien Klin Wochenschr. 1997;109:562–568. [PubMed] [Google Scholar]
- Clausen HF, Fedder J, Drasbek M, Nielsen PK, Toft B, Ingerslev HJ, et al. Serological investigation of Mycoplasma genitalium in infertile women. Hum Reprod. 2001;16:1866–1874. doi: 10.1093/humrep/16.9.1866. [DOI] [PubMed] [Google Scholar]
- Cohen CR, Manhart LE, Bukusi EA, Astete S, Brunham RC, Holmes KK, et al. Association between Mycoplasma genitalium and acute endometritis. Lancet. 2002;359:765–766. doi: 10.1016/S0140-6736(02)07848-0. [DOI] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criss AK, Kline KA, Seifert HS. The frequency and rate of pilin antigenic variation in Neisseria gonorrhoeae. Mol Microbiol. 2005;58:510–519. doi: 10.1111/j.1365-2958.2005.04838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallo SF, Baseman JB. Adhesin gene of Mycoplasma genitalium exists as multiple copies. Microb Pathog. 1991;10:475–480. doi: 10.1016/0882-4010(91)90113-o. [DOI] [PubMed] [Google Scholar]
- Donelson JE. Mechanisms of antigenic variation in Borrelia hermsii and African trypanosomes. J Biol Chem. 1995;270:7783–7786. doi: 10.1074/jbc.270.14.7783. [DOI] [PubMed] [Google Scholar]
- Dybvig K, Voelker LL. Molecular biology of mycoplasmas. Annu Rev Microbiol. 1996;50:25–57. doi: 10.1146/annurev.micro.50.1.25. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, et al. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- Glass JI, Assad-Garcia N, Alperovich N, Yooseph S, Lewis MR, Maruf M, et al. Essential genes of a minimal bacterium. Proc Natl Acad Sci USA. 2006;103:425–430. doi: 10.1073/pnas.0510013103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J, Hazlewood GP, Huskisson NS, Durrant AJ, Gilbert HJ. Conserved serine-rich sequences in xylanase and cellulase from Pseudomonas fluorescens subspecies cellulosa: internal signal sequence and unusual protein processing. Mol Microbiol. 1989;3:1211–1219. doi: 10.1111/j.1365-2958.1989.tb00271.x. [DOI] [PubMed] [Google Scholar]
- Hamasuna R, Osada Y, Jensen JS. Isolation of Mycoplasma genitalium from first-void urine specimens by coculture with Vero cells. J Clin Microbiol. 2007;45:847–850. doi: 10.1128/JCM.02056-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li BC, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjorth SV, Bjornelius E, Lidbrink P, Falk L, Dohn B, Berthelsen L, et al. Sequence-based typing of Mycoplasma genitalium reveals sexual transmission. J Clin Microbiol. 2006;44:2078–2083. doi: 10.1128/JCM.00003-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard MB, Ekborg NA, Taylor LE, Hutcheson SW, Weiner RM. Identification and analysis of polyserine linker domains in prokaryotic proteins with emphasis on the marine bacterium Microbulbifer degradans. Protein Sci. 2004;13:1422–1425. doi: 10.1110/ps.03511604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu PC, Schaper U, Collier AM, Clyde WA, Horikawa M, Huang YS, Barile MF. A Mycoplasma genitalium protein resembling the Mycoplasma pneumoniae attachment protein. Infect Immun. 1987;55:1126–1131. doi: 10.1128/iai.55.5.1126-1131.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamine JM, Loechel S, Collier AM, Barile MF, Hu PC. Nucleotide sequence of the MgPa (mgp) operon of Mycoplasma genitalium and comparison to the P1 (mpp) operon of Mycoplasma pneumoniae. Gene. 1989;82:259–267. doi: 10.1016/0378-1119(89)90051-6. [DOI] [PubMed] [Google Scholar]
- Iverson-Cabral SL, Astete SG, Cohen CR, Rocha EP, Totten PA. Intrastrain heterogeneity of the mgpB gene in Mycoplasma genitalium is extensive in vitro and in vivo and suggests that variation is generated via recombination with repetitive chromosomal sequences. Infect Immun. 2006;74:3715–3726. doi: 10.1128/IAI.00239-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen JS. Mycoplasma genitalium infections. Diagnosis, clinical aspects, and pathogenesis. Dan Med Bull. 2006;53:1–27. [PubMed] [Google Scholar]
