Abstract
Sequestration of parasitized erythrocytes in the microcirculation of tissues is thought to be important in the pathogenesis of severe falciparum malaria. A major variant surface antigen, var/Plasmodium falciparum erythrocyte membrane protein 1, expressed on the surface of the infected erythrocyte, mediates cytoadherence to vascular endothelium. To address the question of tissue-specific accumulation of variant types, we used the unique resource generated by the clinicopathological study of fatal paediatric malaria in Blantyre, Malawi, to analyse var gene transcription in patients dying with falciparum malaria. Despite up to 102 different var genes being expressed by P. falciparum populations in a single host, only one to two of these genes were expressed at high levels in the brains and hearts of these patients. These major var types differed between organs. However, identical var types were expressed in the brains of multiple patients from a single malaria season. These results provide the first evidence of organ-specific accumulation of P. falciparum variant types and suggest that parasitized erythrocytes can exhibit preferential binding in the body, supporting the hypothesis of cytoadherence-linked pathogenesis.
Introduction
The capacity of Plasmodium falciparum to cause severe and fatal disease is believed to be in part due to its ability to sequester in post-capillary venules. The process of cytoadherence is mediated by a variety of host endothelial receptors and by P. falciparum antigens expressed on the surface of the host erythrocyte. The best studied of these is the P. falciparum erythrocyte membrane protein 1 (PfEMP1), encoded by the var multigene family (Baruch et al., 1995). Each mature asexual parasite expresses a dominant PfEMP1 type that can be stably inherited through successive cell cycles or can switch to expression of a different gene (Scherf et al., 1998).
PfEMP1 proteins are composed of adhesive domains, termed Duffy-binding-like (DBL), constant (C2) and cysteine-rich interdomain region. These domains can be sorted into subgroups by sequence motifs and are characterized by distinctive binding properties such that specific domains determine to which endothelial receptors each P. falciparum-infected erythrocyte (pRBC) adheres (Buffet et al., 1999; Smith et al., 2000). pRBC expressing PfEMP1 with defined binding specificities can be selected from a mixed population by adhesion to particular endothelial receptors.
var genes have been classified into various groups (A–E) based on coding and non-coding sequence motifs and domain arrangements. Most genes contain a DBLα domain at their N-terminus which can be further subgrouped into α (sometimes labelled DBLα0) and α1 types that contain two or four conserved cysteines residues respectively (Robinson et al., 2003). DBLα1 domains are characteristic of type A and B/A var genes that do not adhere to CD36 but some of which can mediate rosetting, the binding of pRBC to uninfected erythrocytes (Rowe et al., 1997; Russell et al., 2005). Type A var genes have been associated with severe or cerebral in peripheral populations although such studies have produced conflicting results (Kirchgatter and del Portillo, 2002; Jensen et al., 2004; Kaestli et al., 2004; 2006; Bull et al., 2005; Kyriacou et al., 2006; Rottmann et al., 2006).
Genetically variant isolates of P. falciparum contain overlapping but generally distinct contingents of var genes and sequence relatedness is independent of geographic origin and strain type, apart from some areas of low transmission (Kyes et al., 1997; Albrecht et al., 2006; Barry et al., 2007). A few unusual var, such as the var2csa gene implicated in malaria in pregnancy, are highly conserved in different P. falciparum populations (Salanti et al., 2002; Trimnell et al., 2006; Kraemer et al., 2007). P. falciparum in monoclonal infections express single or a few dominant var genes in circulating populations (Peters et al., 2002; Bull et al., 2005; Lavstsen et al., 2005; Kyriacou et al., 2006). A longitudinal study of asymptomatic hosts demonstrated that var gene expression changes dramatically over time, with minimal overlap in var repertoire between samples taken at 2 week intervals despite few changes in P. falciparum genetic types in the infecting population (Kaestli et al., 2004).
One of the limitations of these previous studies is that it has only been possible to study the dynamics of var expression in the peripheral blood. We have been conducting a clinicopathological study of fatal paediatric malaria in Blantyre, Malawi, since 1996 (Taylor et al., 2004). Using this resource, we recently analysed the distribution of pRBC in the peripheral blood, five sites in the brain and seven other organs by genotyping the merozoite surface protein 1 and 2 (msp1/2) alleles, a commonly used technique for identifying genetically distinct P. falciparum types (Snounou et al., 1999; Farnert et al., 2001). Types amplified from the peripheral blood tended to be detected throughout the body but infections in the organs were more complex than in the peripheral blood. We compared infections in fatal cerebral malaria (CM) patients with parasitaemic children who had non-malarial causes of death (Montgomery et al., 2006). Relative to parasitaemic controls, CM patients had less complex infections, and genetic types were distributed more homogenously throughout the organs. msp type was not associated with the site of sequestration.
