Abstract
An unbiased photo–cross-linking approach was used to probe the “molecular path” of a growing nascent Escherichia coli inner membrane protein (IMP) from the peptidyl transferase center to the surface of the ribosome. The nascent chain was initially in proximity to the ribosomal proteins L4 and L22 and subsequently contacted L23, which is indicative of progression through the ribosome via the main ribosomal tunnel. The signal recognition particle (SRP) started to interact with the nascent IMP and to target the ribosome–nascent chain complex to the Sec–YidC complex in the inner membrane when maximally half of the transmembrane domain (TM) was exposed from the ribosomal exit. The combined data suggest a flexible tunnel that may accommodate partially folded nascent proteins and parts of the SRP and SecY. Intraribosomal contacts of the nascent chain were not influenced by the presence of a functional TM in the ribosome.
Introduction
The signal recognition particle (SRP) is a conserved ribonucleoprotein complex that directs the cotranslational targeting of membrane and secretory proteins to the ER membrane in eukaryotes and to the cytoplasmic or inner membrane in prokaryotes (Koch et al., 2003). Hydrophobic targeting signals in nascent proteins are specifically recognized by the SRP as they emerge from the ribosome. The SRP–ribosome–nascent chain complex then interacts with the SRP receptor (SR), leading to the transfer of the signal peptide into the translocation channel and, subsequently, to the dissociation of the SRP–SR complex. This vectorial process is controlled by GTPase activities in the subunits of the SRP and SR.
Recent evidence indicates that the ribosomal tunnel is more than a passive conduit for the nascent chain. It plays an important regulatory role at several stages in SRP-mediated targeting and membrane integration. First, the eukaryotic ribosomal proteins L23a and L35 have been shown to constitute the ribosome attachment site for SRP54, the signal sequence-binding component of the mammalian SRP (Pool et al., 2002; Halic et al., 2004). These ribosomal proteins are located close to the putative main exit site for nascent chains and, thus, position SRP54 to scan emerging polypeptides for the presence of targeting signals. It is interesting to note that the eukaryotic SRP has a higher affinity for active, translating ribosomes than for those that are inactive, even before a nascent chain emerges at the ribosomal surface (Flanagan et al., 2003). Second, there is evidence that the nature of passing polypeptides is already sensed in the ribosomal tunnel between the peptidyl transferase center (PTC) and the exit site (Liao et al., 1997; Nakatogawa and Ito, 2002). In particular, a transmembrane domain (TM) in a nascent membrane protein that is completely buried in the ribosomal tunnel was shown to induce conformational changes in the Sec–translocation complex in the ER membrane (Liao et al., 1997). These changes may be transduced by specific interactions of ribosomal proteins with a TM inside the ribosome (Woolhead et al., 2004). Third, the exit tunnel is more dynamic than previously anticipated and may expand during protein synthesis, allowing significant portions of the nascent polypeptide to fold and accumulate in the ribosome (Berisio et al., 2003; Gilbert et al., 2004).
Although most structural and mechanistic characteristics of pro- and eukaryotic SRP–SR targeting systems seem conserved, there are some notable differences. The substrate specificity of the Escherichia coli SRP is restricted to inner membrane proteins (IMPs) and a limited number of secretory proteins, whereas the eukaryotic SRP has a more generic function in the targeting of both secretory and membrane proteins. The E. coli SRP is structurally less complex than its eukaryotic counterpart, as it is composed of only one protein: Ffh (54 homologue) and a relatively small 4.5S RNA. In contrast to the eukaryotic SRP, the E. coli SRP does not seem to impose a translational arrest upon binding to a substrate nascent polypeptide (Raine et al., 2003). Although the docking site for the E. coli SRP is conserved (L23, the prokaryotic homologue of L23a; Gu et al., 2003; Ullers et al., 2003), it is shared with trigger factor (TF; Kramer et al., 2002; Ferbitz et al., 2004), which is a chaperone and prolyl isomerase that does not have a homologue in the eukaryotic cytosol. TF has been found in the proximity of nascent cytosolic, secretory, and membrane proteins, and its role in protein targeting and folding is debated (Beck et al., 2000; Bernstein and Hyndman, 2001; Ullers et al., 2003). It has been proposed that the nature of a nascent polypeptide is already sensed in the ribosome, which may influence the binding or positioning of SRP and TF at L23 near the nascent chain exit site (Gu et al., 2003; Ullers et al., 2003).