- Kapiga SH, Sam NE, Mlay J, Aboud S, Ballard RC, Shao JF, Larsen U. The epidemiology of HIV-1 infection in northern Tanzania: results from a community-based study. AIDS Care. 2006;18:379–387. doi: 10.1080/09540120500465012. [DOI] [PubMed] [Google Scholar]
- Kenri T, Taniguchi R, Sasaki Y, Okazaki N, Narita M, Izumikawa K, et al. Identification of a new variable sequence in the P1 cytadhesin gene of Mycoplasma pneumoniae: evidence for the generation of antigenic variation by DNA recombination between repetitive sequences. Infect Immun. 1999;67:4557–4562. doi: 10.1128/iai.67.9.4557-4562.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokotovic B, Friis NF, Jensen JS, Ahrens P. Amplified-fragment length polymorphism fingerprinting of Mycoplasma species. J Clin Microbiol. 1999;37:3300–3307. doi: 10.1128/jcm.37.10.3300-3307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson G, Gutman GA. Slipped-strand mispairing: a major mechanism for DNA sequence evolution. Mol Biol Evol. 1987;4:203–221. doi: 10.1093/oxfordjournals.molbev.a040442. [DOI] [PubMed] [Google Scholar]
- Lysnyansky I, Rosengarten R, Yogev D. Phenotypic switching of variable surface lipoproteins in Mycoplasma bovis involves high-frequency chromosomal rearrangements. J Bacteriol. 1996;178:5395–5401. doi: 10.1128/jb.178.18.5395-5401.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Martin DH. Single-nucleotide polymorphisms in the rRNA operon and variable numbers of tandem repeats in the lipoprotein gene among Mycoplasma genitalium strains from clinical specimens. J Clin Microbiol. 2004;42:4876–4878. doi: 10.1128/JCM.42.10.4876-4878.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Kutty G, Jia Q, Kovacs JA. Characterization of variants of the gene encoding the p55 antigen in Pneumocystis from rats and mice. J Med Microbiol. 2003;52:955–960. doi: 10.1099/jmm.0.05131-0. [DOI] [PubMed] [Google Scholar]
- Manhart LE, Critchlow CW, Holmes KK, Dutro SM, Eschenbach DA, Stevens CE, Totten PA. Mucopurulent cervicitis and Mycoplasma genitalium. J Infect Dis. 2003;187:650–657. doi: 10.1086/367992. [DOI] [PubMed] [Google Scholar]
- Maniloff J. The minimal cell genome: ‘on being the right size’. Proc Natl Acad Sci USA. 1996;93:10004–10006. doi: 10.1073/pnas.93.19.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena L, Wang X, Mroczkowski TF, Martin DH. Mycoplasma genitalium infections in asymptomatic men and men with urethritis attending a sexually transmitted diseases clinic in New Orleans. Clin Infect Dis. 2002;35:1167–1173. doi: 10.1086/343829. [DOI] [PubMed] [Google Scholar]
- Moller BR, Taylor-Robinson D, Furr PM. Serological evidence implicating Mycoplasma genitalium in pelvic inflammatory disease. Lancet. 1984;1:1102–1103. doi: 10.1016/s0140-6736(84)92511-x. [DOI] [PubMed] [Google Scholar]
- Morrison-Plummer J, Lazzell A, Baseman JB. Shared epitopes between Mycoplasma pneumoniae major adhesin protein P1 and a 140-kilodalton protein of Mycoplasma genitalium. Infect Immun. 1987;55:49–56. doi: 10.1128/iai.55.1.49-56.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musatovova O, Dhandayuthapani S, Baseman JB. Transcriptional starts for cytadherence-related operons of Mycoplasma genitalium. FEMS Microbiol Lett. 2003;229:73–81. doi: 10.1016/S0378-1097(03)00789-4. [DOI] [PubMed] [Google Scholar]
- Musatovova O, Herrera C, Baseman JB. Proximal region of the gene encoding cytadherence-related protein permits molecular typing of Mycoplasma genitalium clinical strains by PCR-restriction fragment length polymorphism. J Clin Microbiol. 2006;44:598–603. doi: 10.1128/JCM.44.2.598-603.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noormohammadi AH, Markham PF, Kanci A, Whithear KG, Browning GF. A novel mechanism for control of antigenic variation in the haemagglutinin gene family of Mycoplasma synoviae. Mol Microbiol. 2000;35:911–923. doi: 10.1046/j.1365-2958.2000.01766.x. [DOI] [PubMed] [Google Scholar]
