Abstract
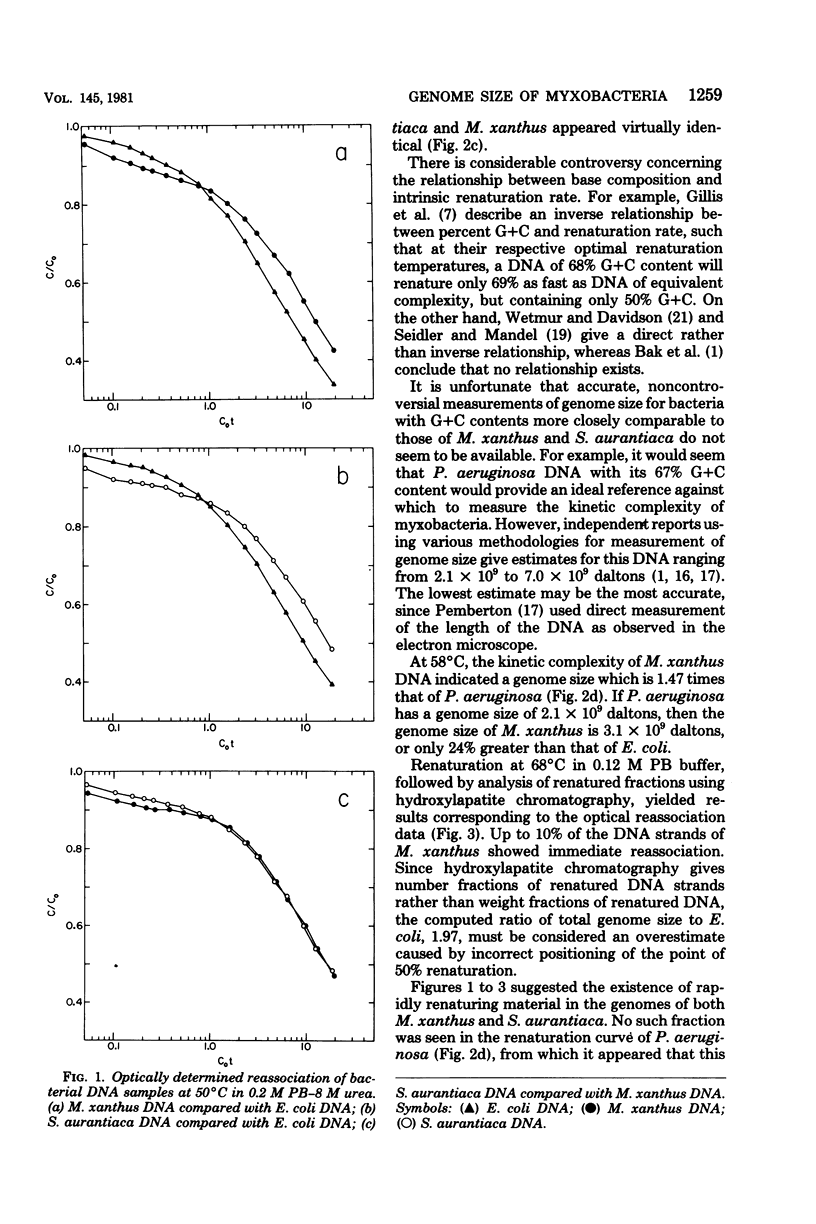
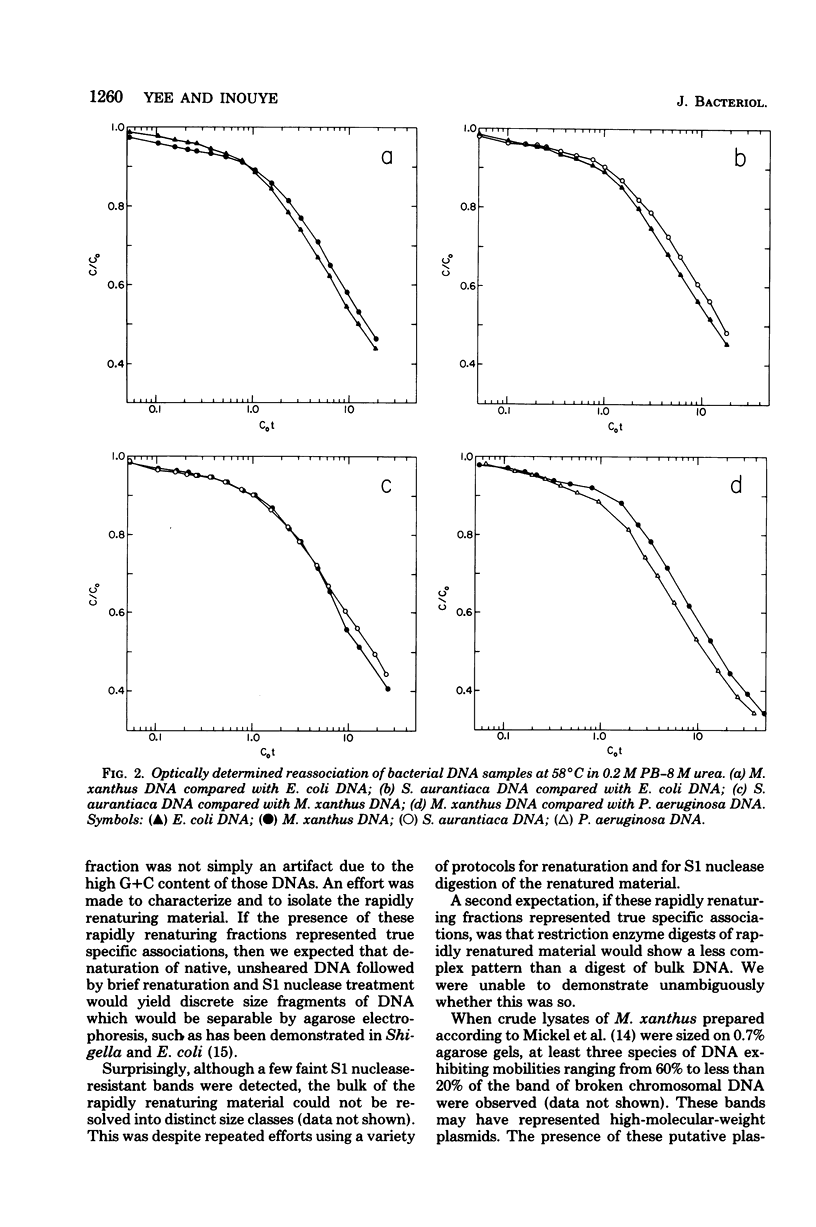
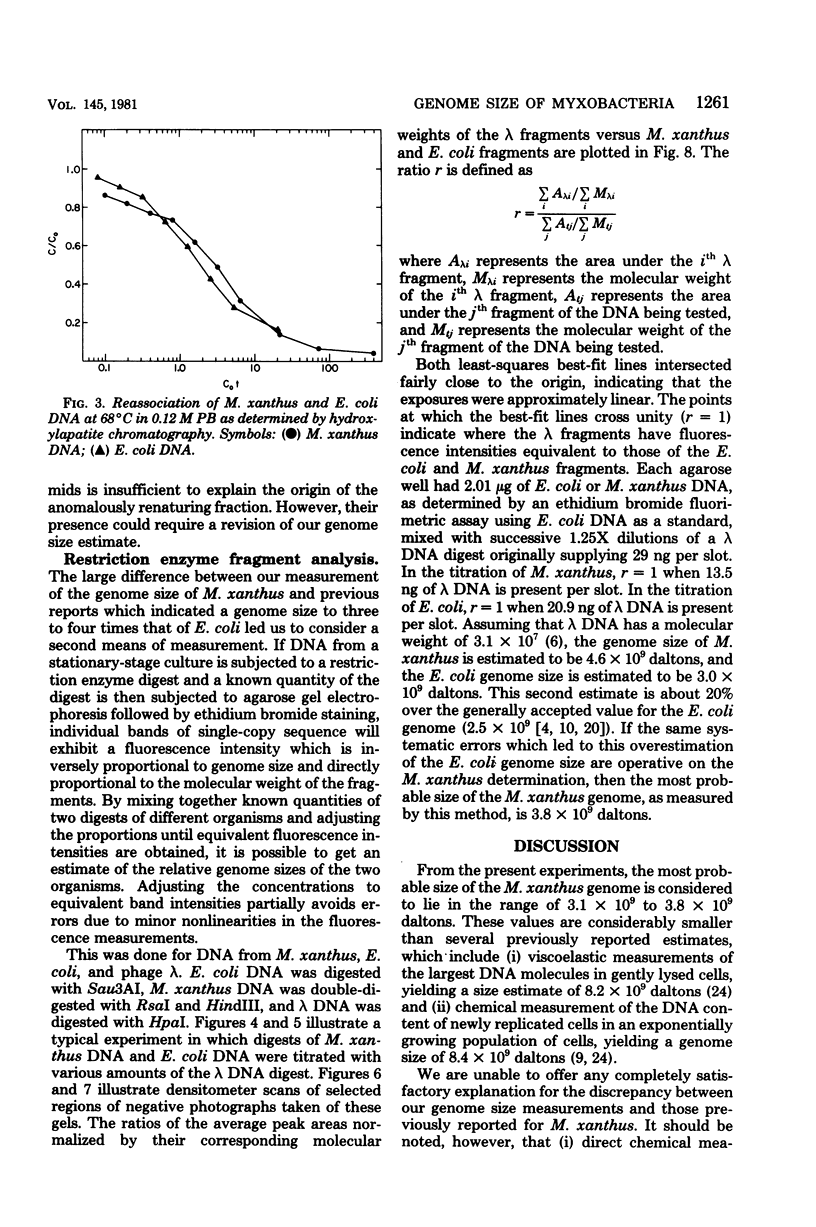
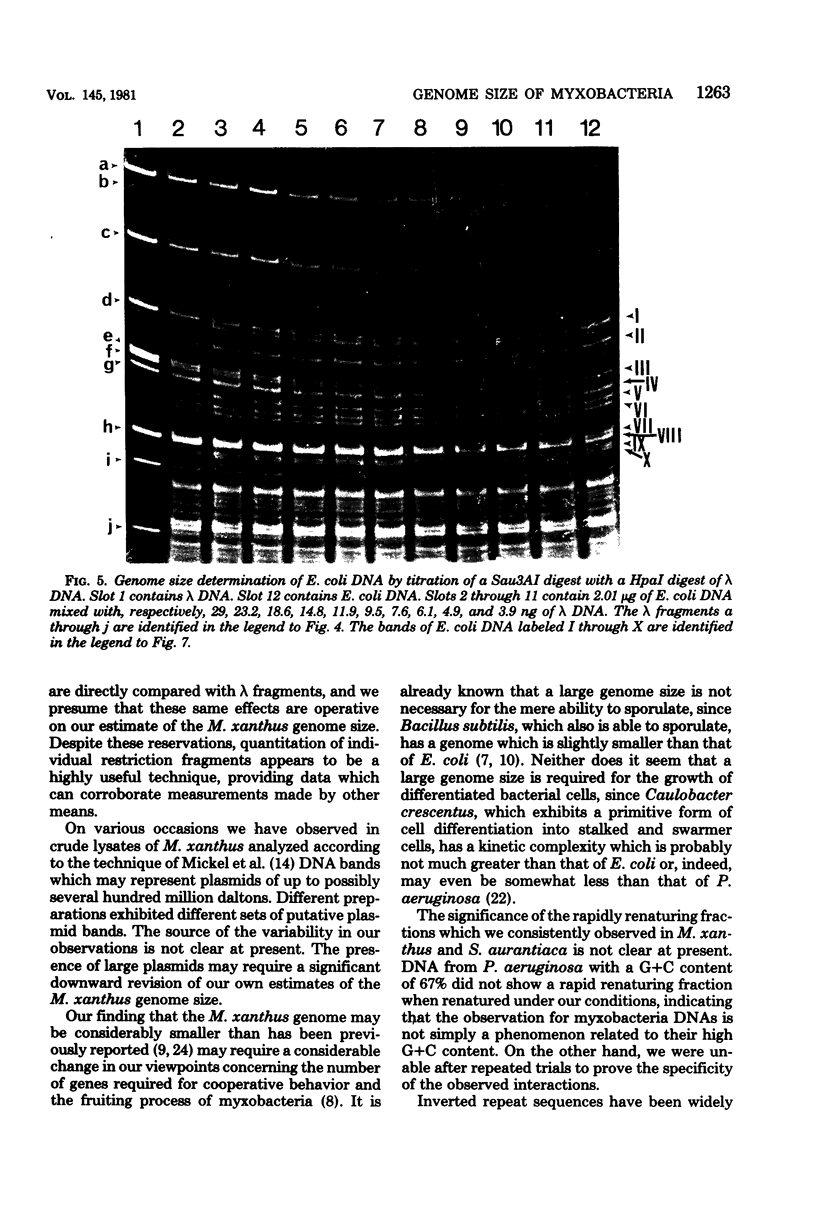
The genome sizes of two myxobacteria, Myxococcus xanthus and Stigmatella aurantiaca, were measured by renaturation analysis and also by a new method involving the quantitation of individual restriction fragments. In contrast to several previous reports, which indicate that M. xanthus has a genome size which is three to four times that of Escherichia coli, the present measurements indicated that the M. xanthus genome is only about 24 to 53% larger than that of E. coli. S. aurantiaca had a genome size nearly identical to that of M. xanthus. Of possible significance is the fact that the renaturation curves of M. xanthus and S. aurantiaca deoxyribonucleic acid both exhibited significant fractions which renatured with rapid, unimolecular kinetics. However, we were unable to establish that these fractions represented inverted repeats of repetitive sequences.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bak A. L., Christiansen C., Stenderup A. Bacterial genome sizes determined by DNA renaturation studies. J Gen Microbiol. 1970 Dec;64(3):377–380. doi: 10.1099/00221287-64-3-377. [DOI] [PubMed] [Google Scholar]