In this study, we describe interactions of a nascent E. coli IMP that is stalled at distinct early stages in protein synthesis. Interactions with ribosomal proteins, cytosolic chaperones, targeting factors, and translocase components were probed in a homologous in vitro translation system using an unbiased photo–cross-linking approach. These analyses were designed to determine the molecular path of the nascent IMP within and outside the exit tunnel and to investigate the likelihood of (partial) folding in the ribosome. The dynamics, timing, and specificity of the observed interactions with ribosomal proteins, TF, and SRP were analyzed in relation to the presence of a functional targeting sequence in the nascent chain.
Results
Model protein and experimental approach
Photo–cross-linking was used to study the contacts of a nascent IMP, leader peptidase (Lep), during its progression through the ribosome and its first interactions with chaperones and targeting factors. Lep has been used previously as a model protein to study various aspects of targeting and integration of membrane proteins in both the prokaryotic inner membrane and eukaryotic ER membrane. Both in vivo and in vitro experiments have indicated that in E. coli, Lep follows what is considered the main pathway for IMP biogenesis: targeting to the inner membrane by the SRP and integration via the Sec–YidC insertion machinery (de Gier et al., 1996; Houben et al., 2000, 2002; Samuelson et al., 2000).
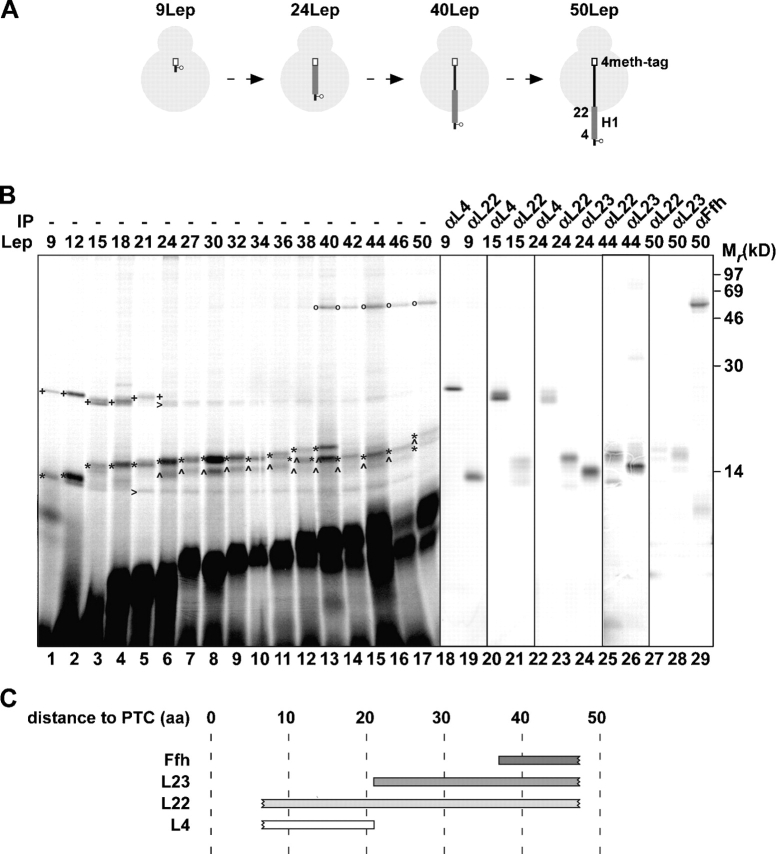
Lep nascent chains with a length of 9–50 amino acids were prepared by in vitro translation in the presence of [35S]methionine using truncated mRNAs that included a COOH-terminal sequence containing four methionines (4meth-tag; GlySer(Met)4) to increase the labeling efficiency (Fig. 1 A). 50Lep was chosen as the longest translation intermediate because it represents the shortest intermediate known to integrate into the membrane at the Sec–YidC insertion site (Houben et al., 2002). To enable site-directed photo–cross-linking, a TAG (stop) codon was introduced at position 3 just upstream of the first TM of Lep (H1). The TAG codon was suppressed during translation by adding a suppressor tRNA that carries a phenylalanine coupled to a photoreactive cross-linking (Tmd) probe (see Materials and methods). After translation, the probe was activated by UV irradiation to covalently link nascent Lep to any molecules that are in close proximity in the E. coli translation lysate. Cross-linked ribosome–nascent chain complexes were then sedimented through a high salt sucrose cushion and were further analyzed. The Lep 48mer failed to be synthesized and was not included in these studies. It should be noted that this cross-linking technique makes use of artificially arrested nascent polypeptides. It cannot be formally excluded that these chains adopt conformations that are less likely in vivo.
