Abstract
An application of flow cytometric sorting is used for isolation of Saccharomyces cerevisiae mutants that mislocalize vacuolar vital dyes. This screen is based on the ability of a lipophilic styryl compound, N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino)phenyl)hexatrienyl)pyridinium dibromide (FM4–64), to label endocytic intermediates from the plasma membrane to the vacuole membrane at 15°C. Cells stained at 15°C for both FM4–64 and carboxydichlorofluorescein diacetate (a vacuolar luminal vital stain), had a pronounced shift in red/green fluorescence from cells stained at 30° or 38°C. Flow cytometric selection based on this characteristic shift allowed the isolation of 16 mutants. These comprised 12 complementation groups, which we have designated SVL for styryl dye vacuolar localization. These groups were put into three classes. Class I mutants contain very large vacuoles; class II mutants have very fragmented vacuoles; and class III mutants show the strongest svl phenotype with punctate/diffuse FM4–64 staining. Limited genetic overlap was observed with previously isolated mutants, namely svl2/vps41, svl6/vps16, and svl7/fab1. The remaining svl mutants appear to represent novel genes, two of which showed temperature-sensitive vacuole staining morphology. Another mutant, svl8, displayed defects in uptake and sorting of phosphatidylcholine and phosphatidylethanolamine. Our flow cytometric strategy may be useful for isolation of other mutants where mislocalization of fluorescent compounds can be detected.
The composition and function of organelles in eukaryotic cells often imposes limitations to subcellular structure. Organelles like the nucleus, mitochondrion, or lysosomes have such diverse roles that their relative spatial distribution within the cytoplasm is critical for normal cell physiology. This arrangement is most important for those cellular functions where vesicle-mediated transport needs to perpetually shuttle membrane-bound cargo between organelles. These mechanisms are a hallmark of both the secretory and endocytic pathways, initially elucidated by using electron microscopy in fixed cells and tissues (1).
Vital staining with fluorescent probes reveals subcellular dynamics and spatial relationships of organelles that cannot be observed in fixed cells. A wide variety of cell-permeant compounds specifically stain such structures as mitochondria, lysosomes, endoplasmic reticulum, and the Golgi complex in live cells (2). A relatively new vital dye, N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino)phenyl)hexatrienyl)pyridinium dibromide (FM4–64), has been used to study the lysosome-like vacuole in the yeast Saccharomyces cerevisiae (3). This aminostyryl pyridinium compound ultimately localizes to the vacuolar membrane but also interacts with the plasma membrane and cytoplasmic intermediates via constitutive endocytic mechanisms. Recently, a flow sorting-based screen was employed for the isolation of two yeast mutants dim1 and dim2 with defective endocytic uptake of FM4–64 (4).
In this report, we describe an innovative flow cytometric approach to isolate new yeast mutants based on morphology differences resulting from mislocalization of intracellular FM4–64. To isolate cells with this rare morphological characteristic required the ability to detect differences in intracellular location among populations with insignificant changes in overall fluorescence intensity. By using a double fluor strategy we uncovered 12 complementation groups, which are designated SVL for styryl dye vacuole localization. The intracellular mislocalization of two fluorescent compounds with different spectral emission not only revealed new genes, but also should be applicable to a wide variety of cell types.
MATERIALS AND METHODS
Media, Strains, and Mutant Screening.
All yeast strains were maintained on YPD media (1% yeast extract/2% peptone/2% dextrose/2.5% bactoagar). The yeast strains used in this study include: SEY6210 (5); SEY6210.1 (isogenic to SEY6210 except Mata); TVY6212 (diploid from crossing SEY6210 × SEY6210.1). All svl mutant alleles were derived from either SEY6210 or SEY6210.1. Mutagenesis with ethyl methanesulfonate (Aldrich) was carried out as described (6). After flow sorting (see below), cells were grown on yeast extract/peptone/dextrose (YPD) plates (1–3 × 103 per plate) at 25°C for 2–3 days and then replicated onto two new plates for growth at 25° and 38°C. After 16 h, the plates were digitized on a flatbed scanner, the 25°C image was inverted, and overlaid on the 38°C image. Colonies that were temperature sensitive (ts) for growth appeared dark gray with this procedure. The ts colonies were streaked for single colonies at 25°C and the ts growth phenotype was reconfirmed in single ≈1-cm streaks. Each ts mutant was then inoculated into 50 μl of YPD in 96-well microtiter plates, grown overnight at room temperature, shifted to 38°C for 2 h, stained with FM4–64 (1 μM) for 1h, and examined with a epifluorescence microscope (see below).
Vital Staining and Radiolabeling.
Staining with CDCFDA (Molecular Probes) and quinacrine (Sigma) was performed as described (7) except for the use of an increased loading concentration (50 μM) and incubation time (30 min) at 15°C for carboxydichlorofluorescein diacetate (CDCFDA). At elevated temperatures (30° and 38°C) FM4–64 (Molecular Probes) was loaded into cells at 10–40 μM for 15 min followed with one wash to remove free dye and a chase period for 45–120 min. When preparing reference samples for flow cytometry, all cells were harvested, resuspended in 20 mM Hepes⋅KOH (pH 7.0), 150 mM potassium acetate, and 5 mM magnesium acetate, and held on ice. To stain with lipids, all strains were grown in YPD to ≈0.5 OD600, 0.1 OD cells were harvested and washed once with YPD. Cells were then resuspended in 90 μl YPD and stained with a 10 μl mixture (1:1) of 0.4 mM BODIPY-labeled phospholipid, 2-(4, 4-difluoro-5-methyl-4-bora-3a,4a-diaza-s-indacene-3-dodecanoyl)-1-hexadecanoyl-sn-glycero-3-phosphocholine (β-BODIPY 500/510 C12–HPC) or 2-(4,4-difluoro-5,7-dimethyl- 4-bora-3a,4a-diaza-s-indacene-3-dodecanoyl)-1-hexadecanoyl-sn-glycero-3-phosphoethanolamine (β-BODIPY FL C12–HPE) (Molecular Probes) and unlabeled phospholipid (Sigma) in YPD. After 1 h incubation at 30°C, cells were washed twice with YP and examined by epifluorescence microscopy. Turnover of Ste3p was examined as described (8) at 25°C with a 10 min pulse, 5 min chase (0 min) and 10, 20, and 60 min chase time points thereafter.
