SUMMARY
Recognition of signal sequences by cognate receptors controls the entry of virtually all proteins to export pathways. Despite its importance, this process remains poorly understood. Here, we present the solution structure of a signal peptide bound to SecA, the 204-kDa ATPase motor of the Sec translocase. Upon encounter, the signal peptide forms an α-helix that inserts into a flexible and elongated groove in SecA. The mode of binding is bimodal, with both hydrophobic and electrostatic interactions mediating recognition. The same groove is used by SecA to recognize a diverse set of signal sequences. Impairment of the signal peptide binding to SecA results in significant translocation defects. The C-terminal tail of SecA occludes the groove and inhibits signal peptide binding, but autoinhibition is relieved by the SecB chaperone. Finally, it is shown that SecA interconverts between two conformations in solution suggesting a simple mechanism for polypeptide translocation.
INTRODUCTION
Although all proteins are synthesized in the cytoplasm, more than a third is transported to other subcellular compartments or regions, inside a membrane or to the extracellular milieu (Wickner and Schekman, 2005). The entry of virtually all proteins to the export pathway, both in eukaryotes and prokaryotes, is controlled by a short signal sequence (15-30 residue long), which is typically encoded at the N terminus of the exported protein (Hegde and Bernstein, 2006). The Sec translocase is a highly conserved multipartite protein machinery responsible for handling the vast majority of bacterial and ER exported proteins (Osborne et al., 2005). In bacteria, protein targeting to the membrane-embedded SecYEG protein-conducting channel is the result of the specific recognition of the signal sequence by either the signal recognition particle (SRP) or SecA, the highly conserved and essential ATPase motor of the translocase (Mitra et al., 2006). How signal sequences interact with any of these receptors is entirely unknown.
Soluble secretory preproteins emerging from the ribosome are captured in an unfolded, translocation–competent state by the SecB chaperone (Randall et al., 1997). The complex is then targeted to SecA, which binds both the signal sequence and the mature domain of the preprotein, as well as SecB (Lill et al., 1990; Randall and Hardy, 2002). SecA couples the export of preproteins through the SecYEG translocon with the expenditure of metabolic energy provided by ATP binding and hydrolysis.
SecA is a large (204-kDa) homodimeric protein (Figure 1A) consisting of 901 residues per protomer (Figure 1B). Its helicase-like motor is located at the N terminus and is assembled by the discontinuous nucleotide binding domain (NBD) and the intramolecular regulator of ATP hydrolysis-2 domain (IRA2). The nucleotide binds at a cleft formed at the interface of NBD and IRA2 (Sianidis et al., 2001; Hunt et al., 2002). Cross-linking and biochemical experiments have suggested that preproteins and signal sequences interact with the preprotein-binding domain (PBD), which ‘sprouts’ out of the body of NBD through an antiparallel β-sheet (Kimura et al., 1991; Kourtz and Oliver, 2000; Papanikou et al., 2005; Musial-Siwek et al., 2007). A number of grooves formed between PBD and other domains have been proposed as potential binding sites for the signal peptide on the basis of structural data of unliganded SecA (Hunt et al., 2002; Osborne et al., 2004). The C-terminal domain of SecA encompasses four substructures: the long α-helical scaffold domain (SD), the IRA1 hairpin, the winged domain (WD) and a flexible, mostly crystallographically unresolved, region (Figure 1B).
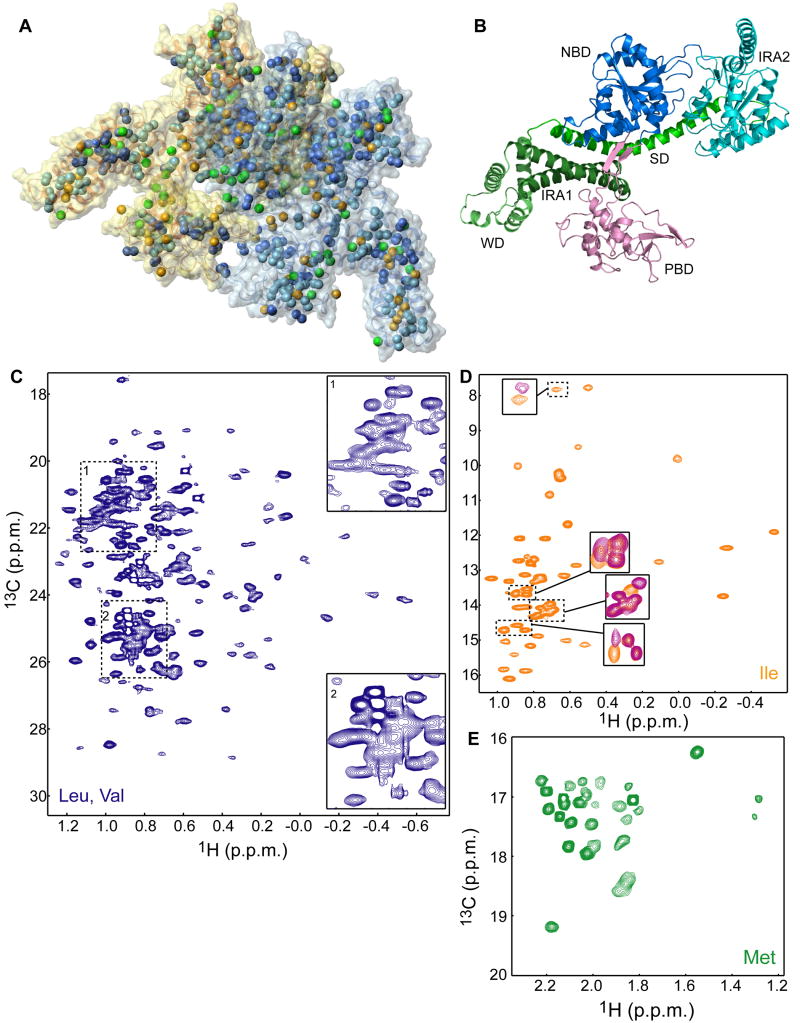
Figure 1. Methyl-TROSY spectra of the full-length 204-kDa SecA.
(A) Structural model of dimeric E. coli SecA (PDB 2FSF) displayed as semi-transparent solvent-accessible surface. The methyl groups are displayed as spheres. The color code is as follows; Ile, orange (54 residues); Leu, light blue (82 residues); Val, dark blue (59 residues); Met, green (33 residues). The two protomers are colored differently.
(B) Structural model of one of the protomers of dimeric E. coli SecA colored according to domain organization.
(C-E) 1H-13C HMQC spectra of SecA (C) U-[2H,12C],Val,Leu-[13CH3,12CD3], (D) U-[2H,12C],Ile-δ1-[13CH3], and (E) U-[2H,12C],Met-[13CH3]. In (C), expanded views of the crowded areas of the spectrum are shown. In (D), regions of the spectrum of unliganded SecA (orange) are overlaid with the spectrum of SecA bound to the KRR-LamB signal sequence (magenta).
Specific interaction between a signal sequence and SecA is a decisive step in correctly sorting secretory from non-secretory proteins. As such, this binding interaction must be of extreme fidelity. One of the most intriguing aspects of SecA is its capacity to recognize hundreds of different signal sequences characterized by the lack of any consensus in their primary sequence. Their only common characteristic is a stretch of hydrophobic residues preceded by positively charged residues in the N terminus (von Heijne, 1985; Gierasch 1989). To date, how SecA accomplishes this challenging task of promiscuous binding remains a conundrum.
Here we have used NMR spectroscopy to structurally characterize the interaction of full-length SecA with functional signal peptides. By exploiting recent advances in isotope labeling and NMR methodology we have been able to observe and assign the methyl side chains of a large number of SecA residues. Structure determination of the SecA-signal peptide complex demonstrates that the peptide forms an α-helix and binds using both its hydrophobic and charged regions into a flexible and elongated groove in SecA. Signal peptide binding to SecA is restricted by an autoinhibitory mechanism, which is relieved by the SecB chaperone. Interestingly, SecA appears to undergo a large conformational change in solution that may potentially be coupled to the protein translocation mechanism. The combined data not only explain how SecA may achieve the promiscuous recognition of a large set of signal sequences, but also provides insight into how the Sec nanomachinery may ultimately be assembled.