Figure 1.
Progression of nascent Lep through the ribosome. (A) Schematic representation of the 9, 24, 40, and 50Lep constructs with a cross-linking probe at position 3. The transmembrane regions (H1) and methionine tags are presented as thick gray lines and white bars, respectively. (B) In vitro translation of nascent 9–50LepTAG3 constructs was performed in the presence of (Tmd)Phe-tRNAsup. After translation, samples were irradiated with UV light to induce cross-linking, and the ribosome–nascent chain complexes were purified and analyzed by SDS-PAGE. UV-irradiated 9, 15, 24, 44, and 50LepTAG3 were immunoprecipitated with antisera as indicated. (C) Schematic representation of the cross-links observed in B. Numbers indicate the distance in amino acids of the cross-linking probe to the PTC. Images in different panels represent different parts of the gel or different exposure times. +, L4 cross-link; *, L22 cross-link; ^, L23 cross-link; o, Ffh cross-link; >, L4 and L22 cross-link to a truncated translation product.
Progression of nascent Lep through the ribosome
Upon synthesis and analysis of nascent Lep with a length of nine amino acids (9LepTAG3), two clear cross-linking products of ∼25 and 14 kD could be detected (Fig. 1 B, lane 1). Immunoprecipitation identified the ribosomal proteins L4 and L22, respectively, as the cross-linking partners in these products (Fig. 1 B, lanes 18 and 19). The diffuse signal at ∼10 kD was also visible without UV irradiation, indicating it is not a specific cross-linked product (unpublished data). Cross-linking to L4 was observed up to a nascent chain length of 24 amino acids. In contrast, L22 continued to be cross-linked up to 50Lep, which was the longest construct studied (Fig. 1 B, lanes 20–23, 25, and 27). Nascent Lep of 24–50 amino acids generated an additional cross-linking product of ∼14–16 kD, which could be immunoprecipitated with anti-L23 (Fig. 1 B, lanes 24, 26, and 28). Finally, an ∼55-kD cross-linking product appeared using nascent Lep of 40 amino acids and longer, which could be immunoprecipitated using anti-Ffh (Fig. 1 B, lane 29). The weak cross-linking adducts of ∼25 and 14 kD to almost all nascent chains (Fig. 1 B, carrats) contained L4 and L22, respectively (verified by immunoprecipitations of 15, 24, and 44Lep). These adducts apparently represented cross-linking to a truncated translation product of about nine amino acids because they ran at the same position as L4 and L22 cross-linked to 9LepTAG3.
In conclusion, these cross-linking data show that Lep progresses through the ribosome via the main ribosomal tunnel, where it contacts or is adjacent to successively L4, L23, and Ffh during nascent chain elongation, whereas L22 is near all tested nascent chain lengths (Fig. 1 C). L4, L22, and L23 decorate, together with the 23S RNA, the main ribosomal tunnel wall (Harms et al., 2001). L4 is only exposed at the most constricted part of the tunnel, not far from the PTC, whereas a long β-hairpin of L22 lies more or less parallel to the tunnel axis. L23 penetrates with a tail into the lower part of the tunnel wall, whereas its globular domain is located at the surface of the ribosome near the ribosomal exit site and functions in the docking of SRP (see Fig. 5).
Figure 5.