We have now examined var gene expression by P. falciparum parasites in the brain, lung, heart and spleen of fatal paediatric malaria patients. Because of the extremely complex nature of the expression of this gene family, a small number of patients were studied in detail. We found dominant expression of particular var genes within a tissue population, and that the dominant form varied between organs. Some of these dominant var types were detected in the same organs of other patients from the same malaria season. This finding provides preliminary evidence that the repertoire of var genes mediating organ-specific sequestration within a season may be limited.
Results and discussion
Pilot study
We analysed the diversity of var transcripts expressed by parasites in the organs of six cases of fatal falciparum malaria. RNA was extracted from the brain, lung, heart and spleen, cDNA was synthesized and DBL1α sequence amplified, cloned and sequenced. The samples consisted primarily of human genetic material and, for this reason, a single polymerase chain reaction (PCR) did not consistently amplify P. falciparum nucleic acid and so a previously described nested PCR was utilized (Duffy et al., 2002). These primers were previously optimized for minimal bias towards individual DBL1α sequences and bias was calculated in that study at less than 2.5% (Duffy et al., 2002). var sequences were considered identical when 99–100% similar, which corresponds to a maximum of 4 bp changes over the 340–450 bp sequence.
Twenty var clones were sequenced from each organ. There was a high degree of diversity, with up to 14 different var sequences identified from a single organ. Cloning was repeated from the tissues of one patient and the frequencies of individual var types were compared between the two reactions. While mainly the same transcripts were detected in both reactions, the proportions in each organ differed widely. We concluded that the number of clones examined was inadequate to study the observed level of diversity in var gene expression. The study design was altered to examine a larger number of transcripts in a subset of these patients to ensure a comprehensive analysis and to reduce potential PCR bias.
Complex var expression in the organs of fatal malaria patients
Three cases of fatal paediatric malaria were chosen for the present study: PM30, who died of severe malarial anaemia (SMA) in the absence of coma; PM32, with a diagnosis of CM and SMA; and PM55, with CM alone (Table 1). DBL1α sequences were amplified from the brain, heart, lung and spleen of the three patients in two separate PCR reactions for each sample. The first three organs were chosen as they are major sites of P. falciparum sequestration. Unfortunately, peripheral blood samples were not collected from these patients at the time of autopsy. We chose to also analyse the spleen, in which pRBC are eliminated from the circulation or ‘pitted’ (removal of parasite without erythrocyte destruction; Angus et al., 1997; Chotivanich et al., 2002). pRBC found within the spleen may therefore be representative of the circulating population.
Table 1.
Clinical details of patients.
| Diagnosis | Age (months) | Time to death (h : min) | Admission parasitaemia (parasites μl−1) | Final parasitaemia (parasites μl−1) | Date of admission | |
|---|---|---|---|---|---|---|
| PM30 | SMA | 7 | 01:40 | 302 400 | 302 400 | February 1999 |
| PM32 | CM + SMA | 18 | 02:40 | 572 880 | 572 880 | March 1999 |
| PM55 | CM | 52 | 23:00 | 286 650 | 97 306 | March 2001 |
| Additional patients from pilot study | ||||||
| PM36 | CM + SMA | 21 | 11:00 | 11 399 | 6350 | April 1999 |
| PM39 | CM | 18 | 17:40 | 6 030 | 100 | May 1999 |
| PM78 | CM | 15 | 02:00 | 637 000 | 637 000 | May 2003 |
Ninety-six products were cloned from each reaction, sequenced and aligned, with comparable frequencies of individual var types observed in the duplicate reactions. Sequences from the pilot study were also included in analyses. A total of 133–202 clones were obtained from each organ sample with 2020 clones overall. Hereafter, ‘clone’ will refer to each sequenced RT-PCR product, and ‘type’ will refer to each different DBL1α sequence identified. A median of 26 (range 11–49) different var types were amplified from each organ of the three patients. Figure 1 illustrates the cloning frequency of individual var types and the overlap between organs and patients. Half of the 248 var types were detected a single time in one organ only, accounting for only 6% of clones examined. All other var types were cloned more than once from a single organ or were detected in multiple organs and/or cases. The single copy clones were grouped together in Fig. 1 (1× var) and excluded from motif analysis.