RESULTS
Methyl transverse relaxation optimized spectroscopy (TROSY) of the 204-kDa SecA
To tackle the 204-kDa SecA, we employed recently developed specific labeling schemes (Sprangers et al., 2007) tailored to overcome resonance broadness and overlap. Specifically, we have produced samples with the methyl groups of Ile, Leu, Val, and Met residues of SecA protonated, in an otherwise completely deuterated background (Figure 1A). The methyl groups of these four residues are excellent probes as they are abundant (220 out of the total 901 residues per protomer) and are distributed throughout SecA (Figure 1A). Methyl-TROSY spectroscopy (Sprangers and Kay, 2007) was used to optimize both sensitivity and resolution. The recorded 1H-13C heteronuclear multiple quantum (HMQC) spectra are of exceptional quality for all four residues (Figure 1C-E).
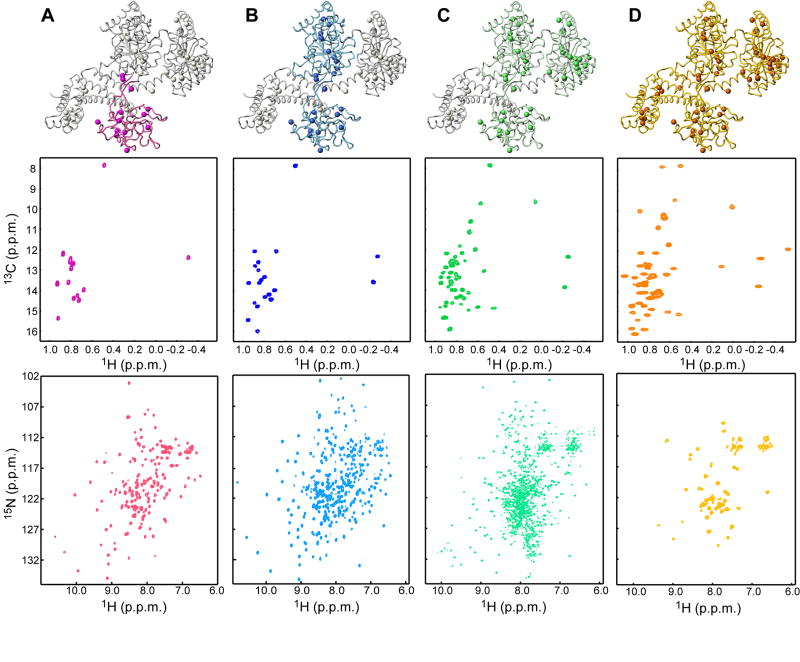
The large size of SecA precludes the use of traditional assignment protocols. For this reason we followed a domain-parsing strategy. Virtually all domains of SecA and a number of fragments comprising contiguous domains have been isolated and characterized by NMR (Figure 2). Assignment of the methyl groups on these relatively small domains was straightforward using standard methodologies (Keramisanou et al., 2006). Comparison of the 1H-13C HMQC spectra of the various domains with that of the full-length SecA demonstrates very good resonance correspondence (Figure 2, S1). Therefore, the majority of the assignment of methyl cross-peaks performed in the isolated domains and fragments could be readily transferred to the full-length SecA. Assignment was completed (Figure S2) using 3D NOESY spectra recorded on full-length SecA (Figure S1C). Mutagenesis was also used to resolve ambiguities that primarily existed for residues located at the interface of domains (Figure S1A).
Figure 2. Strategy for the assignment of methyl correlations of SecA.
Each column in the figure displays a structural model of one of the protomers of SecA with the domain or fragment studied in isolation being highlighted, along with the corresponding 1H-13C HMQC of Ile-δ1 methyls (displayed as spheres in the model) and the backbone 1H-15N HSQC.
(A) PBD (residues 220-379).
(B) SecAΔC/ΔIRA2 (residues 1-420, comprising NBD and PBD).
(C) SecAΔC (residues 1-610, comprising NBD, PBD and IRA2).
(D) Full-length SecA (residues 1-901). Only few resonances for the backbone of the full-length SecA are visible (Figure S9).
Structure determination of the SecA-signal peptide complex by NMR
The well-studied functional signal peptide KRR-LamB, derived from the LamB porin (Triplett et al., 2001; Chou and Gierasch, 2005), was used. The peptide binds specifically and stoichiometrically to SecA (one peptide per SecA subunit; Figure S3A). The peptide suppresses the ATPase activity and inhibits translocation (Figure S3B,C), as expected for a functional signal peptide (Lill et al., 1990). To determine the structure of the complex, we used simulated annealing of the signal peptide in the presence of the X-ray structure of free E. coli SecA (PDB code: 2FSF) using structural information derived from NMR spectroscopy. To obtain distance restraints between SecA and the signal peptide we used site-directed spin labeling (SDSL). SDSL combined with NMR-detected paramagnetic relaxation enhancement (PRE) rates has been frequently used to determine high-resolution structures of proteins and their complexes with various ligands (Battiste and Wagner, 2000; Gross et al., 2003; Roosild et al., 2005). A nitroxide spin label was introduced to select positions in the peptide and methyl-TROSY experiments were used to observe the distance-dependent broadening of the methyl resonances of SecA in the complex (Figure S4). Changes in resonance intensity induced by peptide binding (Figure S5A) were then converted to distances (see Experimental Procedures). Because of the large density of methyl probes close to the peptide binding site (within ~30 Å), a large number of SecA-signal peptide distance restraints were obtained (Figure S5B).
Spin labels were engineered at different positions in the signal peptide by converting a single amino acid to cysteine, where a nitroxide spin label was attached. NMR and isothermal titration calorimetry (ITC) titration of SecA with the mutant and reduced nitroxide-derivatized peptides were used to assess the effect of the mutations and the presence of the label on peptide binding to SecA (see also below). Placement of the spin label in the helix resulted in altered binding energetics, presumably because of steric clash with the protein. Two positions that were observed to give the most reliable data were K7C at the N terminus and Q25C at the C terminus (Figure 4A). Placement of the spin label in these positions, rather than closer to the termini, significantly decreased the mobility of the spin label resulting in reduction of nonspecific broadening. The placement of the spin label in two different positions in the N and C terminus of the peptide not only increased the number of distance restraints by roughly two-fold, but also contributed to the better determination of the relative orientation of the peptide bound to SecA.
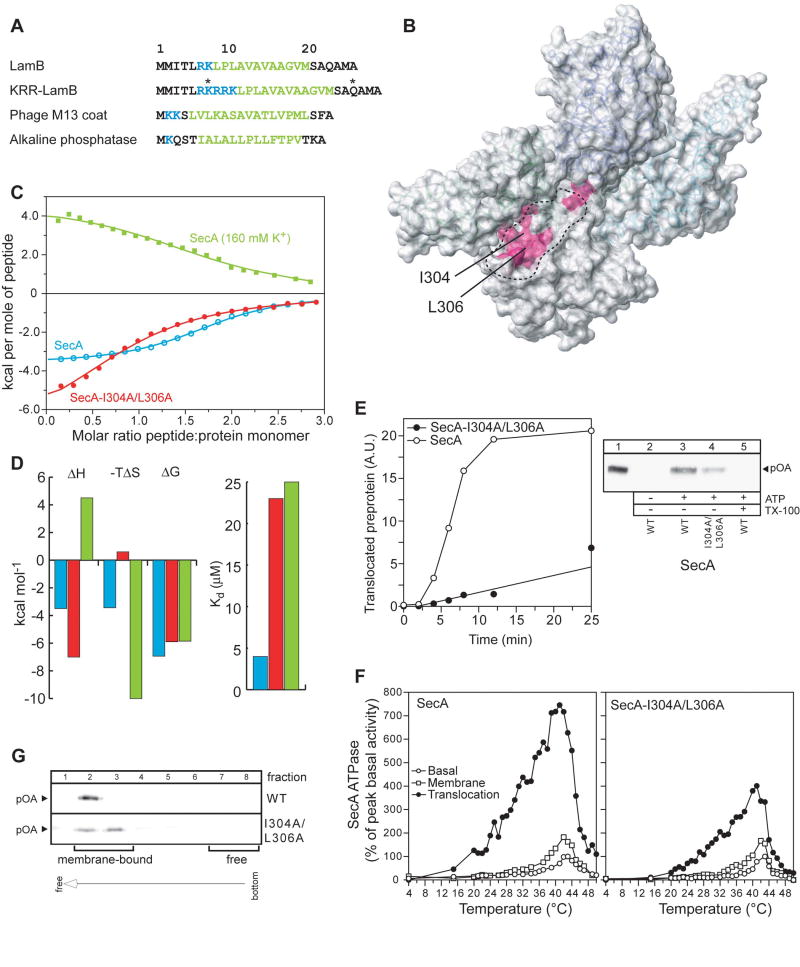
Figure 4. Signal peptide binding to SecA and the effect of its impairment on the function of SecA.