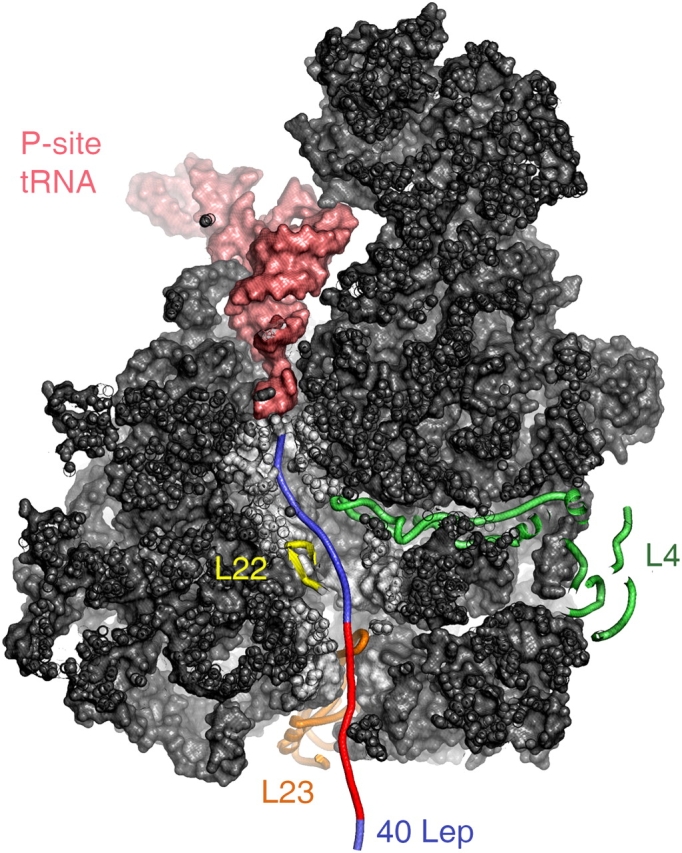
Tunnel view of the large ribosomal subunit from Deinococcus radiodurans. The 40–amino acid nascent Lep (blue) and TM (red) is stretched from P-site tRNA (salmon pink) to the ribosomal exit tunnel and makes contacts with ribosomal proteins L22 (yellow), L4 (green), and L23 (orange).
The initial targeting and insertion steps of Lep
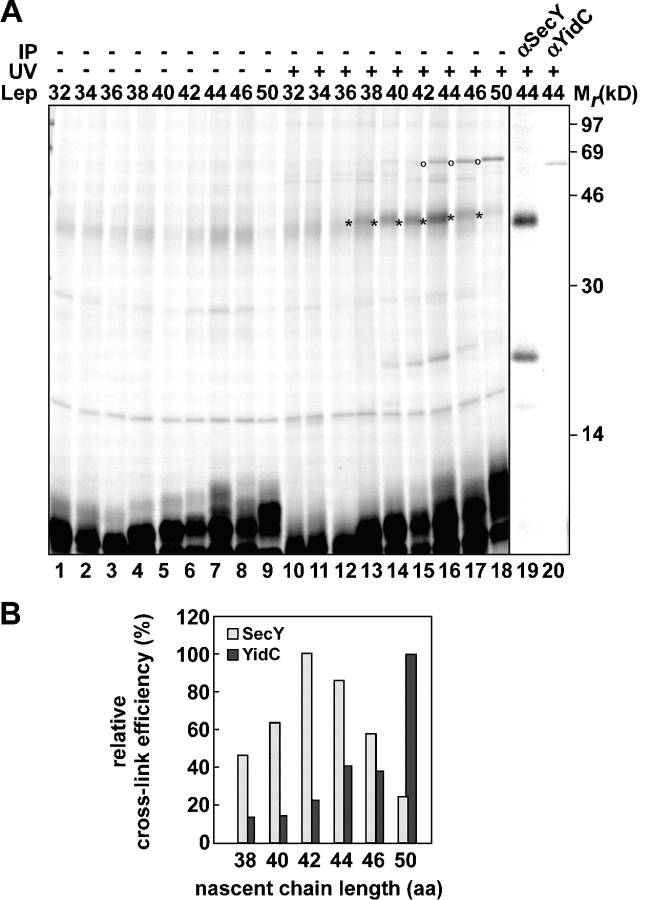
A previous cross-linking study has shown that nascent Lep with a length of 50 amino acids can be targeted to the membrane (Houben et al., 2002). H1 in 50Lep already cross-linked SecY and YidC, although full membrane integration was observed only with constructs that were longer than 70 amino acids (Houben et al., 2002). The remarkably early interaction of nascent Lep with the targeting factor SRP (see previous section) prompted us to reevaluate early contacts of nascent Lep with membrane components. In short, 32–50LepTAG3 were produced in the presence of inverted inner membrane vesicles (IMVs). After translation, one half of each sample was UV irradiated, whereas the other half was kept in the dark to serve as a control. Membranes were treated with sodium carbonate, and membrane-integrated material was collected by centrifugation (Fig. 2 A).