- Bendich A. J., Anderson R. S. Characterization of families of repeated DNA sequences from four vascular plants. Biochemistry. 1977 Oct 18;16(21):4655–4663. doi: 10.1021/bi00640a020. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Graham D. E., Neufeld B. R. Analysis of repeating DNA sequences by reassociation. Methods Enzymol. 1974;29:363–418. doi: 10.1016/0076-6879(74)29033-5. [DOI] [PubMed] [Google Scholar]
- CAIRNS J. The bacterial chromosome and its manner of replication as seen by autoradiography. J Mol Biol. 1963 Mar;6:208–213. doi: 10.1016/s0022-2836(63)80070-4. [DOI] [PubMed] [Google Scholar]
- Campos J. M., Geisselsoder J., Zusman D. R. Isolation of bacteriophage MX4, a generalized transducing phage for Myxococcus xanthus. J Mol Biol. 1978 Feb 25;119(2):167–178. doi: 10.1016/0022-2836(78)90431-x. [DOI] [PubMed] [Google Scholar]
- Gillis M., De Ley J., De Cleene M. The determination of molecular weight of bacterial genome DNA from renaturation rates. Eur J Biochem. 1970 Jan;12(1):143–153. doi: 10.1111/j.1432-1033.1970.tb00831.x. [DOI] [PubMed] [Google Scholar]
- Kaiser D., Manoil C., Dworkin M. Myxobacteria: cell interactions, genetics, and development. Annu Rev Microbiol. 1979;33:595–639. doi: 10.1146/annurev.mi.33.100179.003115. [DOI] [PubMed] [Google Scholar]