Flow Cytometry.
Samples were analyzed on a Coulter EPICS Model 753 equipped with the Cyclops High-Speed Sort System (Cytomation, Ft. Collins, CO). The argon ion laser (Coherent, Palo Alto, CA) was tuned to 514 nm, operated at 100 mW, traveled through a confocal lens block creating a beam height of 16 μm, and then through a 3× quartz SortSense flow cell tip. Detection sensitivity on the EPICS 753 was modified as described previously (9). Emitted light was collected through a series of optical filters including a 514-nm longpass laser blocking filter, a 550-nm longpass dichroic filter to reflect the FM4–64 emission to a 590-nm longpass filter and divert the CDCFDA signal to a 525 nm bandpass emission filter. To compensate for fluorescence overlap between the two fluors, 65% of CDCFDA signal was subtracted from the FM4–64 signal and 25% of the FM4–64 signal was subtracted from the CDCFDA signal. This compensation was always monitored by using reference samples of cells stained separately with the appropriate dyes for each experiment. Cell samples were suspended in 20 mM Hepes⋅KOH (pH 7.0), 150 mM potassium acetate, and 5 mM magnesium acetate to a concentration of 1 × 106 to 9 × 106 cells/ml. Flow rates were typically 1,000–1,500 cells/s. Forward-angle light scatter was collected as a linear signal, and all fluorescence emissions were collected on a four-decade logarithmic scale.
Fluorescence Microscopy.
Cells grown in suspension were concentrated to ≈20 OD600 units/ml in YPD and applied to slides coated with ConA (Sigma). All micrographs were digitally captured on a Zeiss Axioskop epifluorescence microscope by using an Optronics (Goleta, CA) DEI-750 color CCD camera with Adobe premiere software (Adobe Systems, Mountain View, CA), a TARGA 2000 video board (Truevision, Santa Clara, CA), and a PowerPC Macintosh 9500 (Apple Computer). All images were edited with Adobe Systems photoshop software. Quantitation was performed with National Institutes of Health’s image software on inverted grayscale images from the same exposure and background subtraction settings.
RESULTS
Cellular Energy Is Required to Transport FM4–64 between Endocytic Intermediates and the Vacuole Membrane.
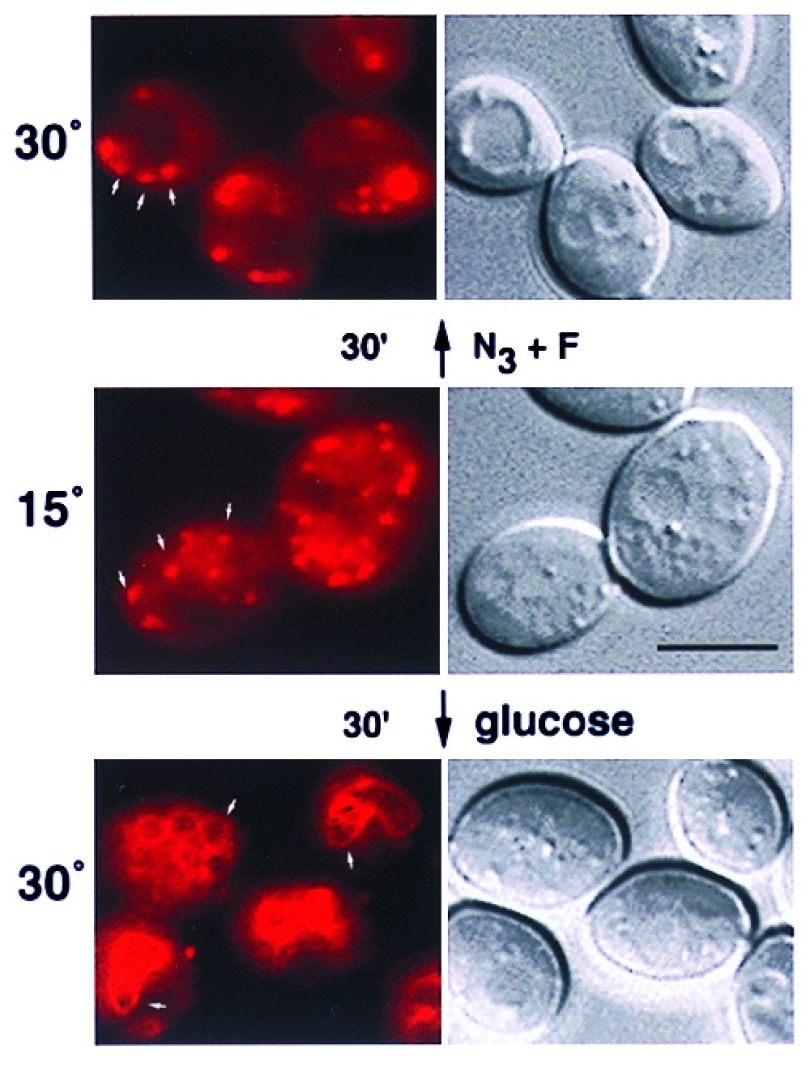
As a vital dye, FM4–64 is useful for examining vacuolar membrane dynamics and bulk-phase endocytosis (3). One property of FM4–64 is its ability to be trapped in punctate cytoplasmic structures at reduced temperatures, which suggests that it accumulates in endocytic intermediates. To characterize these putative intermediates, we determined whether cellular energy was required for their disappearance. After incubating diploid yeast cells at 15°C for 30 min, they were stained with FM4–64 for a continuous pulse of 30 min. Approximately 10–15 distinct spots of red fluorescence existed in the cytoplasm of the cells (Fig. 1), which has been observed previously (3). To examine if energy was required to localize FM4–64 from the cytoplasmic punctate structures to the vacuolar membrane, the cells were washed extensively to remove free dye and then incubated at 30°C with or without energy poisons. In the absence of NaN3 and NaF, FM4–64 staining was confined to the vacuolar membrane after 30 min (Fig. 1). In contrast, the presence of NaN3 and NaF blocked the vacuolar membrane staining of FM4–64 and it remained punctate during the 30-min incubation (Fig. 1). Although not shown, the punctate staining gave way to vacuolar membrane staining (over an additional 30 min) when the energy poisons were washed away and the cells were supplied with fresh glucose. This indicated that the energy block was not simply due to cell death. These results indicated that endosomal intermediates stained with FM4–64 at 15°C required energy-dependent cellular processes to transport membrane material to the vacuole.