Figure 2.
The initial targeting and insertion steps of Lep. (A) In vitro translation of nascent 32–50LepTAG3 was performed in the presence of IMVs and (Tmd)Phe-tRNAsup. After translations, samples were kept in the dark or UV irradiated and were subsequently extracted with sodium carbonate and spun down. UV-irradiated pellet fractions of 44LepTAG3 were immunoprecipitated as indicated. (B) Quantifications of SecY and YidC cross-linking to 38–50LepTAG3 (A, lanes 13–18) relative to the amount of nonirradiated carbonate-resistant nascent chains (A, lanes 4–9). Highest values for cross-linking efficiency were taken as 100%. Images in different panels represent different parts of the gel or different exposure times. *, SecY cross-link; o, YidC cross-link.
32–36LepTAG3 generated no cross-linking products in the membrane fractions, suggesting that these short nascent chains were not targeted to the membrane-embedded Sec–YidC complex. In contrast, 38–50Lep gave rise to a cross-linking product of ∼45 kD (Fig. 2 A, lanes 13–18) that represented cross-linking to SecY (verified by immunoprecipitation of 44Lep; Fig. 2 A, lane 19). In addition, a ∼68-kD cross-linking product could be observed with 38–42LepTAG3 and more clearly with 44–50LepTAG3 (Fig. 2 A, lanes 16–18). Immunoprecipitation of 44Lep identified YidC as the cross-linking partner (Fig. 2 A, lane 20). Finally, a ∼20-kD cross-linking adduct was detected that could be immunoprecipitated with anti-SecY and most likely represents a breakdown product of the SecY adduct, as observed previously (Urbanus et al., 2001; Houben et al., 2002). The release of 40 and 44Lep nascent chains from the ribosome by puromycin or EDTA treatment before UV irradiation abolished cross-linking to Ffh, SecY, and YidC (unpublished data). This confirms that the interaction of the nascent chains with SRP and their subsequent targeting to Sec–YidC occurred strictly in the context of the ribosome.
Quantification of the cross-link efficiencies (Fig. 2 B) revealed an optimum for SecY cross-linking at a nascent chain length of 42 amino acids. Cross-linking to YidC increased considerably, concomitant with a decreased SecY cross-linking from 44 to 50 LepTAG3. By interpreting these results in the context of the dynamic translation process, it can be concluded that position 3 in Lep is initially targeted to the Sec–translocon but moves rapidly to YidC during nascent chain elongation.
Together, the data suggest that targeting of Lep to the Sec–YidC insertion site starts at a nascent chain length of 38 amino acids and is mediated by the SRP. Without membranes, position 3 in 38Lep might be oriented away from Ffh, explaining the absence of Ffh cross-linking to 38LepTAG3. Remarkably, even in a fully extended conformation of the nascent chain, less than half (about eight residues) of H1 in 38Lep is exposed outside the ribosomal exit site (concluded from molecular modeling; see Fig. 5). This suggests that domains of both SRP and SecY may have access to the interior of the translating ribosome. Alternatively, the presence of H1 is sensed in the ribosome, which increases the affinity of ribosomal proteins near the exit site and emerging nascent chain for SRP and SecY.
Cross-linking to L4, L22, and L23 is not dependent on the nature of the nascent polypeptide
Recruitment of SRP to the ribosomal exit site might be indirectly induced by specific interactions of nascent Lep within the ribosomal tunnel. In this scenario, H1 of Lep that is still buried inside the tunnel would be expected to play a decisive discriminatory role. Because we now have a detailed map of the interactions of nascent Lep with L4, L22, and L23 as it moves through the ribosomal tunnel, we decided to examine if these interactions depend on a functional H1 domain. H1 in LepTAG3 constructs was “knocked out” by replacing four hydrophobic Leu residues with charged residues (Glu, Arg, and Lys) and a helix-breaking residue (Pro; Fig. 3 A). 15, 24, 34, and 44mers of the resulting Lep knockout (LepKOTAG3) were synthesized, cross-linked, and analyzed (Fig. 3 B).