- Perez G, Skurnick JH, Denny TN, Stephens R, Kennedy CA, Regivick N, et al. Herpes simplex type II and Mycoplasma genitalium as risk factors for heterosexual HIV transmission: report from the heterosexual HIV transmission study. Int J Infect Dis. 1998;3:5–11. doi: 10.1016/s1201-9712(98)90088-1. [DOI] [PubMed] [Google Scholar]
- Peterson SN, Bailey CC, Jensen JS, Borre MB, King ES, Bott KF, et al. Characterization of repetitive DNA in the Mycoplasma genitalium genome: possible role in the generation of antigenic variation. Proc Natl Acad Sci USA. 1995;92:11829–11833. doi: 10.1073/pnas.92.25.11829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosengarten R, Wise KS. Phenotypic switching in mycoplasmas: phase variation of diverse surface lipoproteins. Science. 1990;247:315–318. doi: 10.1126/science.1688663. [DOI] [PubMed] [Google Scholar]
- Ruland K, Himmelreich R, Herrmann R. Sequence divergence in the ORF6 gene of Mycoplasma pneumonia. J Bacteriol. 1994;176:5202–5209. doi: 10.1128/jb.176.17.5202-5209.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoyo G, Romero D. Gene conversion and concerted evolution in bacterial genomes. FEMS Microbiol Rev. 2005;29:169–183. doi: 10.1016/j.femsre.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Seifert HS, So M. Genetic mechanisms of bacterial antigenic variation. Microbiol Rev. 1988;52:327–336. doi: 10.1128/mr.52.3.327-336.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serkin CD, Seifert HS. Frequency of pilin antigenic variation in Neisseria gonorrhoeae. J Bacteriol. 1998;180:1955–1958. doi: 10.1128/jb.180.7.1955-1958.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svenstrup HF, Jensen JS, Gevaert K, Birkelund S, Christiansen G. Identification and characterization of immunogenic proteins of Mycoplasma genitalium. Clin Vaccine Immunol. 2006;13:913–922. doi: 10.1128/CVI.00048-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Robinson D. Mycoplasma genitalium– an up-date. Int J STD AIDS. 2002;13:145–151. doi: 10.1258/0956462021924776. [DOI] [PubMed] [Google Scholar]
- Taylor-Robinson D, Furr PM, Tully JG, Barile MF, Moller BR. Animal models of Mycoplasma genitalium urogenital infection. Isr J Med Sci. 1987;23:561–564. [PubMed] [Google Scholar]
- Tully JG, Taylor-Robinson D, Cole RM, Rose DL. A newly discovered mycoplasma in the human urogenital tract. Lancet. 1981;i:1288–1291. doi: 10.1016/s0140-6736(81)92461-2. [DOI] [PubMed] [Google Scholar]
- Tully JG, Rose DL, Baseman JB, Dallo SF, Lazzell AL, Davis CP. Mycoplasma pneumoniae and Mycoplasma genitalium mixture in synovial fluid isolate. J Clin Microbiol. 1995;33:1851–1855. doi: 10.1128/jcm.33.7.1851-1855.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RY, Grandinetti T, Shih JW, Weiss SH, Haley CL, Hayes MM, Lo SC. Mycoplasma genitalium infection and host antibody immune response in patients infected by HIV, patients attending STD clinics and in healthy blood donors. FEMS Immunol Med Microbiol. 1997;19:237–245. doi: 10.1111/j.1574-695X.1997.tb01093.x. [DOI] [PubMed] [Google Scholar]
- Wise KS. Adaptive surface variation in mycoplasmas. Trends Microbiol. 1993;1:59–63. doi: 10.1016/0966-842X(93)90034-O. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Woude MW, Baumler AJ. Phase and antigenic variation in bacteria. Clin Microbiol Rev. 2004;17:581–611. doi: 10.1128/CMR.17.3.581-611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JR, Norris SJ. Genetic variation of the Borrelia burgdorferi gene vlsE involves cassette-specific, segmental gene conversion. Infect Immun. 1998;66:3698–3704. doi: 10.1128/iai.66.8.3698-3704.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QY, DeRyckere D, Lauer P, Koomey M. Gene conversion in Neisseria gonorrhoeae: evidence for its role in pilus antigenic variation. Proc Natl Acad Sci USA. 1992;89:5366–5370. doi: 10.1073/pnas.89.12.5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.