Figure 1.
Cellular energy is required for localization of FM4–64 to the vacuolar membrane from endosomal intermediates. Diploid yeast cells were stained with FM4–64 at 15°C for 30 min in rich media containing glucose (2%). The cells were placed on ice, washed twice with rich media (no glucose), and then incubated at 30°C for 30 min with glucose (2%) or NaN3 and NaF (10 mM each), as indicated. The cells were then viewed with epifluorescence microscopy by using a Texas red filter set and DIC optics. Arrows point to punctate intermediates and vacuole membrane staining. (Bar = 5 μm.)
A Genetic Screen for Isolation of Mutants That Accumulate FM4–64 in Endosomal Intermediates.
To further understand the molecular mechanism of FM4–64 endocytosis, we sought ways to isolate yeast mutants that accumulated this vital dye specifically in the endosomal intermediates. A strategy using flow cytometry (FCM) was developed because of its high throughput screening and cell sorting capabilities. The general basis for previous screens using FCM is sorting and collecting cells with reduced fluorescence of FM4–64, a relatively simple task (4, 10). However, conventional FCM will not detect differences based solely on intracellular location of a single fluorescence signal (see Fig. 2 D and E), which was needed to sort cells with punctate FM4–64 fluorescence from cells with vacuolar membrane fluorescence. We reasoned that a second fluor, with a uniform cellular location but different excitation/emission spectra than FM4–64 (515/640 nm), might allow flow cytometric discrimination if the emission was simultaneously monitored at two wavelengths. CDCFDA is a standard vital stain for the luminal space of the yeast vacuole with excitation and emission wavelengths of 480 and 550 nm, respectively (7). This dye is membrane permeable until nonspecific esterases hydrolyse the acetate groups to form more highly charged, less membrane-permeable compounds (11). Moreover, unlike FM4–64, CDCFDA localization is temperature and energy-independent.
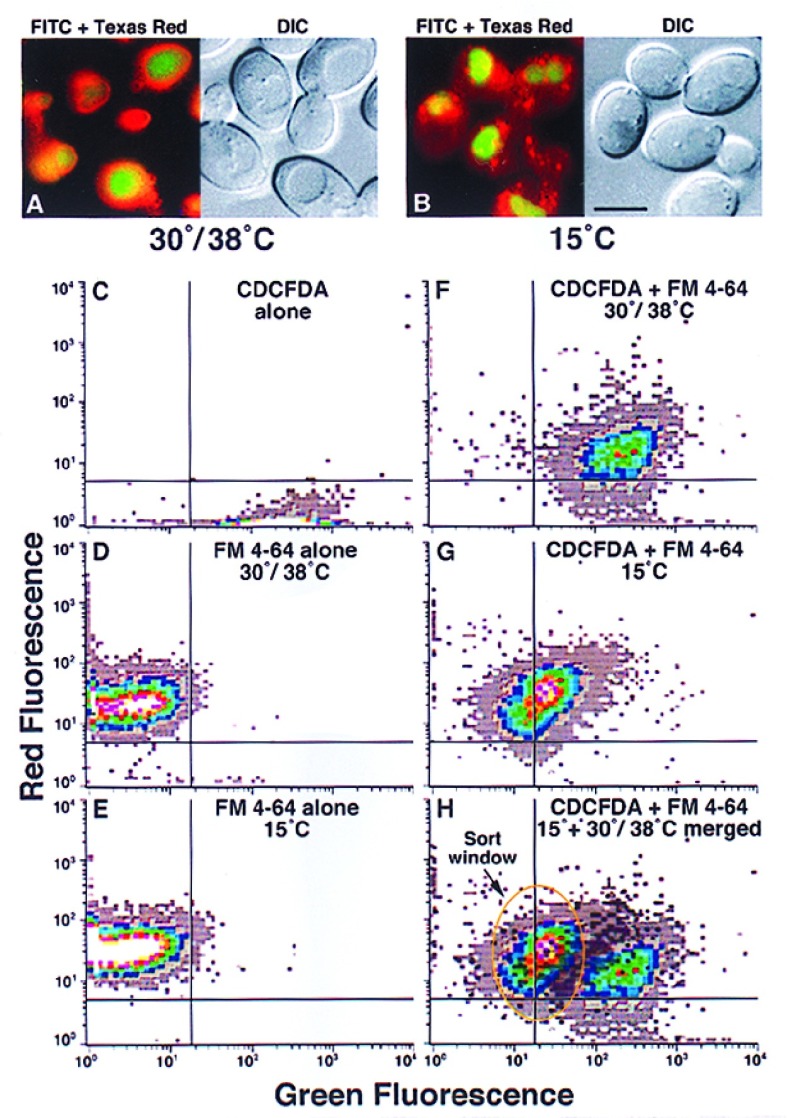
Figure 2.
Flow cytometry can distinguish cells with punctate FM4–64 staining morphology from cells with vacuolar membrane staining morphology. Diploid yeast cells were stained with FM4–64 (10 μM) and CDCFDA (10 μM) at the indicated temperatures. At 30°C/38°C (A), the cells were pulsed with FM4–64 for 30 min and chased for 60 min. After 60 min, CDCFDA was added (at pH 4) for 15 min. At 15°C (B), the cells were simultaneously pulsed with both FM4–64 and CDCFDA (50 μM) for 30 min. For flow cytometry, haploid yeast cells (SEY6210) were stained with CDCFDA alone (C), FM4–64 alone at 30°C (D), and 15°C (E), or stained with both CDCFDA and FM4–64 at 38°C (F) or 15°C (G). The fluorescence emission was simultaneously monitored at 525 (green) and 590 (red) nm. After overlaying the patterns from double-staining cells at 38° and 15°C, a sort window was defined for putative mutants. Each flow cytometry sample represents ≈2–5 × 104 cells. The white and light gray areas represent the most and least events, respectively.