Fig. 1.
Frequency of var types in the organs of paediatric malaria patients. Each graph represents one patient (labelled above with final diagnosis in parentheses) with organs denoted by shading. var types are listed on the y-axis by accession number and the frequency of cloning is adjusted for the number of complete sequences analysed from each organ. Single copy var types are represented together and labelled 1× var.
The distribution of mature pRBC and P. falciparum msp1/2 genetic types has been previously described (Montgomery et al., 2006). There was no correlation between either of these factors and the diversity of var types detected in the organs of these patients. The homogeneous distribution of msp genetic types throughout the organs of CM patients is in contrast to the current findings, where up to 102 different var types are expressed by P. falciparum parasites in a single patient but only one to two types are expressed at high levels in brain and heart microvasculature.
The infections consisted of both mature and ring stage pRBC; however, immature asexual parasites have been shown to transcribe the same dominant var transcript as the mature stages that express PfEMP1 protein (Peters et al., 2002). Mature pRBC were not observed in the lungs, heart and spleen of PM55, although they were present at high number in the brain (Montgomery et al., 2006). Dense accumulations of pigment in these organs provided evidence of recent sequestered populations. However, this child had a high circulating parasitaemia at the time of death and the var expression observed here is assumed to be from immature stages transiently present in organs other than the brain, where their expression would be overwhelmed by the sequestered population.
A subset of var types are expressed at high levels in the brain and heart
The number of var types amplified in each organ varied, with less diversity in the brain and heart (6.7 and 5.0 types/genotype respectively) than in the lung (8.4 types/genotype) and spleen (9.2 types/genotype). In most samples, one to two var types were detected at far higher frequency than other types in the same organ (Fig. 1 and Fig. S1). This was particularly obvious in all three brain samples, where the dominant types made up a third to a half of all clones detected in this tissue but were present in other organs only at low levels. These findings suggest organ-specific sequestration of particular var types and support the hypothesis that PfEMP1 type determines the site of cytoadherence.
Many of the less abundant var clones were detected in multiple organs within a patient; between 16% and 65% (median 47%) of var types detected in one tissue were detected in one or more other organs of the same patient. When the frequency at which each clone was detected was considered, the median overlap in detection of var types shifted to 77% (range 37–93%). There were no clones detected in all four organs of PM30. Four to six per cent of the var types amplified in PM32 and PM55 were detected in all organs, or up to 40% of clones. These types may represent pRBC sequestered in multiple organs, or var expression by circulating forms.
Major overlap in expression of var types between the lung and spleen was common in all three patients, accounting for 90% of lung and 80% of spleen clones in PM55, presumably mainly expressed by non-sequestered parasites. In PM32, 58% of brain var types were also detected in the lung, which accounted for 90% of all clones detected in the brain but only 57% of those from the lung. Despite these overlaps, the dominance in expression of particular var types in the brain and other organs strongly suggests that particular var/PfEMP1 types mediate sequestration in these tissues.
Organ-specific var expression is observed in multiple patients from a single malaria season
We investigated if any var types were shared between the three patients. This was not expected as previous studies have shown minimal overlap in the expressed var repertoire between patients (Kaestli et al., 2004; Bull et al., 2005; Kyriacou et al., 2006). There was only one var type shared between PM55 and PM32, detected at two copies in the heart of PM55 and in all four organs of PM32. There were no var types shared between PM30 and PM55. Surprisingly, there was substantial overlap in the var types detected in PM30 and PM32, with 20 DBLα/var types detected in both cases (Fig. 1). These sequences were identical between the two patients. It is important to note that shared DBLα sequence does not necessarily imply that that entire var genes represented by each tag are identical. However, we will continue to refer to these as ‘var types’ for continuity.
The shared var types accounted for 20% of PM30 var types and 26% of PM32 var types, compromising 61% and 32% of all clones detected in each patient respectively. This overlap was particularly striking in the brain, with 26% of PM30 brain var types (90% of clones) also detected in the brain of PM32, including all of the dominant types in both cases. There was also major overlap in the heart of both patients, accounting for 42% of all PM30 var types in the heart (89% of clones) but only 14% of PM32 heart var types (24% of clones).