- Kimchi A., Rosenberg E. Linkages between deoxyribonucleic acid synthesis and cell division in Myxococcus xanthus. J Bacteriol. 1976 Oct;128(1):69–79. doi: 10.1128/jb.128.1.69-79.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz L. C., Zimm B. H. Size of DNA determined by viscoelastic measurements: results on bacteriophages, Bacillus subtilis and Escherichia coli. J Mol Biol. 1972 Dec 30;72(3):779–800. doi: 10.1016/0022-2836(72)90191-x. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Mandel M. Deoxyribonucleic acid base composition in the genus Pseudomonas. J Gen Microbiol. 1966 May;43(2):273–292. doi: 10.1099/00221287-43-2-273. [DOI] [PubMed] [Google Scholar]
- Mandel M., Leadbetter E. R. Deoxyribonucleic acid base composition of myxobacteria. J Bacteriol. 1965 Dec;90(6):1795–1796. doi: 10.1128/jb.90.6.1795-1796.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickel S., Arena V., Jr, Bauer W. Physical properties and gel electrophoresis behavior of R12-derived plasmid DNAs. Nucleic Acids Res. 1977;4(5):1465–1482. doi: 10.1093/nar/4.5.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park I. W., De Ley J. Ancestral remnants in deoxyribonucleic acid from Pseudomonas and Xanthomonas. Antonie Van Leeuwenhoek. 1967;33(1):1–16. doi: 10.1007/BF02045528. [DOI] [PubMed] [Google Scholar]