(A) The four different signal sequences used in the present study are shown. In KRR-LamB, the asterisks indicate the positions where a single Cys mutation was introduced, followed by the incorporation of the nitroxide spin label. Basic and hydrophobic residues are colored blue and green, respectively.
(B) Chemical shift mapping of the interaction of the signal peptides shown in (A) with SecA. The magenta colored region indicates the common residues whose chemical shift is significantly affected upon signal peptide binding (see text for details).
(C) Binding isotherms of the calorimetric titration of the KRR-LamB signal peptide to SecA (open, cyan circles), SecA-I304A/L306A (filled, red circles), and SecA in 160 mM K+ buffer (green squares). In the first two cases, SecA is in 40 mM K+ buffer. Ile304 and Leu306, whose position within the groove is indicated in (B), were mutated to assess the relative contribution of hydrophobic interactions in SecA-signal peptide binding, whereas higher buffer salt was used to assess the contribution of electrostatic interactions.
(D) Thermodynamic parameters of the calorimetric titrations in (C) displayed as bars. The color code is as in (C). Weakening of the electrostatic interactions results in hydrophobic interactions being the dominant driving force for complex formation, as suggested by the observation that the reaction becomes enthalpically unfavorable, but strongly entropically favorable.
(E) ATP-driven in vitro translocation of proOmpA-His into SecYEG-containing IMVs catalyzed by SecA or SecA-I304A/L306A (right panel). Lane 1: 5% of undigested proOmpA-His input. Lane 5: membranes were dissolved with Triton x-100 (1%) prior to proteinase K addition. Proteins were TCA-precipitated, analyzed by SDS-PAGE and immunostained with α-His antibody. Left panel: Time-course of proOmpA translocation kinetics. 20 A.U. corresponds to 1.6 pmol of translocated proOmpA.
(F) kcat values (pmoles Pi/pmol SecA protomer per min) of basal, membrane and translocation ATPase activities of SecA as a function of temperature. Averaged data of three repetitions are shown.
(G) IMV flotation assay showing weaker binding of proOmpA to SecYEG-bound SecA- I304A/L306A than to wild-type SecA.
To determine the structure of the signal peptide in complex with SecA, we used transferred nuclear Overhauser effect spectroscopy (trNOESY) and differential line broadening experiments (Matsuo et al., 1999). The conformation of the peptide bound to SecA was probed by measuring NOEs within the peptide in the presence of a small amount of SecA. In an effort to collect the maximum number of inter-peptide NOEs we extended previous insightful studies (Chou and Gierasch, 2005) by recording NOESY spectra on a 900 MHz spectrometer equipped with a cryoprobe to increase resolution and sensitivity. The data, in agreement with the previous study (Chou and Gierasch, 2005), show that while the free peptide is unstructured, its hydrophobic region (residues Leu13-Val21) adopts an αhelical conformation when interacting with SecA. The structure of the peptide was calculated on the basis of 50 intramolecular NOEs. The family of the best 10 structures of the peptide was then used for the determination of the complex using the crystal structure of E. coli SecA (Papanikolau et al., 2007) as the starting conformation. The structure of the complex was determined on the basis of 162 intermolecular distance restraints. The lowest-energy structure of the complex is shown in Figure 3, while the ensemble of the 10 best structures is shown in Figure S6.
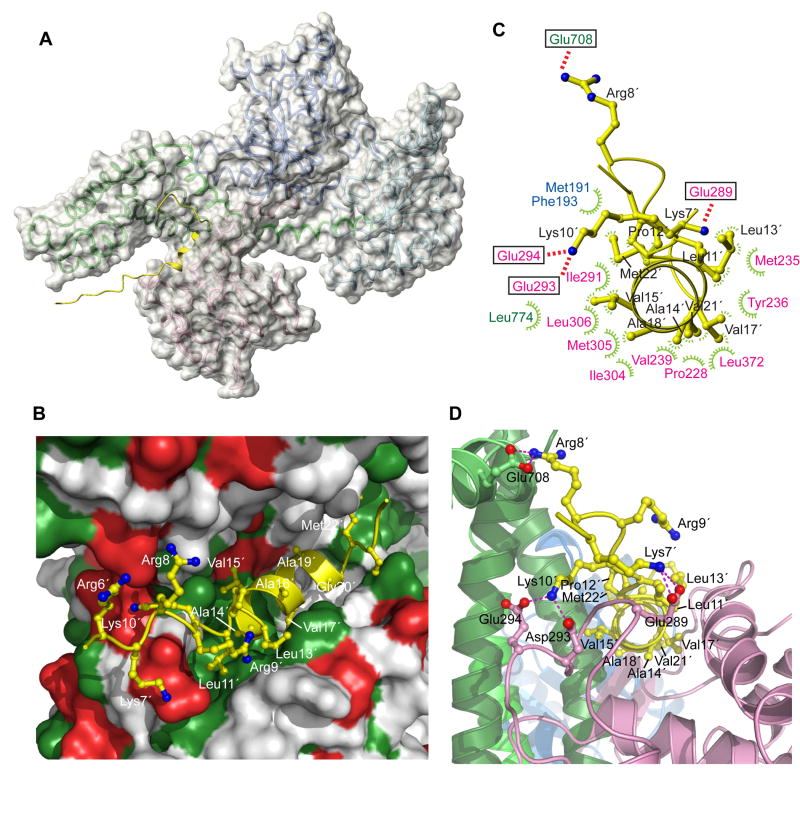
Figure 3. Structural basis for signal peptide recognition by SecA.
(A) The lowest-energy structure of SecA bound to the KRR-LamB signal peptide is shown. SecA is displayed as a semi-transparent solvent-accessible surface and the signal peptide is shown in yellow. A ribbon model is displayed below the surface (color code is as in Figure 1B).
(B) Closer view of the groove bound to the signal peptide. Green and red surface indicates hydrophobic and acidic residues, respectively. Peptide is shown as a ribbon ball-and-stick representation and most of its residues are numbered.
(C) Contacts between the peptide (shown in yellow) and SecA residues. Electrostatic and hydrophobic interactions are indicated with red and green lines, respectively. SecA residues are colored according to the domain they are located at.
(D) A view of the groove bound to the signal peptide, wherein SecA is shown in ribbons. The peptide orientation is similar to that in (C). Dotted lines indicate electrostatic interactions between basic peptide residues and acidic SecA residues. Primed numbers indicate peptide residues.
Signal peptide binds into an expandable and elongated groove using both hydrophobic and electrostatic interactions
The structural data demonstrate that the signal peptide binds into a relatively large groove formed at the interface of two domains: the PBD and the IRA1 hairpin (Figure 3). The groove consists primarily of PBD residues with a number of IRA1 residues lining only one side of the groove. The groove is mostly hydrophobic, but is surrounded by a number of polar and charged residues (Figure 3B). The hydrophobic character of the groove is highly conserved, although the sequence identity of the residues that make up the groove is not (Figure S7).
The entire length of the α-helical hydrophobic region of the peptide (consisting of two turns) inserts into the groove, which is relatively long (~28 Å) and, thus, it can easily accommodate even longer α-helices (Figure 3A,B). As expected, hydrophobic interactions between the peptide and SecA dominate the 2,500 Å2 (1,800 Å non-polar and 700 Å polar) surface area buried at the SecA-peptide interface. Four short-chain nonpolar aliphatic residues of the peptide (Ala14, Val17, Ala18, and Val21) project toward the center of the hydrophobic face presented by SecA and are completely buried at the interface (Figure 3B-D). In contrast, longer aliphatic residues (Leu11, Leu13, Met22, along with Val15) project from the sides of the helix and are partially solvent exposed (Figure 3B,D). Together, these residues create a hydrophobic ridge running along the peptide helix axis, forming intimate associations with the predominantly hydrophobic surface of the groove. The face of the peptide α-helix that projects away consists exclusively of short-chain residues, which do not appear to make interactions. These data are in agreement with the differential line broadening results showing that the resonances of these residues do not become as broad as those of the other residues of the α-helix.