Why there should be such overlap in the var types expressed by parasites in these two infections is intriguing. PM30 and PM32 were hospitalized a month apart and were from widely separated villages, whereas PM55 was admitted to the study 2 years later. Data from the pilot study showed that two of the most highly transcribed var types in the brain of PM32, DQ519140 and DQ519141, also expressed in PM30, were additionally detected in the brain of a third child from the same season, PM36. This child was from the same district as PM30 but was admitted to the ward 2 months later (Table 1). None of these infections were comprised of similar msp1 and 2 genetic types (Montgomery et al., 2006). var expression from the infection of PM39, a fourth child from this season who was admitted 6 weeks subsequent to PM36, did not display any overlap in var types detected in the brains of the other cases. However, 15% of var types from the additional three cases in the pilot study were also detected in other patients, including each other and the three patients from the main study. The distribution of these shared types did not exhibit any tissue tropism.
As striking as the overlap in var types between patients is the fact that the organ localization is conserved. The potential of PfEMP1 as a vaccine candidate has been questionable due to its antigenically variant nature, although there is evidence that the var repertoire may be more restricted than originally thought (Kraemer and Smith, 2003; Bull et al., 2005). Using this molecule as a vaccine in malaria in pregnancy appears to be more promising in view of the very restricted and conserved nature of the PfEMP1 types involved in placental sequestration (Duffy et al., 2005). However, our finding that not only are a limited number of var types expressed in a dominant fashion in the brain (and heart) of malaria patients, but also that these types are shared between patients, suggests that some form of organ-specific var expression such as seen in placental malaria may also occur in severe paediatric malaria.
The patients described in this study carried genetically complex P. falciparum infections with up to seven genotypes detected in a single organ (Montgomery et al., 2006). We have shown here that these parasites expressed up to 49 var types in a single organ, and between 68 and 102 var types in an individual patient. It is possible that had we amplified more clones, we would have detected additional minor var types, although the dominance of particular types in some organs was evident (Fig. S1). That parasites can express over 100 var types in a single host, but only one to two types at high frequency in the brain, suggests that particular var genes are responsible for adherence in this organ.
It must be questioned whether the dominance of particular var types within an organ truly reflects differential expression levels, or whether this is due to biased amplification or cloning, or to contamination of circulating stages. For either of these to be true, we would expect that the same bias or contamination would be observed in all samples within a patient and therefore we would not see different var expression profiles between organ populations. There is disagreement regarding var expression in early asexual stages, with some researchers finding relaxed transcription of var types in early stages shifting to expression of a single var transcript in late asexual stages, and another study finding relaxed transcription even in late stages (Chen et al., 1998; Scherf et al., 1998; Kyes et al., 2000; Duffy et al., 2002). A recent paper demonstrated that the placental malaria-related gene, var2csa, appears to be the only var transcript expressed throughout the asexual life cycle (Schieck et al., 2007). However, as this is an atypical var gene, it is unclear how much this data can be extrapolated to the entire multigene family. Taken as a whole, the fact that the dominant var types observed in this study vary between organ populations argues strongly that they are not an artefact of bias or contamination by early asexual stages.
DBL sequence motifs are not associated with organ localization
A general phylogenetic analysis of the var sequences did not display clustering associated with their detection in any of the four host organs (J. Montgomery, unpublished). Most of our sequences contained four cysteines with only 12% of var types (excluding those detected a single time) containing only two cysteine residues, a motif previously associated with severe malarial disease in infants (Kirchgatter and del Portillo, 2002). None of the major transcripts in any organ were of the two cysteine/DBLα1 sequence group except in the spleen of PM32 (DQ519244).
The sequences were then classified according to previously identified DBLα motifs such as the number of cysteine residues and positions of limited variance (PoLV; Bull et al. 2005). We have previously shown that the distribution of PoLV groups among these Malawian var types correspond well to other populations in Africa, Asia and South America (Bull et al., 2007). We now investigated whether var types consisting of particular PoLV groups were differentially distributed between tissues.