When yeast cells were double-stained with CDCFDA and FM4–64 at 30° or 38°C they had green vacuole luminal fluorescence surrounded by red vacuole membrane fluorescence (Fig. 2A). By contrast, double-staining at 15°C resulted in green vacuole luminal fluorescence but the cytoplasm had red punctate endosomal intermediates (Fig. 2B). These two cellular fluorescence conditions mimicked the appearance of wild-type (30°/38°C) or “mutant” (15°C) yeast cells with defective transport from endosomal intermediates to the vacuolar membrane. Cells stained at these two temperatures with just FM4–64 were nearly identical. The mean fluorescence intensity (MFI) at 30°/38°C was 33 (590 nm, red) and 5 (525 nm, green) and at 15°C the MFI was 41 (590 nm) and 3 (525 nm) (Fig. 2 C and D). Double-staining with CDCFDA at 30° or 38°C increased the MFI at 525 nm to 252 but reduced the MFI at 590 nm to 19 (Fig. 2F). In contrast, double-staining cells with CDCFDA at 15°C increased the MFI at 525 nm to only 32, whereas the MFI at 590 nm was not changed, 39 (Fig. 2G). Although not shown, when cells were stained with just CDCFDA at 30°/38° or 15°C they were very similar suggesting that the punctate FM4–64 fluorescence in the cytoplasm at 15°C affected detection of vacuole luminal fluorescence. Therefore, merging the 30°/38° and 15°C data sets defined a window with which to sort putative mutant cells that accumulated FM4–64 in cytoplasmic intermediates based on the idea that the 15°C fluorescence profile generated a “mutant” phenotype (Fig. 2H).
To isolate mutants, haploid yeast cells were subjected to mutagenesis with ethylmethane sulfonate such that 30–40% remained viable. The mutagenized cells were grown for several generations at 25°C and then shifted to 38°C for 3 h to screen for ts mutations. After double-staining the cells with CDCFDA and FM4–64, they were sorted. This typically resulted in a 100–500-fold enrichment. After plating the sorted cells for viability at 25°C, they were tested for ts growth at 38°C, which was in the range of 0.3–0.5%. The final screen was examining the cells individually under the microscope after preshifting them to 38°C for 2 h and staining with FM4–64. Mutants that showed morphology different than wild-type, which at this stage in the enrichment was ≈1–3% of the ts mutants, were identified and further characterized. Therefore, the overall frequency of isolating a mutant with the appropriate defective phenotype was ≈1 × 10−7, a very large enrichment factor.
Characterization of the svl Mutants.
To begin genetic characterization of the mutants, they were first crossed back to haploid wild-type strains three consecutive times. In most cases, after meiosis the ts growth phenotype cosegregated with the defective FM4–64 morphology phenotype. This suggested that a single mutation most likely caused both phenotypes. All isolated mutants proved to be recessive to wild-type when heterozygous diploid strains were constructed and could be put into 12 complementation groups (Table 1). Because in wild-type yeast cells styryl dyes like FM4–64 localize to the vacuole, we have named these complementation groups SVL for styryl dye vacuolar localization. The svl mutants were further classified into three groups based on FM4–64 staining morphology. Class I mutants had very large vacuoles, class II mutants had fragmented vacuoles, and finally class III mutants had punctate/diffuse FM4–64 staining without vacuole fragmentation, the true svl mutant phenotype (Table 1).
Table 1.
svl mutant complementation groups
| Group | Class/allele | proCPY | proAPI | Ste3p | PC | PE |
|---|---|---|---|---|---|---|
| SVL | − | <5% | <5% | ≈25 min | 1.00 | 1.00 |
| svl2 | II/vps41 | ++++ | ++++ | >60 min | 0.81 | 0.49 |
| svl3 | I | <5% | <5% | ≈20 min | 1.60 | 0.97 |
| svl4 | I | ND | ND | ≈46 min | 1.01 | 1.35 |
| svl5 | III | ++ | ++++ | ≈24 min | 0.95 | 2.10 |
| svl6 | II/vps16 | ++++ | ++++ | >60 min | 2.36 | 2.53 |
| svl7 | I/fab1 | ND | ND | ≈28 min | 1.16 | 1.17 |
| svl8 | III | <5% | +++ | ≈58 min | 2.80* | 4.93* |
| svl9 | II | +++ | ++++ | ND | 2.69 | 1.25 |
| svl10 | I | ++++ | ++++ | >60 min | 1.17 | 1.53 |
| svl11 | I | +++ | ++++ | >60 min | 1.24 | 1.48 |
| svl12 | III | <5% | ++ | ≈15 min | 1.18 | 1.14 |
Mutant class I, large vacuoles; class II, fragmented vacuoles; and class III, punctate/diffuse structures. Alleles of previous mutants are indicated. For accumulation of proCPY and proAPI, each + sign represents 25% of the total from Western blot analysis. The turnover of Ste3p was performed as described in Materials and Methods and is reported in half times. Uptake of both phosphatidylcholine (PC) and phosphatidylethanolamine (PE) was performed using BODIPY-labeled lipids and 10–30 cells were quantitated with digital image analysis. All values were expressed relative to SVL cells arbitrarily set to 1.00.
All svl mutants showed wild-type morphology for both phosphatidylcholine and phosphatidylethanolamine except svl8, which showed accumulation at the plasma membrane (see Fig. 3B).