The positively charged N-terminal residues of the peptide (Lys7, Arg8, Arg9, and Lys10) engage in the formation of salt bridges with acidic residues of SecA located adjacent to the hydrophobic groove (Figure 3B-D). These residues are seen to interact electrostatically with various acidic residues on SecA in the ensemble of the structures. In any single structure, multiple salt bridges are present suggesting an important role of these electrostatic interactions in the recognition process. Therefore, the present structural data reveal a dual binding mode for the peptide, which is capable of using both its positively charged N-terminus and the α-helical hydrophobic region to interact with the same groove in SecA.
The C-terminal region of the peptide (residues 24-28), which contains the signal peptidase cleavage site, remains unstructured. This region is not involved in intimate interactions with SecA and is solvent exposed. This picture is in agreement with NOE and differential broadening data showing that this region remains relatively flexible when bound to SecA.
SecA uses a unique site to recognize a variety of signal sequences
NMR chemical shift mapping was used to address whether SecA uses the same groove to recognize various signal sequences. In addition to the KRR-LamB signal peptide used above, three signal peptides from the following preproteins were used: the wild-type LamB, alkaline phosphatase, and M13 procoat (Figure 4A). In all cases, addition of the signal peptide caused significant shift of only a small set of methyl resonances (Figure 1D) suggesting that peptide binding is localized and does not cause global conformational changes to SecA. For example, only 4 out of the total 54 Ile residues have their chemical shift significantly perturbed upon KRR-LamB signal peptide binding. Similarly, only a small number of the Val, Leu and Met methyls are affected. The common set of residues that exhibit the largest shift upon binding of the various signal peptides consists of Ile225, Met235, Val239, Ile291, Met292, Ile304, Met305, Leu306, Val310 and Leu372 at the PDB, and Leu774, Met810 and Met814 at the IRA1 hairpin of the C-domain. In spite of the diverse amino acid sequence of the peptides, the chemical shift changes effected by all of them map onto the same region of SecA (Figure 4B). Thus, SecA appears to use the same groove to recognize diverse signal sequences.
Hydrophobic and electrostatic interactions contribute to signal peptide recognition by SecA
To test the contribution of the hydrophobic contacts on the stability of the complex, we constructed a double SecA mutant wherein two of the most prominent hydrophobic residues of the groove (Ile304 and Leu306) were mutated to Ala (Figure 3C and 4B). The thermodynamic data show that, despite the relatively small perturbation of the hydrophobic surface of the groove, the signal peptide binds with a 6-fold lower affinity to SecA-I304A/L306A than to wild-type SecA (Figure 4C,D). This affinity decrease is the result of a large entropic penalty that apparently accompanies the disruption of favorable hydrophobic interactions (Figure 4D).
To assess the contribution of the electrostatic interactions to SecA-peptide binding, we measured the energetics of complex formation in a buffer containing higher salt concentration. Electrostatic interactions are weakened due to the ‘screening’ conferred by the salt ions. Indeed, increasing the salt concentration from 40 to 160 mM K+ caused a 7-fold decrease in the affinity of the peptide for SecA (Figure 4C,D). Interestingly, the binding is now purely entropy driven, presumably because it is dominated by hydrophobic interactions. Therefore, both hydrophobic and electrostatic interactions appear to be important for strong binding of the signal peptide to SecA, in accordance with the structural data.
Impairment of the signal peptide binding to SecA results in translocation defects
To assess the effect of an impaired SecA-signal peptide interaction on the biological function of SecA, we employed in vitro and in vivo experiments. An in vivo genetic complementation experiment indicates that the ability of SecA-I304A/L306A to translocate proteins is compromised (Figure S8). Using a standard in vitro translocation assay, SecA-I304A/L306A was found to be less efficient, by a factor of 8, in its ability to translocate the model preprotein proOmpA into the lumen of SecYEG-containing inverted inner membrane vesicles (IMVs) when compared to wild-type SecA (Figure 4E). Similarly, IMVs plus preprotein cannot stimulate the basal ATPase activity of SecA-I304A/L306A to the levels seen with wild-type SecA (Figure 4F). The interaction of SecA-bound preproteins with SecYEG is also less efficient in SecA-I304/L306A (Figure 4G). Collectively, our data demonstrate that compromised binding of a signal peptide to SecA results in significant in vivo and in vitro defects during translocation.
The C-tail of SecA constitutes an autoinhibitory element
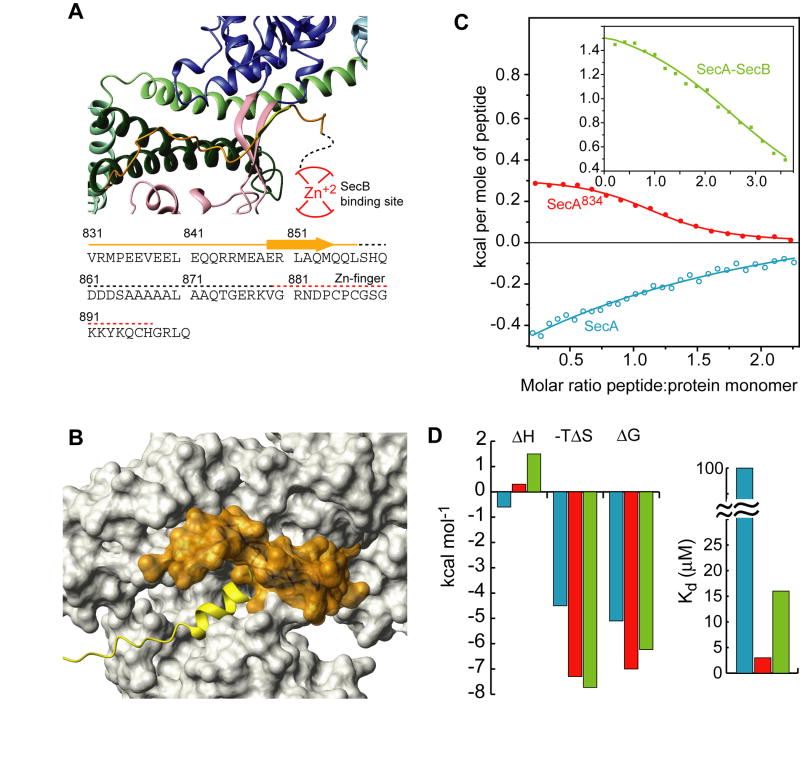
The extreme C-terminal region of SecA (C-tail; residues 834-901 in E. coli) comprises two regions. The first region, consisting of residues 834-855, interacts with the core of SecA and ends up in a β-strand (residues 849-854) that forms a β-sheet with the two antiparallel β-strands linking NBD and PBD (Figure 5A; Hunt et al., 2002). The second region contains a zinc-finger motif that constitutes the primary binding site for the SecB chaperone (Fekkes et al., 1997; Zhou and Xu, 2003). An intriguing aspect of the present structural data is that portion of the peptide binding groove appears to be occluded by the C-tail (Figure 5A,B and S9). Indeed, signal peptide titration to SecA results in displacement of the C-tail region that occludes the groove, as evidenced by 1H-15N HSQC spectra (Figure S10). Thus, the binding sites for the signal peptide and the C-tail are mutually exclusive.
Figure 5. Inhibition of signal peptide binding by the C-tail of SecA.
(A) View of the C-tail of B. subtilis SecA (PDB 1M6N). This is the only crystal structure wherein part of the C-tail is resolved. The E. coli SecA sequence of the C-tail is shown below the model. Dotted lines indicate crystallographically unresolved regions. Red lines indicate the zinc-finger region, which is the primary SecB binding site.
(B) Structural modeling of the interaction of the C-tail in E. coli SecA. The C-tail is shown in orange surface and it partially occludes the peptide binding groove.
(C) Binding isotherms of the calorimetric titration of the wild-type LamB signal peptide to SecA (open, cyan circles), SecA834 (filled, red circles), and SecA bound to SecB (green squares).
(D) Thermodynamic parameters of the calorimetric titrations in (C) displayed as bars. The color code is as in (C).
This observation raises the possibility that the C-tail may prevent efficient binding of the signal sequence. To test this hypothesis, we determined the thermodynamics of signal peptide binding to full-length SecA and SecA834 (residues 1-834), a deletion construct lacking the entire C-tail. The wild-type LamB signal peptide binds to full-length SecA relatively weakly (Kd ~100 μM). Remarkably, removal of the C-tail improves binding by more than 30-fold (Figure 5C,D). The KRR-LamB peptide binds to full-length SecA relatively strongly (Kd ~3 μM), presumably because of the enhanced charged nature of the N terminus; however, its binding is still improved by a factor of ~10 in SecA834. Similarly, binding of the alkaline phosphatase peptide is inhibited by a factor of ~4 (Figure S11). Therefore, the partial occlusion of the peptide binding groove by the C-tail of SecA provides a mechanism to prevent unrestricted access to the signal peptide binding groove.