Figure 3.
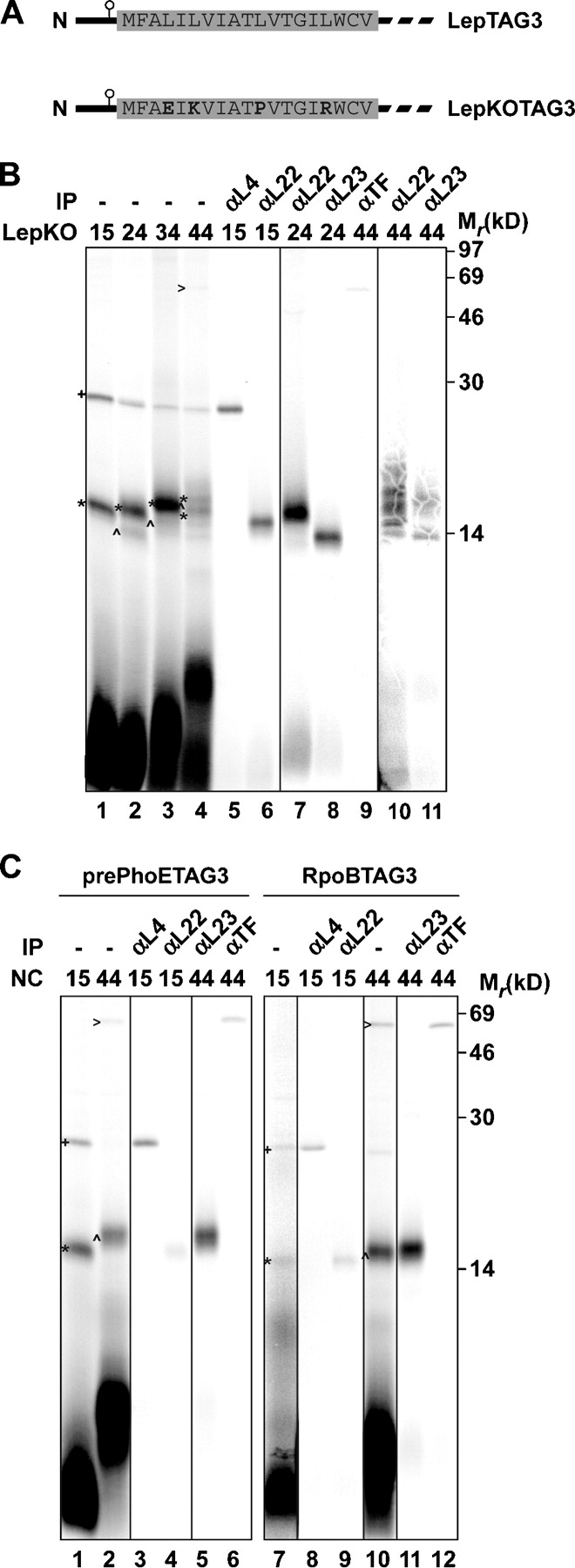
Interactions of nascent LepKO, pre-PhoE, and RpoB with ribosomal proteins and cytosolic chaperones. (A) Schematic representation of LepTAG3 and LepKOTAG3. The sequences of wild-type and mutated H1 are depicted. (B and C) 15–44LepKOTAG3 (B) and 15/44pre-PhoETAG3 and 15/44RpoBTAG3 (C) were produced, cross-linked, purified, and immunoprecipitated as described in Fig. 1. Images in different panels represent different parts of the gel or different exposure times. +, L4 cross-link; *, L22 cross-link; ^, L23 cross-link; >, TF cross-link.
As expected, combined mutations in the KO mutant rendered H1 not sufficiently hydrophobic to sustain cross-linking to Ffh in the 44mer (Fig. 3 B, lane 4). Instead, a weak cross-linked partner of ∼50 kD was observed that could be immunoprecipitated by using antiserum against TF (Fig. 3 B, lane 9), which is a cytosolic chaperone and peptidyl-prolyl isomerase that has been cross-linked to nascent chains of various origin (Valent et al., 1995; Hesterkamp et al., 1996; Beck et al., 2000; Ullers et al., 2004). In addition, although 44LepTAG3 revealed clear interactions with membrane components after translation in the presence of membranes (see The initial targeting and insertion steps of Lep), no such interactions could be observed with 44LepKOTAG3 (unpublished data). These findings confirm that mutated H1 is not a functional TM and, therefore, is not recognized as such by the SRP.