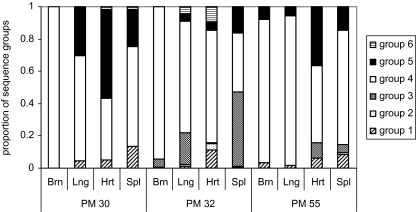
Figure 2 shows the distribution of PoLV groups in our var sequences. None of the groups shows an association with the site of sequestration, even when adjusted for the frequency of cloning (data not shown). Expression levels of PoLV group 1 sequences were found to be negatively associated with the variant antibody repertoire in P. falciparum-infected Kenyan children (Bull et al., 2005). These sequences were found at low frequency in our data; immune regulation may preclude the expression of var types containing these conserved sequences in paediatric infections in areas of high malaria transmission such as Malawi.
Fig. 2.
Frequency of sequence groups in var transcripts from the organs of paediatric malaria patients. Shading represents the sequence groups as identified by Bull et al. (2005), which are characterized by the number of cysteine residues and other semi-conserved motifs known as positions of limited variance. The data are expressed as the percentage of var types within each organ containing the corresponding sequence motifs. Brn, brain; Lng, lung; Hrt, heart; Spl, spleen.
var sequences expressed in the hearts of two Malawian patients are similar to 3D7 var genes
Many of the var types expressed by P. falciparum in heart tissue of PM30 and PM32 were highly similar or identical to 3D7 var types. All of the 11 var types detected in the heart of PM30 displayed greater than 70% identity with var sequences from the 3D7 genome, and eight were greater than 80% identical to these genes. In pRBC from the heart of PM32, seven of 25 multiple copy var types displayed high similarity to 3D7 var types, plus three of the 13 single copy var types. With one exception (DQ519236), these 3D7-similar var types were exclusively expressed in the heart of PM32. The number of 3D7 var genes to which the Malawian isolates showed similarity was limited, with 24 Malawian DBL1α sequences displaying varying levels of similarity to 14 3D7 var genes.
Conclusions
The expression of var genes in the human host is complex; half of the 248 var types were amplified a single time from a single organ, 29% were observed in multiple organs from the same patient, and 9% were detected in two patients. In the brain, at least, there is clear dominance of certain var types and the dominant types vary between organs. The data obtained in this study suggest that the PfEMP1 proteins encoded by only a small number of var genes are responsible for sequestration in brain microvasculature. An additional and intriguing finding is that there appears to be overlap between the var types expressed in the brains of children who have died of malaria within a single season. This finding suggests that the number of var types mediating sequestration in the brain may be limited, and if so, therapies capable of blocking or reversing adhesion of P. falciparum parasites in the brain may be feasible.
Experimental procedures
Clinical
Clinical details, including diagnosis and treatment, have been previously described (Montgomery et al., 2006). An initial pilot study examined four patients from the 1999 malaria season and one patient each from the 2001 and 2003 seasons. The selection of these cases was based on an autopsy-confirmed diagnosis of fatal malaria and a high peripheral parasitaemia at admission. The three patients examined in detail were chosen by high RNA yields from organ samples and their clinical details are outlined in Table 1.
This study was approved by ethics committees at the University of Malawi, Michigan State University and the University of Liverpool.
RNA extraction
Organ samples collected at autopsy were snap frozen in liquid nitrogen in frozen tissue matrix (OCT compound, Tissue-Tek) and stored at −80°C. Approximately 0.5 g of frozen material was ground in liquid nitrogen and transferred to 10× volume of Trizol (Invitrogen), prewarmed to 37°C. Insoluble material was removed by centrifugation at 12 000 g for 10 min followed by incubation at room temperature for 5 min. Extraction was then performed according to manufacturer's instructions. RNA was treated for DNA contamination using a DNA-free RNA kit (Genetix) and complete removal was tested by PCR of DBL1α sequence as described below.
cDNA synthesis, amplification and cloning
cDNA was synthesized from 2 μl of RNA using the Retroscript kit (Ambion) and quality was checked by agarose gel electrophoresis. One microlitre of cDNA initiated a primary PCR of DBL1α sequence using previously described oligonucleotides DBL-fo and DBL-ro (Duffy et al., 2002) at 1 μM final concentration and 1 mM dNTPs, 4 mM Mg2+ and 0.025 U Taq DNA polymerase (Qiagen). One microlitre of primary product was used for nested PCR with the same reaction components, oligonucleotides DBL-fi and DBL-ri and reaction conditions as described (Duffy et al., 2002). Products were purified using a QIAquick PCR purification or gel extraction kit (Qiagen) as required.