SecB relieves C-tail mediated autoinhibition
The poor affinity of the wild-type LamB signal peptide for SecA is clearly unexpected, considering that this naturally-occurring sequence is functional in vivo. It is noteworthy that, in contrast to most other secretory proteins, LamB-preprotein targeting to SecA in vivo is absolutely SecB-dependent (Ureta et al., 2007). As the primary SecB-binding site lies at the extension of the C-tail of SecA, it is conceivable that SecB binding to SecA might displace the C-tail, thereby exposing the binding groove to the incoming LamB signal sequence. To test this hypothesis, we measured the energetics of LamB signal peptide binding to SecB-bound SecA. Interestingly, in the presence of SecB, LamB signal peptide binds to SecA with a much higher affinity (7-fold increase; Figure 5C,D). Thus, SecB appears to counteract the autoinhibitory conformational arrangement, thereby resulting in stronger signal peptide binding.
SecA in solution interconverts between an open and closed conformation
An interesting aspect that has emerged from the various crystal structures of SecA proteins is that PBD can adopt two very different conformations (Figure 6A). In the so-called “closed” conformation, PDB interacts extensively with the C-domain forming a compact structure (Hunt et al., 2002; Sharma et al., 2003; Vassylyev et al., 2006). In contrast, in the “open” conformation PDB undergoes a ~60° rigid-body rotation resulting in minimal interaction with the C-domain and exposing most of PDB surface to the solvent (Osborne et al., 2004; Papanikolau et al., 2007). Crystal packing effects could selectively favor one of the two forms. Currently, it is unknown which is the most stable PBD conformation in solution.
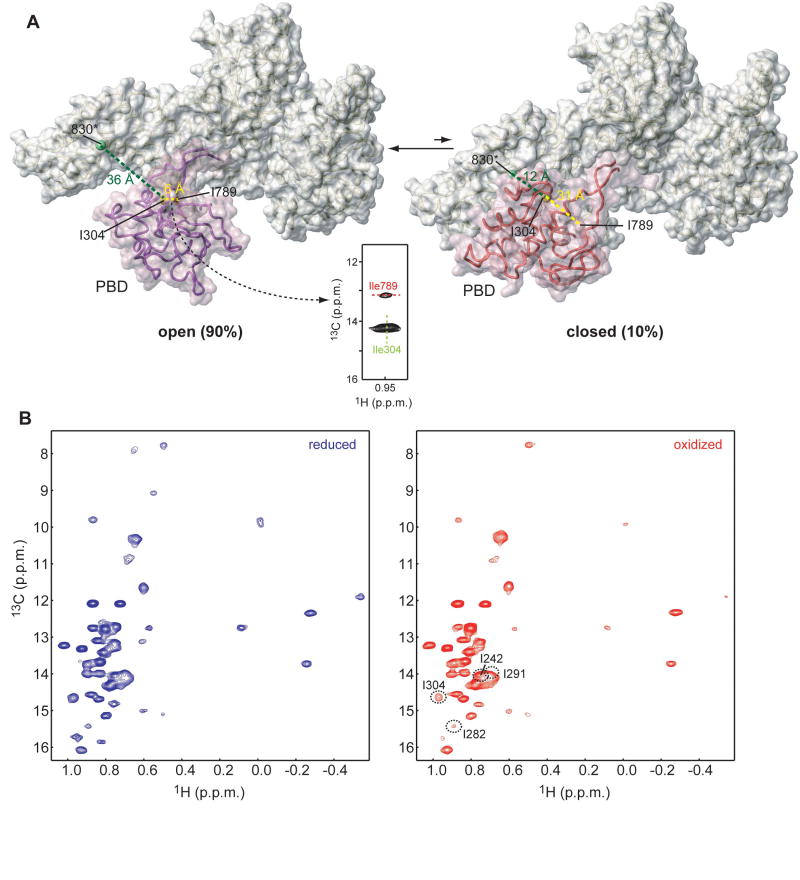
Figure 6. SecA interconverts between an open and closed conformation in solution.
(A) SecA shown in the so-called open (left) and closed (right) conformations. Interconversion between the two conformations requires that PBD undergo a ~60° rigid-body rotation (Osborne et al., 2004). PBD is displayed as semi-transparent surface. The green sphere indicates residue 830, where a paramagnetic spin label was introduced. Residues Ile304 and Ile789 are shown as yellow and red spheres, respectively. Characteristic distances in the two conformations are indicated. A strong NOE between Ile304 and Ile789 was observed demonstrating that SecA adopts predominantly the open conformation in solution.
(B) Overlaid 1H-13C HMQC spectra of SecA bearing a spin label in position 830 in the reduced (blue) and oxidized (red) state. Residues that approach the spin label, even transiently, experience a broadening effect, which is suppressed in the reduced state.
To determine the relative PBD conformational state in solution we examined the NOESY data of SecA. An NOE cross-peak was readily assigned between the methyl groups of Ile 304 and Ile789 (Figure 6A). This is compatible only with the open conformation, wherein the two methyl groups are within 6 Å, and not with the closed conformation, wherein they are located more than 30 Å apart (Figure 6A). Thus, the NOE data provide strong evidence that the PBD of SecA is in the open conformation in solution.
To gain further insight, we used SDSL to determine inter-domain PBD-C domain distances. A spin label was introduced at position 830, located in the end of the IRA1 hairpin (Figure 6A), where the presence of a paramagnetic center would differentially affect the intensity of the methyl probes in the two conformations. For example, the distance to the spin label in position 830 from PBD residues 242, 282, 291, and 304 would be 16 Å vs. 44 Å, 13 Å vs. 43 Å, 15 Å vs. 24 Å, and 12 Å vs. 36 Å, in the closed and open conformation, respectively.
If SecA spent 100% of the time in the open conformation, then the presence of a paramagnetic center at position 830 would have absolutely no effect on the intensity of the above group of methyl resonances of PBD residues. However, even methyls that are more than 40 Å away from the spin label in the open conformation, but very close to the spin label in the closed conformation, are weakly affected (Figure 6B). For example, the intensity of the I304 methyl resonance is reduced by ~30% in the presence of the spin label in position 830, despite the fact that it lies 36 Å apart from the paramagnetic center in the open conformation (Figure 6A,B). Similar changes in the intensity were also observed for residues 242, 282 and 291. The intensity loss is fully recovered in the reduced state suggesting that the intensity decrease of these residues in the paramagnetic sample is due to the, apparently transient, proximity of residue 304 to the spin center at position 830 (Figure 6). As has been elegantly shown recently (Iwahara and Clore, 2006; Volkov et al., 2006; Tang et al., 2007), PRE rates are extremely sensitive to the presence of transiently populated excited states. The estimated PRE rates of the methyl of Ile304 are ~0 and 400 s-1 for the open and closed state, respectively. Therefore, our results suggest that the closed conformation represents ~10% of the total population. Overall, the combined NOE and PRE data argue that SecA interconverts in solution between an open conformation, the major form, and a closed conformation, the minor form.
DISCUSSION
Molecular understanding of the translocation process necessitates determination of the structural and dynamic basis for the assembly of the entire Sec translocase machinery. Towards this goal, we have undertaken a challenging NMR study aiming at the structural characterization of the 204-kDa SecA motor ATPase and its interaction with signal peptides. The specific recognition of the N-terminally fused signal sequence by SecA is arguably the most decisive step in correctly targeting secretory polypeptides. The use of specific labeling schemes (Sprangers et al., 2007) and domain-parsing approaches for resonance assignment, combined with paramagnetic spin labeling and relaxation enhancement measurements enabled the structure determination of this large complex. We anticipate that similar strategies will render other large macromolecular complexes tractable to structural characterization by NMR.