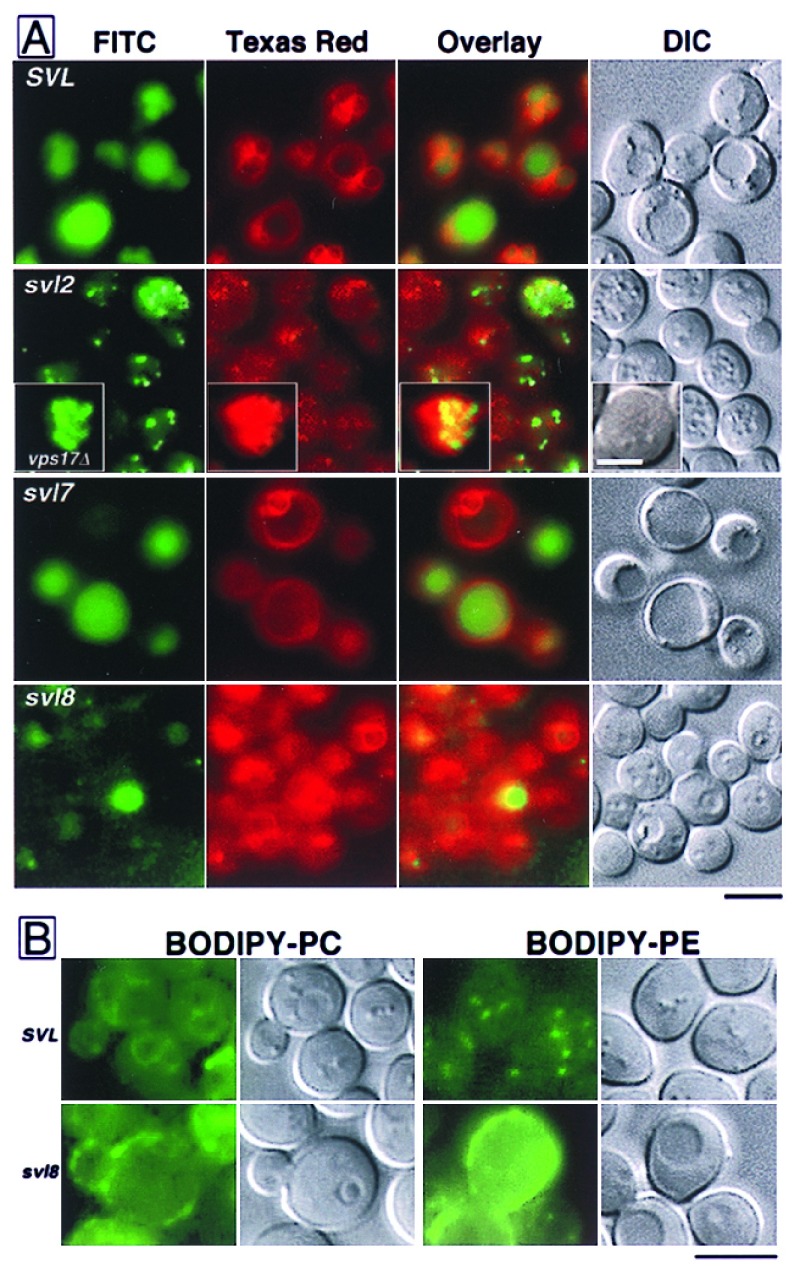
To morphologically characterize the svl mutants, they were stained with CDCFDA and FM4–64. In general, class II and class III mutants did not colocalize these dyes at the vacuole. Staining with CDCFDA allowed for discrimination of class II from class III mutants because class II mutants showed numerous small structures, suggesting fragmented vacuole morphology (Fig. 3A). The class II svl mutants (svl2, svl6, and svl9) also showed punctate/diffuse staining with FM4–64 (Fig. 3A, svl2 shown only). Interestingly, the small, CDCFDA-staining mutant vacuoles in svl2 cells were not surrounded by red ring-like staining (Fig. 3A). This was not due to resolution limits of the microscopy because another mutant with fragmented vacuoles, vps17 (12), had similar sized vacuoles but every one was bound by FM4–64 membrane fluorescence (Fig. 3A, Inset). Even without vital dye staining, DIC optics made the nonfragmented vacuole appearance of class III mutants very easily scored as near wild type, compared with the fragmented vacuole appearance in class II mutants (Fig. 3A). However, double-staining these class III mutants revealed unexpected defects in vacuole luminal distribution of CDCFDA as well as defects in vacuole membrane localization of FM4–64 (Fig. 3A). In particular, svl5 and svl8 mutant cells had significant decreases in the amount of luminal CDCFDA despite the fact that vacuoles were present via DIC optics and the membrane was weakly delineated with FM4–64 fluorescence in certain cases (Fig. 3A, svl8 shown only).
Figure 3.
Morphology of the svl mutants. (A) Yeast cells from wild-type, SVL, or from svl2, svl7, and svl8 cultures were double-stained with CDCFDA and FM4–64 at 25°C, as indicated. Images of the cells were captured with epifluorescence microscopy separately by using a fluorescein isothiocyanate (FITC) or Texas red filter set, and DIC optics, as indicated. The overlay represents a digital blending of the two separate fluorescence captures. The inset in the svl2 panel represents typical vacuole morphology in vps17Δ cells. [Bar = 5 μm (Inset = 2.5 μm).] (B) SVL and svl 8 strains were grown to early-log phase. Cells were stained with BODIPY-PC and BODIPY-PE, and examined by epifluorescence microscopy, as indicated. (Bar = 5 μm.)
To characterize any genetic overlap with previously isolated mutants, we examined the biogenesis of two vacuolar proteins, carboxypeptidase Y (CPY) and aminopeptidase I, in the svl mutants. Accumulation of both proCPY and proaminopeptidase I without detection of their mature wild-type forms was observed in Western blot analyses of extracts from svl2, svl6, and svl10 cells (Table 1). This phenotype indicates that these mutants were defective for vacuolar protein sorting and they may overlap with previously identified vps mutants (5, 13). Some mutants such as svl5, svl8, svl9, and svl12 showed significant amounts of mature CPY but were more defective for the maturation of aminopeptidase I (Table 1). This phenotype indicates that they might have overlapped with previously identified cvt mutants defective for cytosol to vacuole targeting (14). We tested these possibilities by making heterozygous svl/cvt diploid strains and all were wild type, indicating no genetic overlap (data not shown). Furthermore, we examined the FM4–64 staining patterns in cvt2, 3, 5, 7, 9, 10, 11, 12, 13, and 17 mutant cells and none displayed a svl phenotype (data not shown).
By contrast, we did find some overlap with vps mutants. Specifically, svl6 was shown to be allelic with vps16 by heterozygous diploid analysis and svl2 was shown to be allelic with vps41 (15, 16) after complementation cloning with a genomic DNA library. Moreover, the VPS41 gene alone can fully complement the svl2 FM4–64 staining defect when expressed on a centromere-linked plasmid (data not shown). The enlarged vacuole morphology of class I svl mutants was reminiscent of the fab1 mutant phenotype (17). When heterozygous diploids were made between fab1 and svl3, svl4, svl7, and svl10 all but the fab1/svl7 strain was wild type, indicating that svl7 was allelic to fab1 (Table 1). From these genetic and biochemical experiments, we concluded that, at the least, svl3, svl8, and svl12 show novel phenotypes and most likely were not isolated in previous mutant selections or screens.