PCR products were ligated into the pGEM-T Easy vector (Promega) and transformed into Escherichia coli DH5α bacteria. Colonies were grown in liquid media and frozen in 96 well plates. Plasmid purification and DNA sequencing were performed at the Wellcome Trust Sanger Institute.
Sequence analysis
Sequences were aligned using clustalw (http://align.genome.jp). Searches for sequence identity with the P. falciparum 3D7 genome were performed using PlasmoDB (http://www.plasmodb.org). DBL motifs were analysed using a database kindly provided by Peter Bull. These sequence data have been submitted to the DDBJ/EMBL/GenBank databases under accession numbers DQ519104–DQ519354, EF640985 and EF640986.
Acknowledgments
The authors would like to thank the parents and guardians of children admitted to the study, the staff of the paediatric research ward, the Department of Paediatrics, College of Medicine, and mortuary attendants. We also wish to thank Kay Jagels, Sharon Moule and Carol Churcher in the Pathogen Sequencing Unit for performing the sequencing reactions, and Dan Milner for helpful discussions. This work was supported by The Wellcome Trust (042390 and 071376) and the US National Institutes of Health (RO1 AI34969). DNA sequencing was funded by the Wellcome Trust through its support of the Pathogen Sequencing Unit at the Wellcome Trust Sanger Institute.
Supplementary material
The following supplementary material is available for this article:
Frequency of cloning of individual var types within the organs of Malawian paediatric malaria patients PM30 (A-D), PM32 (E-H) and PM55 (I-L). var types are listed in order of decreasing frequency of cloning within each tissue and are identified by accession number. The dashed line indicates 33% frequency of cloning, the cut-off point for dominance of one to two var types.
This material is available as part of the online article from: http://www.blackwell-synergy.com/doi/abs/10.1111/j.1742-4658.2006.05837.x (This link will take you to the article abstract).
Please note: Blackwell Publishing is not responsible for the content or functionality of any supplementary materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Albrecht L, Merino EF, Hoffmann EH, Ferreira MU, de Mattos Ferreira RG, Osakabe AL, et al. Extense variant gene family repertoire overlap in Western Amazon Plasmodium falciparum isolates. Mol Biochem Parasitol. 2006;150:157–165. doi: 10.1016/j.molbiopara.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Angus BJ, Chotivanich K, Udomsangpetch R, White NJ. In vivo removal of malaria parasites from red blood cells without their destruction in acute falciparum malaria. Blood. 1997;90:2037–2040. [PubMed] [Google Scholar]
- Barry AE, Leliwa-Sytek A, Tavul L, Imrie H, Migot-Nabias F, Brown SM, et al. Population genomics of the immune evasion (var) genes of Plasmodium falciparum. PLoS Pathog. 2007;3:e34. doi: 10.1371/journal.ppat.0030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baruch DI, Pasloske BL, Singh HB, Bi X, Ma XC, Feldman M, et al. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- Buffet PA, Gamain B, Scheidig C, Baruch D, Smith JD, Hernandez-Rivas R, et al. Plasmodium falciparum domain mediating adhesion to chondroitin sulfate A: a receptor for human placental infection. Proc Natl Acad Sci USA. 1999;96:12743–12748. doi: 10.1073/pnas.96.22.12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull PC, Berriman M, Kyes S, Quail MA, Hall N, Kortok MM, et al. Plasmodium falciparum variant surface antigen expression patterns during malaria. PLoS Pathog. 2005;1:e26. doi: 10.1371/journal.ppat.0010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull PC, Kyes S, Buckee CO, Montgomery J, Kortok MM, Newbold CI, Marsh K. An approach to classifying sequence tags sampled from Plasmodium falciparum var genes. Mol Biochem Parasitol. 2007 doi: 10.1016/j.molbiopara.2007.03.011. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Fernandez V, Sundstrom A, Schlichtherle M, Datta S, Hagblom P, Wahlgren M. Developmental selection of var gene expression in Plasmodium falciparum. Nature. 1998;394:392–395. doi: 10.1038/28660. [DOI] [PubMed] [Google Scholar]