A central feature of the recognition process is the transition of the peptide hydrophobic region from a random-coil conformation to an α-helix upon its interaction with SecA (Figure 3; Chou and Gierasch, 2005). This helix inserts into the relatively deep groove and almost all of its residues are involved in intimate interactions with the hydrophobic surface of the groove. A similar mechanism appears also to be used by the Tom20 import receptor to bind its signal sequences (Abe et al., 2000). Different signal sequences cause similar chemical shift perturbation patterns (Figure 4A,B), suggesting that SecA uses this groove to recognize the hundreds of different protein substrates. The binding groove has several distinct features: (i) It is quite long (~28 Å), thus, explaining how SecA can accommodate signal sequences with much longer α-helical hydrophobic regions. (ii) It consists of hydrophobic residues and is surrounded by acidic ones, thereby permitting the signal peptide to bind in a dual mode. This observation explains previous results highlighting the important role of both the hydrophobic and positively charged regions of the peptide (Akita et al., 1990; Mori et al., 1997; Wang et al., 2000; Karamyshev and Johnson, 2005). Both recognition modes contribute greatly to the binding since impairment of the hydrophobic or electrostatic contacts between SecA and the signal peptide results in weaker interaction and significant translocation defects (Figure 4C-G and S8). Clearly, the extent to which different signal peptides rely on these recognition modes can vary. (iii) The binding groove is relatively deep and many small pockets are present at its sides. Hence, it can accommodate signal sequences of varying length and bulkiness of side chains. (iv) The groove is lined with several loosely packed methyl groups and Met residues, whose side chain is particularly flexible. These provide a malleable hydrophobic surface that can adapt itself to the binding of signal sequences of varying dimensions. (v) The groove is formed at the interface of two domains, PBD and IRA1. Thus, it is anticipated that the binding site will be quite flexible and expandable by small rearrangement of the domains. Such a rearrangement would give rise to grooves of somehow varying dimensions and expose surfaces of variable amino acid composition. Indeed, inspection of the structural ensemble (Figure S6) suggests that such small rearrangements are energetically allowed. Collectively, the structural plasticity of the binding groove at both the level of the individual side chains and the orientation of the involved domains may be crucial for SecA to recognize its surprisingly diverse range of substrates.
An interesting finding in the present study is that the C-tail of SecA partially occludes the peptide binding groove, thereby forming the basis of an autoinhibitory mechanism. Intriguingly, the binding of the wild-type LamB signal peptide to SecA is strongly inhibited, but sufficient binding is restored in the presence of the SecB chaperone. Apparently, SecB binding to the zinc-finger site located at the extension of the C-tail somehow displaces the C-tail from the binding groove allowing for a stronger SecA-signal peptide interaction. This finding may explain why export of LamB is SecB-dependent (Ureta et al., 2007). Since the C-tail masks only a portion of the binding groove, it is expected that the extent of signal peptide binding inhibition will depend on the exact positioning of the peptide along the elongated groove. This is indeed seen with the alkaline phosphatase peptide, whose binding is inhibited by only a factor of ~4. In this case the signal sequence will counteract the autoinhibitory conformation and no additional factors, such as SecB, are needed to relieve autoinhibition. The role of this autoinhibitory mechanism in SecA may be two-fold. First, it may prevent the untimely interaction of a preprotein with SecA, unless the ternary complex with SecB has formed. Second, it may safeguard against non-specific binding by acting as a selectivity barrier to ensure that only genuine signal sequences on secretory proteins will have the affinity to overcome it.
The combined NOE and PRE NMR data show that SecA interconverts between an open (major form, 90%) and a closed conformation (minor form, 10%) in solution (Figure 6), which correspond to the two alternative conformations seen in crystal structures (Hunt et al., 2002; Osborne et al., 2004; Papanikolau et al., 2007). Intriguingly, although the PDB part of the groove is accessible in both states, the complete peptide binding groove forms only in the open state. It is conceivable that the equilibrium of the two extreme conformations may shift as a result of preprotein or SecYEG binding, and possibly also during the ATPase cycle. In this case, the large conformational change undergone by PBD may be a functional one, linking preprotein binding to the catalytic cycle, thus, presenting a simple translocation mechanism (see below).
On the basis of the present and previous results, we put forward a refined model of SecA-mediated translocation (Figure 7). SecA partitions between the cytosol and the membrane (Mori et al., 1997). In solution, SecA adopts a catalytically inactive, ADP-liganded state, wherein strong domain-domain interactions prevail (Figure 7A; Sianidis et al., 2001; Fak et al., 2004). Binding to SecYEG at the membrane incurs a loosening of these interactions, causing marginal ATPase stimulation. Although how exactly SecA interacts with SecYEG remains to be determined, both the NBD and the C-domain appear to be involved (Mori and Ito, 2006; Osborne and Rapoport, 2007). The peptide binding groove remains partially masked by the C-tail. In the next step (Figure 7C), the SecB-preprotein complex is targeted to SecA. SecB binding to the zinc-finger at the end of the C-tail somehow displaces the C-tail from the groove, thereby relieving autoinhibition. The signal peptide then binds into the groove and adopts an α-helical conformation. Next, SecB is released and the preprotein is transferred entirely to SecA (Figure 7D). Because of the polarity of signal peptide binding to SecA, the mature domain will likely be directed towards the helicase motor and may bind by bridging the two motor domains (Figure 7D and S12). In such a case, we hypothesize that SecA will bind its protein substrate in a way similar to that by which the Vasa helicase binds its RNA substrate (Sengoku et al., 2006). In this mode of binding the preprotein may open further apart the motor domains, explaining why preprotein is required for maximal stimulation of the ATPase activity (Karamanou et al, 2007). In the first step of the actual translocation the signal peptide will dissociate from SecA and insert into SecYEG, in a step that likely requires ATP hydrolysis (Schiebel et al., 1991; Wang et al., 2004). This transfer is quite favorable as the positively charged N terminus of the peptide interacts with the negative charge of the membrane. For the actual translocation process to work (Figure 7E,F), we conjecture that two mechanisms should proceed in synergy. The first one is the closure and opening of the motor, which will be directly regulated by ATP binding, hydrolysis and ADP release (Keramisanou et al., 2006). Such a movement would constantly push the preprotein forward towards the membrane. The motor motion could potentially be coupled to a second mechanism involving the large conformational change undergone by PBD. Due to its structural and physicochemical properties, the peptide binding groove is an excellent candidate for binding also the mature portion of the preprotein. We could envision that the two states, open and closed, would have different affinity for the preprotein, as the binding groove forms only in the open state. Such a simple mechanism may be sufficient to carry out the translocation process. Intimate association of SecA to SecYEG ensures that these SecA motions also direct corresponding movements of SecYEG that will allow the protein-conducting channel constriction (Van den Berg et al., 2004) to tighten and loosen around the translocated preprotein.
Figure 7. Model of the SecA-mediated preprotein translocation.
PBD is shown in the closed state in E and in the open state in all other panels. The C-tail is not shown in D-F for clarity. See text for details.
EXPERIMENTAL PROCEDURES
Protein Preparation
His-tagged E. coli SecA, SecA834, SecAΔC, SecAΔC-ΔIRA2, SecAC (34 kDa, residues 611-901) and isolated PBD and IRA2 domains were constructed as described previously and transformed in BL21DE3/pLysS (Sianidis et al., 2001; Keramisanou et al., 2006). Cultures for full-length SecA and its mutants were grown at 30 °C and protein synthesis was induced by addition of 0.5 mM of IPTG at A600 ~0.3. Cells were harvested at A600 ~ 0.75. For isotope labeling, minimal media containing 15NH4Cl and [2H,12C] or [2H,13C])-glucose in 99.9% 2H2O were used. For the production of U-[2H],Ile-δ1-[13CH3] and Val,Leu-[13CH3,12CD3] samples, 50 mg l-1 of alpha-ketobutyric acid (methyl-13CH3) and 100 mg l-1 of alpha-ketoisovaleric acid (dimethyl-13CH3,12CD3) were added to the culture 1 hour prior to addition of IPTG. Met[13CH3] labeled samples were produced by supplementing the medium with 250 mg l-1 of [13CH3]- methionine (no side chain scrambling takes place with Met). All protein samples were purified over a nickel-chelating column, followed by ion-exchange and gel filtration.
To produce MTSL-derivatized SecA, we constructed the SecA834-Q830C mutation (the SecA834 construct was used to avoid cross-linking to Cys residues of the Zn finger). The only other Cys in SecA (Cys98) is not reactive, as judged by the Elman’s test and NMR, and therefore it was not mutated. After purification the protein was exchanged to phosphate buffer (50mM KPi, 50mM KCl, pH=8.0), free of any reducing agent and it was concentrated (~8 μM). MTSL was added from a concentrated stock in acetonitrile at a ten-fold excess and the reaction was let to proceed at 4 °C for ~12 h. The completion of the reaction was confirmed by mass spectrometry. Excess MTSL was removed by extensive dialysis using an Amicon stirred cell and the pH was corrected to 7.5.