With respect to ribosomal proteins, the cross-link patterns of LepKOTAG3 nascent chains did not significantly differ from their wild-type counterparts (Fig. 1 B). The 15mer showed cross-linking to L4 and L22, whereas the 24, 34, and 44mers were cross-linked to L22 and L23 (Fig. 3 B, lanes 1–4; immunoprecipitations in lanes 5–8, 10, and 11). These results indicated that the order and nature of intraribosomal contacts with L4, L22, and L23 in nascent Lep do not change in response to the presence of a functional downstream TM, at least when probed from position 3 in the nascent polypeptide.
To further investigate the extent to which contacts with ribosomal proteins are dependent on the nature of the nascent polypeptide, intraribosomal interactions of a nascent outer membrane protein (PhoE) and a cytoplasmic protein (RpoB) were examined. PhoE is synthesized with a cleavable signal sequence and is dependent on SecB for efficient targeting to the inner membrane (Kusters et al., 1989). RpoB is the β subunit of the E. coli RNA polymersase. 15 and 44mers of pre-PhoE and RpoB with the photo–cross-linker at position 3 were synthesized, cross-linked, and analyzed (Fig. 3 C).
The cross-link patterns of both pre-PhoE and RpoB were very similar to those obtained with the LepKO constructs of the same lengths and TAG position. The 15mers of pre-PhoE and RpoB showed cross-linking to L4 and L22, whereas the 44mers revealed clear cross-linking to L23 and TF (Fig. 3 C, lanes 1, 2, 7, and 8; immunoprecipitations in lanes 3–6 and 9–12). These results indicate that pre-PhoE and RpoB follow the same path through the ribosome as Lep. Most important, the nature and sequence of contacts with L4, L22, and L23 inside the ribosome appear to be independent of the future destination of the nascent polypeptide. In addition, the combined data suggest that TF is adjacent to all (partially) exposed sequences that are not sufficiently hydrophobic to constitute a TM.
SRP is not oriented toward the ribosomal exit before it is able to interact with H1
It has been suggested that structural information can be sensed in the exit tunnel and transduced to the ribosomal surface to influence the binding of SRP and TF near the exit site (Gu et al., 2003; Ullers et al., 2003). Although differences in intraribosomal cross-linking between nascent Lep derivatives with a functional or compromised TM were not detected (see the previous section), subtle changes in ribosomal contacts and conformation in response to a functional TM cannot be excluded. To more directly examine a possible recruitment of SRP at the nascent chain exit site by Lep H1 present in the ribosome interior, we designed and executed the following experiment. 30Lep, in which H1 is completely buried in the ribosomal tunnel, was extended at its NH2 terminus with an arbitrary hydrophilic peptide, including a cross-link probe to monitor a possible recruitment of SRP near the ribosomal exit site. The extension comprises the 18 NH2-terminal residues of the phage coat protein Pf3 with a TAG codon at position 7 (Fig. 4 A, 48Pf3LepTAG7). This peptide does not interfere with the targeting and insertion of full-length Lep in vivo (Lee et al., 1992) and is translocated across the inner membrane like the natural Lep NH2-terminal three amino acids. The position of the TAG is chosen such that it has the same spacing (41 amino acids) to the PTC as TAG3 in the 44Lep construct that was shown to cross-link strongly to Ffh (Fig. 4 A). As a control, H1 in 48Pf3LepTAG7 was subjected to knockout mutagenesis (described in previous section) to abolish its hydrophobic character without altering the spacing between the PTC and the cross-linking probe.
Figure 4.