- Chotivanich K, Udomsangpetch R, McGready R, Proux S, Newton P, Pukrittayakamee S, et al. Central role of the spleen in malaria parasite clearance. J Infect Dis. 2002;185:1538–1541. doi: 10.1086/340213. [DOI] [PubMed] [Google Scholar]
- Duffy MF, Brown GV, Basuki W, Krejany EO, Noviyanti R, Cowman AF, Reeder JC. Transcription of multiple var genes by individual, trophozoite-stage Plasmodium falciparum cells expressing a chondroitin sulphate A binding phenotype. Mol Microbiol. 2002;43:1285–1293. doi: 10.1046/j.1365-2958.2002.02822.x. [DOI] [PubMed] [Google Scholar]
- Duffy MF, Byrne TJ, Elliott SR, Wilson DW, Rogerson SJ, Beeson JG, et al. Broad analysis reveals a consistent pattern of var gene transcription in Plasmodium falciparum repeatedly selected for a defined adhesion phenotype. Mol Microbiol. 2005;56:774–788. doi: 10.1111/j.1365-2958.2005.04577.x. [DOI] [PubMed] [Google Scholar]
- Farnert A, Arez AP, Babiker HA, Beck HP, Benito A, Bjorkman A, et al. Genotyping of Plasmodium falciparum infections by PCR: a comparative multicentre study. Trans R Soc Trop Med Hyg. 2001;95:225–232. doi: 10.1016/s0035-9203(01)90175-0. [DOI] [PubMed] [Google Scholar]
- Jensen AT, Magistrado P, Sharp S, Joergensen L, Lavstsen T, Chiucchiuini A, et al. Plasmodium falciparum associated with severe childhood malaria preferentially expresses PfEMP1 encoded by group A var genes. J Exp Med. 2004;199:1179–1190. doi: 10.1084/jem.20040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaestli M, Cortes A, Lagog M, Ott M, Beck HP. Longitudinal assessment of Plasmodium falciparum var gene transcription in naturally infected asymptomatic children in Papua New Guinea. J Infect Dis. 2004;189:1942–1951. doi: 10.1086/383250. [DOI] [PubMed] [Google Scholar]
- Kaestli M, Cockburn IA, Cortes A, Baea K, Rowe JA, Beck HP. Virulence of malaria is associated with differential expression of Plasmodium falciparum var gene subgroups in a case-control study. J Infect Dis. 2006;193:1567–1574. doi: 10.1086/503776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchgatter K, del Portillo HA. Association of severe noncerebral Plasmodium falciparum malaria in Brazil with expressed PfEMP1 DBL1 alpha sequences lacking cysteine residues. Mol Med. 2002;8:16–23. [PMC free article] [PubMed] [Google Scholar]
- Kraemer SM, Smith JD. Evidence for the importance of genetic structuring to the structural and functional specialization of the Plasmodium falciparum var gene family. Mol Microbiol. 2003;50:1527–1538. doi: 10.1046/j.1365-2958.2003.03814.x. [DOI] [PubMed] [Google Scholar]
- Kraemer SM, Kyes SA, Aggarwal G, Springer AL, Nelson SO, Christodoulou Z, et al. Patterns of gene recombination shape var gene repertoires in Plasmodium falciparum: comparisons of geographically diverse isolates. BMC Genomics. 2007;8:45. doi: 10.1186/1471-2164-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyes S, Taylor H, Craig A, Marsh K, Newbold C. Genomic representation of var gene sequences in Plasmodium falciparum field isolates from different geographic regions. Mol Biochem Parasitol. 1997;87:235–238. doi: 10.1016/s0166-6851(97)00071-6. [DOI] [PubMed] [Google Scholar]
- Kyes S, Pinches R, Newbold C. A simple RNA analysis method shows var and rif multigene family expression patterns in Plasmodium falciparum. Mol Biochem Parasitol. 2000;105:311–315. doi: 10.1016/s0166-6851(99)00193-0. [DOI] [PubMed] [Google Scholar]
- Kyriacou HM, Stone GN, Challis RJ, Raza A, Lyke KE, Thera MA, et al. Differential var gene transcription in Plasmodium falciparum isolates from patients with cerebral malaria compared to hyperparasitaemia. Mol Biochem Parasitol. 2006;150:211–218. doi: 10.1016/j.molbiopara.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavstsen T, Magistrado P, Hermsen CC, Salanti A, Jensen AT, Sauerwein R, et al. Expression of Plasmodium falciparum erythrocyte membrane protein 1 in experimentally infected humans. Malar J. 2005;4:21. doi: 10.1186/1475-2875-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery J, Milner DA, Tse MT, Njobvu A, Kayira K, Dzamalala CP, et al. Genetic analysis of circulating and sequestered populations of Plasmodium falciparum in fatal paediatric malaria. J Infect Dis. 2006;194:115–122. doi: 10.1086/504689. [DOI] [PubMed] [Google Scholar]