Peptide Preparation
All peptides were chemically synthesized by GeneScript (Piscataway, NJ). To cross link the KRR-LamB cysteine mutants with MTSL, the peptides were dissolved in phosphate buffer (25 mM KPi, 5mM KCl, pH=8.0) at a concentration of ~0.1 mM and MTSL was added in a ten-fold excess. Complete cross-linking was verified by mass spectrometry. Excess MTSL was removed by extensive dialysis using an Amicon stirred cell and the pH was corrected to 7.5.
NMR Spectroscopy
NMR experiments were performed on Varian 600- and 800-MHz and Bruker 900-MHz spectrometers. Sequential assignment of the 1H, 13C and 15N protein backbone chemical shifts for isolated domains and fragments was achieved by means of through-bond heteronuclear scalar correlations using standard pulse sequences. Methyl group assignment was accomplished using 3D (H)C(CO)NH, 3D H(C)(CO)NH and 3D 15N- or 13C-edited NOESY-HMQC spectra. All NMR samples were prepared in 50 mM KCl, 50 mM potassium phosphate, 1 mM DTT and 1 g l-1 NaN3 (pH 7.5). Concentrations were 0.3 mM for full-length SecA and 0.3-0.8 mM for the various constructs. Spectra were recorded at 25 °C. Under these conditions, SecA in the presence or absence of the signal peptide is dimeric (Figure S13).
Determination of Distance Restraints from PREs
PRE-derived distances were determined from methyl-TROSY spectra of SecA by measuring peak intensities before (paramagnetic) and after (diamagnetic) reduction of the nitroxide spin label with ascorbic acid. PRE values were then converted to distances by using a modified Solomon-Bloembergen equation for transverse relaxation, as described previously (Battiste and Wagner, 2000). Two sets of restraints were incorporated into subsequent structure calculations. Methyl groups strongly affected by the presence of the spin label in the peptide (Ipara/Idia < 0.15) and whose resonances broaden beyond detection in the paramagnetic spectrum, were restrained with only an upper bound distance estimated from the noise of the spectrum plus 4 Å. Methyl groups whose resonances appear in the paramagnetic spectra (Ipara/Idia < 0.85) were restrained as the calculated distance with ± 4 Å upper/lower bounds. For the peptide with the MTSL at residue position 7 and 25, 66 and 96 (162 it total) inter-molecular restraints were determined (see Supplemental experimental procedures).
Structure Calculation
The structure of the SecA-KRR-LamB signal peptide was calculated using CNS (Brunger et al., 1998), within HADDOCK 2.0 (Dominguez et al., 2003). The crystal structure of E. coli SecA (Papanikolau et al., 2007) was used as the starting conformation of SecA. For the signal peptide, a helical structure was imposed for residues L13 to M22 of the hydrophobic core on the basis of transferred NOESY data of the peptide bound to SecA, while the positively charged N terminus and the polar C terminus were unrestrained. The PRE derived intermolecular distances were introduced as unambiguous restraints. In addition, the chemical shift perturbation data for methyl groups of SecA were used to define ambiguous interaction restraints. The solutions were clustered using a 3.0 Å RMSD cut-off criterion (the RMSD refers to the peptide backbone atoms calculated after fitting on SecA backbone atoms). The twenty lowest-score structures of the lowest score cluster were selected for analysis (see Supplemental experimental procedures).
ITC experiments
All calorimetric titrations were performed on a VP-ITC microcalorimeter (Microcal). Protein samples were extensively dialyzed against the ITC buffer containing 20 mM KPi (pH 7.5), 20 mM KCl, and 1 mM TCEP. The ligand solution was prepared by dissolving peptide in the flow through of the last buffer exchange. Hydrophobic peptides were first dissolved in 100% DMSO and then transferred stepwise to the ITC buffer at a final concentration of 2-4% DMSO. DMSO at the same concentration was also added to the protein solution to match the buffer composition of the peptide. The data were fitted using Origin 7.0 (Microcal).
Supplementary Material
Acknowledgments
We are grateful to B. Pozidis for the MALLS data, S. Backo and O. Uchime for assistance with sample preparation and R. Ghose for useful discussions. This work was supported by US National Institutes of Health grant GM-73854 (to CGK), a Scientist Development Grant by American Heart Association (to CGK), by the European Union (grant LSHG-CT-2005-037586 to AE) and the Greek General Secretariat of Research and the European Regional Development Fund (grants 01AKMON46 and PENED03ED623 to AE). GG is an Onassis Foundation predoctoral fellow.
Footnotes
ACCESSION NUMBERS Structural coordinates are deposited in the Protein Data Bank (ID 2FSF).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe Y, Shodai T, Muto T, Mihara K, Torii H, Nishikawa S, Endo T, Kohda D. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 2000;100:551–560. doi: 10.1016/s0092-8674(00)80691-1. [DOI] [PubMed] [Google Scholar]
- Akita M, Sasaki S, Matsuyama S, Mizushima S. SecA interacts with secretory proteins by recognizing the positive charge at the amino terminus of the signal peptide in Escherichia coli. J Biol Chem. 1990;265:8164–8169. [PubMed] [Google Scholar]
- Battiste JL, Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Bio Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chou YT, Gierasch LM. The conformation of a signal peptide bound by Escherichia coli preprotein translocase SecA. J Biol Chem. 2005;280:32753–32760. doi: 10.1074/jbc.M507532200. [DOI] [PubMed] [Google Scholar]
- Dominguez C, Boelens R, Bonvin AM. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- Fak JJ, Itkin A, Ciobanu DD, Lin EC, Song XJ, Chou YT, Gierasch LM, Hunt JF. Nucleotide exchange from the high-affinity ATP-binding site in SecA is the rate-limiting step in the ATPase cycle of the soluble enzyme and occurs through a specialized conformational state. Biochemistry. 2004;43:7307–7327. doi: 10.1021/bi0357208. [DOI] [PubMed] [Google Scholar]
- Fekkes P, van der Does C, Driessen AJ. The molecular chaperone SecB is released from the carboxy-terminus of SecA during initiation of precursor protein translocation. EMBO J. 1997;16:6105–6113. doi: 10.1093/emboj/16.20.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierasch LM. Signal sequences. Biochemistry. 1989;28:923–930. doi: 10.1021/bi00429a001. [DOI] [PubMed] [Google Scholar]
- Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JE, Wagner G. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell. 2003;115:739–750. doi: 10.1016/s0092-8674(03)00975-9. [DOI] [PubMed] [Google Scholar]
- Hegde RS, Bernstein HD. The surprising complexity of signal sequences. Trends Biochem Sci. 2006;31:563–571. doi: 10.1016/j.tibs.2006.08.004. [DOI] [PubMed] [Google Scholar]
- Hunt JF, Weinkauf S, Henry L, Fak JJ, McNicholas P, Oliver DB, Deisenhofer J. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science. 2002;297:2018–2026. doi: 10.1126/science.1074424. [DOI] [PubMed] [Google Scholar]
- Iwahara J, Clore GM. Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature. 2006;440:1227–1230. doi: 10.1038/nature04673. [DOI] [PubMed] [Google Scholar]
- Karamanou S, Gouridis G, Papanikou E, Sianidis G, Gelis I, Keramisanou D, Vrontou E, Kalodimos CG, Economou A. Preprotein-controlled catalysis in the helicase motor of SecA. EMBO J. 2007 doi: 10.1038/sj.emboj.7601721. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamanou S, Vrontou E, Sianidis G, Baud C, Roos T, Kuhn A, Politou AS, Economou A. A molecular switch in SecA protein couples ATP hydrolysis to protein translocation. Mol Microbiol. 1999;34:1133–1145. doi: 10.1046/j.1365-2958.1999.01686.x. [DOI] [PubMed] [Google Scholar]