We examined the turnover of an endogenous plasma membrane protein, Ste3p, in all svl mutants to determine if they showed defects in constitutive endocytosis. Pulse-chase radiolabeling revealed that the half time for Ste3p turnover in the vacuole was increased from ≈25 min (SVL cells) to >60 min in svl2, 6, 8, 10, and 11 mutants (Table 1). This most likely reflected deficiencies in vacuolar protease activity (i.e., proCPY accumulation) in svl2, 6, 10, and 11. However, svl8 was not significantly defective in CPY maturation so the increase in Ste3p turnover may suggest a defect in plasma membrane internalization. Such a defect was also suggested in svl8 mutant cells after examining the uptake and localization of phospholipids. Both 2-(4,4-difluoro-5-methyl-4-bora-3a,4a-diaza-s-indacene-3-dodecanoyl)-1-hexadecanoyl-sn-glycero-3-phosphocholine (BODIPY-PC) and 2-(4,4-difluoro-5-methyl-4-bora-3a,4a-diaza-s-indacene-3-dodecanoyl)-1-hexadecanoyl-sn-glycero-3-phosphoethanolamine (BODIPY-PE) predominately stained the cell surface in svl8 mutant cells (Fig. 3B). In contrast, BODIPY-PC stained vacuole and nuclear membranes and BODIPY-PE stained discrete spots like mitochondria-related structures and lipid particles in SVL cells (Fig. 3B). This localization of PC and PE agreed well with the lipid sorting mechanisms described previously in yeast (18). The localization of BODIPY-PC and BODIPY-PE was unaffected in all other svl mutants (data not shown). However, some of the svl mutants displayed differences in the amount of BODIPY-PC or BODIPY-PE cellular fluorescence (Table 1). This might suggest that these mutants have defects in lipid catabolism.
ts Vacuolar Vital Staining.
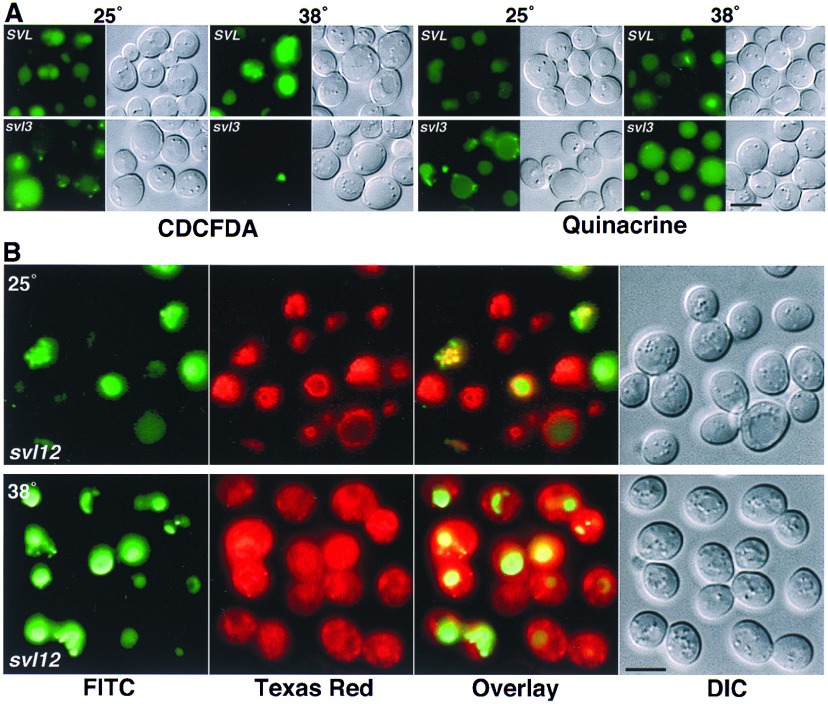
Most svl mutants were temperature insensitive for vacuolar staining morphology, appearing the same at 25° or 38°C. However, the svl3 and svl12 mutants showed near wild-type morphology at 25°C but aberrant staining at 38°C. The single allele of svl3 (a class I mutant) showed ts defects for CDCFDA staining but not FM4–64 staining. At 25°C, CDCFDA stained the enlarged vacuole lumen of svl3 to near wild-type levels (Fig. 4A). Additionally, several relatively small, intensely stained structures also existed in the svl3–1 mutant. After preshifting the cells to 38°C for 60 min, fluorescence was conspicuously absent in the vacuole but not in the small structures (Fig. 4A and Fig. 5). By contrast, vacuolar membrane localization of FM4–64 in the svl3 mutant was temperature insensitive, which appeared nearly identical at 25° (data not shown) and 38°C (see Fig. 5). The decreased CDCFDA staining in the svl3 vacuole at 38°C might have suggested a defect in organelle acidification. However, svl3–1 did not display growth inhibition at 25°C on media buffered to pH 7.5, 8.0 or 9.4, which is a common phenotype of mutants defective for vacuole acidification (19). Moreover, media buffered to pH 4, 7.5, 8.0, and 9.4 failed to rescue growth at 38°C, suggesting that a defect in pH regulation did not cause the svl3 ts growth phenotype. More significantly, quinacrine did not show ts staining in the vacuole lumen of svl3–1 cells (Fig. 4A). Quinacrine is an acidotropic dye that does not require enzymatic activity for its sequestration in low pH organelles (7).
Figure 4.
ts staining with vacuolar vital dyes in two svl mutants. (A) Yeast cells from wild-type (SVL) and the svl3–1 mutant were stained at 25°C or after a 60-min preincubation at 38°C with CDCFDA and quinacrine for 15 min, as indicated. The cells were then viewed with epifluorescence microscopy using a fluorescein isothiocyanate filter set and DIC optics. (B) Cells from the svl12–1 mutant were double-stained with CDCFDA and FM4–64 at 25°C or after a 60 min preincubation at 38°C. Images of the cells were captured with epifluorescence microscopy separately using a fluorescein isothiocyanate, or Texas red filter set, and DIC optics, as indicated. (Bar = 5 μm.)