- Peters J, Fowler E, Gatton M, Chen N, Saul A, Cheng Q. High diversity and rapid changeover of expressed var genes during the acute phase of Plasmodium falciparum infections in human volunteers. Proc Natl Acad Sci USA. 2002;99:10689–10694. doi: 10.1073/pnas.162349899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson BA, Welch TL, Smith JD. Widespread functional specialization of Plasmodium falciparum erythrocyte membrane protein 1 family members to bind CD36 analysed across a parasite genome. Mol Microbiol. 2003;47:1265–1278. doi: 10.1046/j.1365-2958.2003.03378.x. [DOI] [PubMed] [Google Scholar]
- Rottmann M, Lavstsen T, Mugasa JP, Kaestli M, Jensen AT, Muller D, et al. Differential expression of var gene groups is associated with morbidity caused by Plasmodium falciparum infection in Tanzanian children. Infect Immun. 2006;74:3904–3911. doi: 10.1128/IAI.02073-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe JA, Moulds JM, Newbold CI, Miller LH. P. falciparum rosetting mediated by a parasite-variant erythrocyte membrane protein and complement-receptor 1. Nature. 1997;388:292–295. doi: 10.1038/40888. [DOI] [PubMed] [Google Scholar]
- Russell C, Mercereau-Puijalon O, Le Scanf C, Steward M, Arnot DE. Further definition of PfEMP-1 DBL-1alpha domains mediating rosetting adhesion of Plasmodium falciparum. Mol Biochem Parasitol. 2005;144:109–113. doi: 10.1016/j.molbiopara.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Salanti A, Jensen AT, Zornig HD, Staalsoe T, Joergensen L, Nielsen MA, et al. A sub-family of common and highly conserved Plasmodium falciparum var genes. Mol Biochem Parasitol. 2002;122:111–115. doi: 10.1016/s0166-6851(02)00080-4. [DOI] [PubMed] [Google Scholar]
- Scherf A, Hernandez-Rivas R, Buffet P, Bottius E, Benatar C, Pouvelle B, et al. Antigenic variation in malaria: in situ switching, relaxed and mutually exclusive transcription of var genes during intra-erythrocytic development in Plasmodium falciparum. EMBO J. 1998;17:5418–5426. doi: 10.1093/emboj/17.18.5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieck E, Pfahler JM, Sanchez CP, Lanzer M. Nuclear run-on analysis of var gene expression in Plasmodium falciparum. Mol Biochem Parasitol. 2007;153:207–212. doi: 10.1016/j.molbiopara.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Smith JD, Subramanian G, Gamain B, Baruch DI, Miller LH. Classification of adhesive domains in the Plasmodium falciparum erythrocyte membrane protein 1 family. Mol Biochem Parasitol. 2000;110:293–310. doi: 10.1016/s0166-6851(00)00279-6. [DOI] [PubMed] [Google Scholar]
- Snounou G, Zhu X, Siripoon N, Jarra W, Thaithong S, Brown KN, Viriyakosol S. Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans R Soc Trop Med Hyg. 1999;93:369–374. doi: 10.1016/s0035-9203(99)90120-7. [DOI] [PubMed] [Google Scholar]
- Taylor TE, Fu WJ, Carr RA, Whitten RO, Mueller JS, Fosiko NG, et al. Differentiating the pathologies of cerebral malaria by postmortem parasite counts. Nat Med. 2004;10:143–145. doi: 10.1038/nm986. [DOI] [PubMed] [Google Scholar]
- Trimnell AR, Kraemer SM, Mukherjee S, Phippard DJ, Janes JH, Flamoe E, et al. Global genetic diversity and evolution of var genes associated with placental and severe childhood malaria. Mol Biochem Parasitol. 2006;148:169–180. doi: 10.1016/j.molbiopara.2006.03.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Frequency of cloning of individual var types within the organs of Malawian paediatric malaria patients PM30 (A-D), PM32 (E-H) and PM55 (I-L). var types are listed in order of decreasing frequency of cloning within each tissue and are identified by accession number. The dashed line indicates 33% frequency of cloning, the cut-off point for dominance of one to two var types.