- Karamyshev AL, Johnson AE. Selective SecA association with signal sequences in ribosome-bound nascent chains - A potential role for SecA in ribosome targeting to the bacterial membrane. J Biol Chem. 2005;280:37930–37940. doi: 10.1074/jbc.M509100200. [DOI] [PubMed] [Google Scholar]
- Keramisanou D, Biris N, Gelis I, Sianidis G, Karamanou S, Economou A, Kalodimos CG. Disorder-order folding transitions underlie catalysis in the helicase motor of SecA. Nat Struct Mol Biol. 2006;13:594–602. doi: 10.1038/nsmb1108. [DOI] [PubMed] [Google Scholar]
- Kimura E, Akita M, Matsuyama S, Mizushima S. Determination of a region in SecA that interacts with presecretory proteins in Escherichia coli. J Biol Chem. 1991;266:6600–6606. [PubMed] [Google Scholar]
- Kourtz L, Oliver D. Tyr-326 plays a critical role in controlling SecA-preprotein interaction. Mol Microbiol. 2000;37:1342–1356. doi: 10.1046/j.1365-2958.2000.02078.x. [DOI] [PubMed] [Google Scholar]
- Lill R, Dowhan W, Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 1990;60:271–280. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- Matsuo H, Walters KJ, Teruya K, Tanaka T, Gassner GT, Lippard SJ, Kyogoku Y, Wagner G. Identification by NMR spectroscopy of residues at contact surfaces in large, slowly exchanging macromolecular complexes. J Am Chem Soc. 1999;121:9903–9904. [Google Scholar]
- Mitchell C, Oliver D. Two distinct ATP-binding domains are needed to promote protein export by Escherichia coli SecA ATPase. Mol Microbiol. 1993;10:483–497. doi: 10.1111/j.1365-2958.1993.tb00921.x. [DOI] [PubMed] [Google Scholar]
- Mitra K, Frank J, Driessen A. Co- and post-translational translocation through the protein-conducting channel: analogous mechanisms at work? Nat Struct Mol Biol. 2006;13:957–964. doi: 10.1038/nsmb1166. [DOI] [PubMed] [Google Scholar]
- Mori H, Araki M, Hikita C, Tagaya M, Mizushima S. The hydrophobic region of signal peptides is involved in the interaction with membrane-bound SecA. Biochim Biophys Acta. 1997;1326:23–36. doi: 10.1016/s0005-2736(97)00004-7. [DOI] [PubMed] [Google Scholar]
- Mori H, Ito K. Different modes of SecY-SecA interactions revealed by site-directed in vivo photo-cross-linking. Proc Natl Acad Sci USA. 2006;103:16159–16164. doi: 10.1073/pnas.0606390103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musial-Siwek M, Rusch SL, Kendall DA. Selective photoaffinity labeling identifies the signal peptide binding domain on SecA. J Mol Biol. 2007;365:637–648. doi: 10.1016/j.jmb.2006.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne AR, Clemons WM, Jr, Rapoport TA. A large conformational change of the translocation ATPase SecA. Proc Natl Acad Sci USA. 2004;101:10937–10942. doi: 10.1073/pnas.0401742101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne AR, Rapoport TA, van den Berg B. Protein Translocation by the Sec61/Secy Channel. Annu Rev Cell Dev Biol. 2005;21:529–550. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- Osborne AR, Rapoport TA. Protein Translocation Is Mediated by Oligomers of the SecY Complex with One SecY Copy Forming the Channel. Cell. 2007;129:97–110. doi: 10.1016/j.cell.2007.02.036. [DOI] [PubMed] [Google Scholar]
- Papanikolau Y, Papadovasilaki M, Ravelli RB, McCarthy AA, Cusack S, Economou A, Petratos K. Structure of Dimeric SecA, the Escherichia coli Preprotein Translocase Motor. J Mol Biol. 2007;366:1545–1557. doi: 10.1016/j.jmb.2006.12.049. [DOI] [PubMed] [Google Scholar]
- Papanikou E, Karamanou S, Baud C, Frank M, Sianidis G, Keramisanou D, Kalodimos CG, Kuhn A, Economou A. Identification of the preprotein binding domain of SecA. J Biol Chem. 2005;280:43209–43217. doi: 10.1074/jbc.M509990200. [DOI] [PubMed] [Google Scholar]
- Randall LL, Hardy SJ. SecB, one small chaperone in the complex milieu of the cell. Cell Mol Life Sci. 2002;59:1617–1623. doi: 10.1007/PL00012488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randall LL, Topping TB, Hardy SJ, Pavlov MY, Freistroffer DV, Ehrenberg M. Binding of SecB to ribosome-bound polypeptides has the same characteristics as binding to full-length, denatured proteins. Proc Natl Acad Sci USA. 1997;94:802–807. doi: 10.1073/pnas.94.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosild TP, Greenwald J, Vega M, Castronovo S, Riek R, Choe S. NMR structure of Mistic, a membrane-integrating protein for membrane protein expression. Science. 2005;307:1317–1321. doi: 10.1126/science.1106392. [DOI] [PubMed] [Google Scholar]
- Schiebel E, Driessen AJ, Hartl FU, Wickner W. Delta mu H+ and ATP function at different steps of the catalytic cycle of preprotein translocase. Cell. 1991;64:927–939. doi: 10.1016/0092-8674(91)90317-r. [DOI] [PubMed] [Google Scholar]
- Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- Sharma V, Arockiasamy A, Ronning DR, Savva CG, Holzenburg A, Braunstein M, Jacobs WR, Jr, Sacchettini JC. Crystal structure of Mycobacterium tuberculosis SecA, a preprotein translocating ATPase. Proc Natl Acad Sci USA. 2003;100:2243–2248. doi: 10.1073/pnas.0538077100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sianidis G, Karamanou S, Vrontou E, Boulias K, Repanas K, Kyrpides N, Politou AS, Economou A. Cross-talk between catalytic and regulatory elements in a DEAD motor domain is essential for SecA function. EMBO J. 2001;20:961–970. doi: 10.1093/emboj/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprangers R, Kay LE. Quantitative dynamics and binding studies of the 20S proteasome by NMR. Nature. 2007;445:618–622. doi: 10.1038/nature05512. [DOI] [PubMed] [Google Scholar]
- Sprangers R, Velyvis A, Kay LE. Solution NMR of supramolecular complexes: providing new insights into function. Nat Methods. 2007;4:697–703. doi: 10.1038/nmeth1080. [DOI] [PubMed] [Google Scholar]
- Tang C, Iwahara J, Clore GM. Visualization of transient encounter complexes in protein-protein association. Nature. 2006;444:383–386. doi: 10.1038/nature05201. [DOI] [PubMed] [Google Scholar]
- Triplett TL, Sgrignoli AR, Gao FB, Yang YB, Tai PC, Gierasch LM. Functional signal peptides bind a soluble N-terminal fragment of SecA and inhibit its ATPase activity. J Biol Chem. 2001;276:19648–19655. doi: 10.1074/jbc.M100098200. [DOI] [PubMed] [Google Scholar]
- Ureta AR, Endres RG, Wingreen NS, Silhavy TJ. Kinetic analysis of the assembly of the outer membrane protein LamB in Escherichia coli mutants each lacking a secretion or targeting factor in a different cellular compartment. J Bacteriol. 2007;189:446–454. doi: 10.1128/JB.01103-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Mori H, Vassylyeva MN, Tsukazaki T, Kimura Y, Tahirov TH, Ito K. Crystal Structure of the Translocation ATPase SecA from Thermus thermophilus Reveals a Parallel, Head-to-Head Dimer. J Mol Biol. 2006;364:248–258. doi: 10.1016/j.jmb.2006.09.061. [DOI] [PubMed] [Google Scholar]
- Volkov AN, Worrall JA, Holtzmann E, Ubbink M. Solution structure and dynamics of the complex between cytochrome c and cytochrome c peroxidase determined by paramagnetic NMR. Proc Natl Acad Sci USA. 2006;103:18945–18950. doi: 10.1073/pnas.0603551103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- Zhou J, Xu Z. Structural determinants of SecB recognition by SecA in bacterial protein translocation. Nat Struct Biol. 2003;10:942–947. doi: 10.1038/nsb980. [DOI] [PubMed] [Google Scholar]
- Wang L, Miller A, Rusch SL, Kendall DA. Demonstration of a specific Escherichia coli SecY-signal peptide interaction. Biochemistry. 2004;43:13185–13192. doi: 10.1021/bi049485k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Miller A, Kendall DA. Signal peptide determinants of SecA binding and stimulation of ATPase activity. J Biol Chem. 2000;275:10154–10159. doi: 10.1074/jbc.275.14.10154. [DOI] [PubMed] [Google Scholar]
- Wickner W, Schekman R. Protein translocation across biological membranes. Science. 2005;310:1452–1456. doi: 10.1126/science.1113752. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.