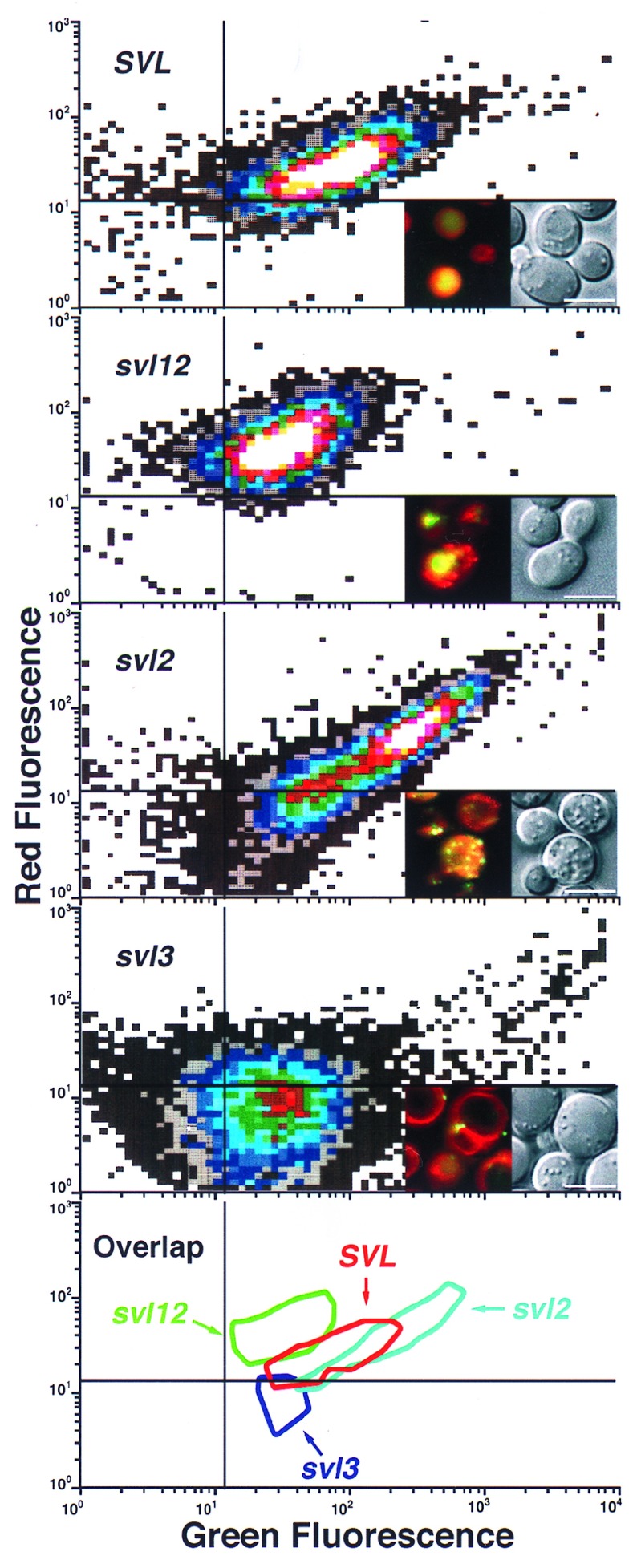
Figure 5.
Flow cytometry of three representative svl mutants. Yeast cells from wild-type, SVL, or the svl12, svl2, and svl3 mutants were double-stained with CDCFDA and FM4–64 after a 60-min preincubation at 38°C, as indicated. The cells were then subjected to flow cytometry using the same conditions as Fig. 2. The white and dark gray areas represent the most and least events, respectively. Representative cells from each sample are shown with digital blending of the two separate fluorescein isothiocyanate and Texas Red filter sets plus DIC optics. The bottom panel is an overlay representing >75% of the population from each strain. (Bar = 5 μm.)
In contrast to svl3–1, the single allele of svl12 (a class III mutant) showed ts defects in morphology for FM4–64 staining but not for CDCFDA staining. At 25°C, both dyes stained the vacuole of svl12–1 with characteristics indistinguishable from wild-type cells (Fig. 4). After preshifting the svl12–1 cells to 38°C for 60 min, FM4–64 fluorescence did not colocalize with the vacuole membrane and was diffusely distributed in the cytoplasm, which also contained well-defined punctate structure (Fig. 4). Unlike svl3–1, the vacuoles in svl12–1 cells maintained the ability to stain with CDCFDA at 38°C (Fig. 4). Of all six class III mutants, the morphology of svl12–1 after double-staining with CDCFDA and FM4–64 at 38°C (Fig. 4) most closely resembled the morphology of wild-type cells double stained at 15°C (Fig. 2B).
A formal test of our flow cytometric conditions that discriminated between cells with mislocalization of CDCFDA and FM4–64 was to examine individual svl mutants via FCM. Representative members of each svl mutant class not only showed a distinct pattern relative to wild-type cells, but also to each other. The MFI at 525 nm for the svl12 mutant (class III) was decreased 2.5-fold compared with SVL cells (48 vs. 120) while the MFI at 590 nm was increased only 1.5-fold (52 vs. 35, Fig. 5). By comparison, the MFI at 525 nm for the svl2 mutant (class II) was increased about 2.4-fold compared with SVL (287 vs. 120) cells with no significant increase in MFI at 590 nm (39 vs. 35, Fig. 5). The MFI at 525 and 590 nm for the svl3 mutant (class I) was decreased 2.7 and 1.2-fold respectively to SVL cells (Fig. 5).
Because of the compensation used for each wavelength necessary to obtain two parameter histograms with both red and green emission on their respective axis, these values only approximate the actual fluorescence intensity of the cells. For example, digital quantitation demonstrated that svl3 was decreased nearly 8-fold in CDCFDA signal and increased 1.6-fold in FM4–64 signal compared with SVL cells. The svl12 mutant was increased nearly 5-fold in FM4–64 signal and only 1.5-fold in CDCFDA signal compared with SVL cells. The svl2 mutant was increased ≈2-fold for both CDCFDA and FM4–64 compared with SVL cells. Each of the four FCM patterns showed some overlap when they were merged but were largely distinct (Fig. 5). These results suggested that differences not only in fluorescence intensity, but more importantly, in the intracellular morphology after double-staining with CDCFDA and FM4–64 produced populations of yeast cells with discrete characteristics.
DISCUSSION
The overall significance of this work rests on an important advancement. Developing a flow sorting-based method to distinguish the disparate localization of two intracellular fluors is an original accomplishment, which may be a forerunner for future applications in a variety of cell sorting issues. A major conclusion from this approach is that fluorescence detection from two compounds with different emission spectra can be affected by intracellular localization. Obvious factors that likely contribute to this phenomenon for CDCFDA and FM4–64 are their relative intracellular amounts and potential shifts in emission spectra when FM4–64 interacts with different membranes. However, the sorting pattern by using our conditions does not absolutely correlate with fluorescent intensity, suggesting the involvement of additional elements. These might include parameters such as the relative volume, shape, or spatial arrangement of the two fluors within the cell. More experimentation is required to prove or disprove the idea that intracellular fluorescence in three dimensions using multiple fluors can be detected and resolved with FCM. Nevertheless, even without a precise understanding for the basis of our cell sorting strategy, we achieved the desired goal of isolating mutants.