- Pemberton J. M. Size of the chromosome of Pseudomonas aeruginosa PAO. J Bacteriol. 1974 Sep;119(3):748–752. doi: 10.1128/jb.119.3.748-752.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts T. M., Lauer G. D., Klotz L. C. Physical studies on DNA from "primitive" eucaryotes. CRC Crit Rev Biochem. 1975;3(4):349–449. [PubMed] [Google Scholar]
- Seidler R. J., Mandel M. Quantitative aspects of deoxyribonucleic acid renaturation: base composition, state of chromosome replication, and polynucleotide homologies. J Bacteriol. 1971 May;106(2):608–614. doi: 10.1128/jb.106.2.608-614.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D. C., Morowitz H. J. Genome size and evolution. Chromosoma. 1973;40(2):121–126. doi: 10.1007/BF00321457. [DOI] [PubMed] [Google Scholar]
- Wetmur J. G., Davidson N. Kinetics of renaturation of DNA. J Mol Biol. 1968 Feb 14;31(3):349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]
- Wood N. B., Rake A. V., Shapiro L. Structure of Caulobacter deoxyribonucleic acid. J Bacteriol. 1976 Jun;126(3):1305–1315. doi: 10.1128/jb.126.3.1305-1315.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zusman D. R., Krotoski D. M., Cumsky M. Chromosome replication in Myxococcus xanthus. J Bacteriol. 1978 Jan;133(1):122–129. doi: 10.1128/jb.133.1.122-129.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]