The genetic overlap of some svl mutants reveals new phenotypes of previously isolated vps mutants. For example, our screen uncovered that svl2/vps41 mutant cells (also known as vam2, cvt8, and fet2) do not colocalize CDCFDA and FM4–64. This suggests that the fragmented vacuoles may be a defective target for fusion with internalized intermediates from the plasma membrane or the intermediates may be aberrantly directed to another destination. The allelism of svl6 with vps16 is particularly intriguing because we isolated two independent alleles of svl6 from different populations of mutagenized cells. The three other class C vps mutants, vps11, 18, and 33 were not found in our screen despite their strong overall similarity to vps16 in vacuole morphology. Perhaps this means that vps16 has defects in targeting to the vacuole via endocytosis, giving it morphological distinction from the other class C mutants.
Some of the SVL complementation groups represent a new class of yeast genes that are involved with vacuolar function. In particular, the svl3 mutant has very large, acidic vacuoles that do not exhibit a vps or cvt phenotype yet shows ts staining with CDCFDA. We know of no other mutant affecting vacuolar function that displays this phenotype. The small structures in svl3 mutants that do stain with CDCFDA are reminiscent of what is observed in class C vps mutants, but they all lack any larger, vacuole-like organelles. One that explanation for the svl3 phenotype might be loss of the requisite esterase(s) or lipase(s) activity for CDCFDA cleavage in the vacuole. At present, the identity of these enzymes is completely unknown and efforts are underway to isolate their genes.
Acknowledgments
We thank Brian Davis for suggesting svl as our mutant nomenclature; Beverly Wendland and Scott Emr for sharing results before publication, and for providing us with dim1, dim2, and fab1 mutant yeast strains; Sidney Scott and Daniel Klionsky for providing their cvt mutant collection and antisera against pro-aminopeptidase I; Yoh Wada for providing us with a VPS41 plasmid; and Eric Bensen and Greg Payne for the gift of antiserum to Ste3p as well as advice on turnover experiments. Daniel Klionsky made critical comments on the manuscript. This work was supported by National Institutes of Health-National Institute of General Medical Sciences Grant GM52092 to T.V..
ABBREVIATIONS
- CDCFDA
carboxydichlorofluorescein diacetate
- FM4–64
N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino)phenyl)hexatrienyl)pyridinium dibromide
- BODIPY-PC
2-(4,4-difluoro-5-methyl-4-bora-3a,4a-diaza-s-indacene-3-dodecanoyl)-1-hexadecanoyl-sn-glycero-3-phosphocholine
- BODIPY-PE
2-(4,4-difluoro-5-methyl-4-bora-3a,4a-diaza-s-indacene-3-dodecanoyl)-1-hexadecanoyl-sn-glycero-3-phosphoethanolamine
- YPD
yeast extract/peptone/dextrose
- ts
temperature sensitive
- FCM
flow cytometry
- MFI
mean fluorescence index
References
- 1. Palade G. Science. 1975;189:347–357. doi: 10.1126/science.1096303. [DOI] [PubMed] [Google Scholar]
- 2.Haugland R P. Methods Cell Biol. 1994;42 B:641–663. doi: 10.1016/s0091-679x(08)61100-0. [DOI] [PubMed] [Google Scholar]
- 3.Vida T A, Emr S D. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wendland B, McCaffery J M, Xiao Q, Emr S D. J Cell Biol. 1996;135:1485–1500. doi: 10.1083/jcb.135.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson J S, Klionsky D J, Banta L M, Emr S D. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lawrence C W. Methods Enzymol. 1991;194:273–281. doi: 10.1016/0076-6879(91)94021-4. [DOI] [PubMed] [Google Scholar]
- 7.Roberts C J, Raymond C K, Yamashiro C T, Stevens T H. Methods Enzymol. 1991;194:644–661. doi: 10.1016/0076-6879(91)94047-g. [DOI] [PubMed] [Google Scholar]
- 8.Roth A F, Davis N G. J Cell Biol. 1996;134:661–674. doi: 10.1083/jcb.134.3.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis D E, Schober W, Murrell S, Nguyen D, Scott J, Boinoff J, Simpson J L, Bischoff F Z, Elias S. Cytometry. 1996;23:218–227. doi: 10.1002/(SICI)1097-0320(19960301)23:3<218::AID-CYTO5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y X, Zhao H, Harding T M, Gomes de Mesquita D S, Woldringh C L, Klionsky D J, Munn A L, Weisman L S. Mol Biol Cell. 1996;7:1375–1389. doi: 10.1091/mbc.7.9.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breeuwer P, Drocourt J L, Bunschoten N, Zwietering M H, Rombouts F M, Abee T. Appl Environ Microbiol. 1995;61:1614–1619. doi: 10.1128/aem.61.4.1614-1619.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohrer K, Emr S D. J Biol Chem. 1993;268:559–569. [PubMed] [Google Scholar]
- 13.Raymond C K, Howald-Stevenson I, Vater C A, Stevens T H. Mol Biol Cell. 1992;3:1389–402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harding T M, Morano K A, Scott S V, Klionsky D J. J Cell Biol. 1995;131:591–602. doi: 10.1083/jcb.131.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radisky D C, Snyder W B, Emr S D, Kaplan J. Proc Natl Acad Sci USA. 1997;94:5662–5666. doi: 10.1073/pnas.94.11.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamura N, Hirata A, Ohsumi Y, Wada Y. J Biol Chem. 1997;272:11344–11349. doi: 10.1074/jbc.272.17.11344. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto A, DeWald D B, Boronenkov I V, Anderson R A, Emr S D, Koshland D. Mol Biol Cell. 1995;6:525–539. doi: 10.1091/mbc.6.5.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kean L S, Grant A M, Angeletti C, Mahe Y, Kuchler K, Fuller R S, Nichols J W. J Cell Biol. 1997;138:255–270. doi: 10.1083/jcb.138.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banta L M, Robinson J S, Klionsky D J, Emr S D. J Cell Biol. 1988;107:1369–1383. doi: 10.1083/jcb.107.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]