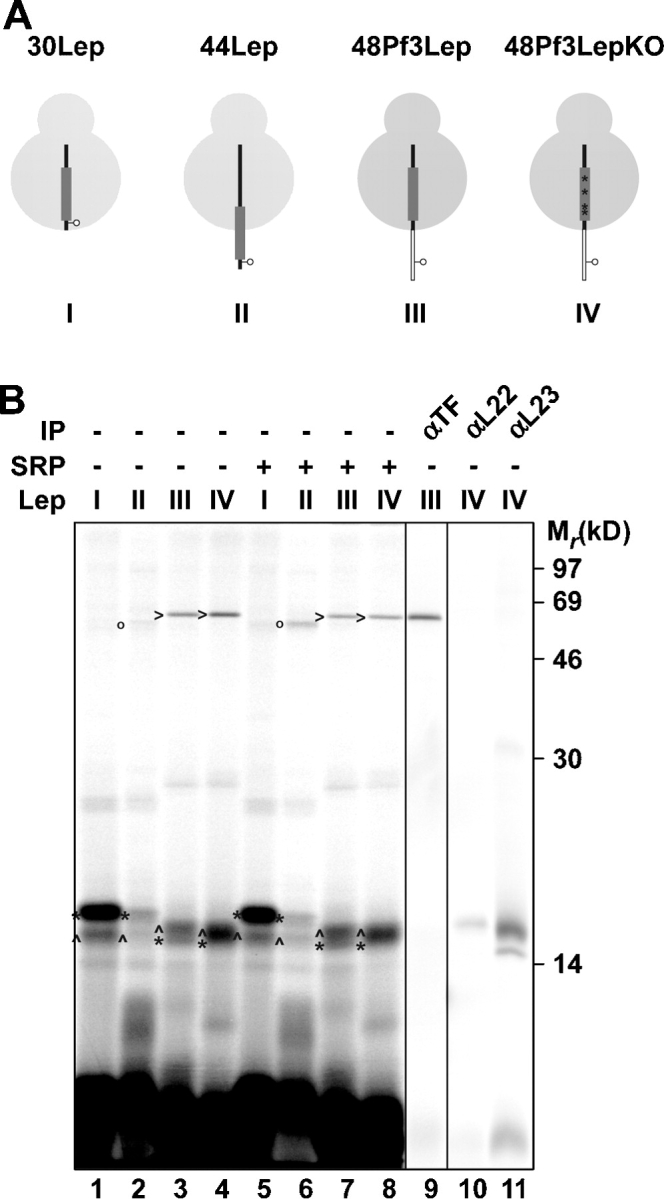
SRP is not oriented toward the ribosomal exit before it is able to interact with H1. (A) Schematic representation of 30 and 44Lep with a cross-linking probe at position 3 and 48Pf3Lep and 48Pf3LepKO with a cross-linking probe at position seven. H1 and the Pf3 extension are depicted as a thick gray line and a thin white bar, respectively. The four mutations in H1 to obtain the Pf3LepKO construct are the same as in Fig. 3 and are indicated here with four asterisks. (B) The constructs shown in A were translated in vitro with and without the addition of 350 nM of purified SRP. After translation, samples were cross-linked, purified, and immunoprecipitated as described in Fig. 1. Images in different panels represent different parts of the gel or different exposure times. *, L22 cross-link; ^, L23 cross-link; o, Ffh cross-link; >, TF cross-link.
30LepTAG3 and 44LepTAG3 revealed cross-linking to L22/L23 and L22/L23/Ffh, respectively, as observed above (Fig. 4 B, lanes 1 and 2). Both nascent 48Pf3LepTAG7 and 48Pf3LepKOTAG7, however, revealed cross-linking to L22, L23, and TF but not to Ffh (Fig. 4 B, lanes 3, 4, 9, and 10). Even when a saturating amount of SRP (350 nM; Ullers et al., 2003) was added during translation, which enhanced the cross-linking of 44LepTAG3 to Ffh (Fig. 4 B, lane 6), Ffh cross-linking to the nascent Pf3Lep constructs was not observed, and TF cross-linking remained unaffected (Fig. 4 B, lane 7). This clear result argues against the hypothesis that the presence of a TM is sensed in the ribosome and transduced to the ribosomal surface to recruit SRP near the exit site. Rather, the result is consistent with a default attendance of TF at this location, which is only replaced by SRP when (part of) the TM is exposed outside the ribosome.
Discussion
The E. coli IMP Lep is targeted cotranslationally by the SRP to the Sec–YidC insertion site in the membrane (de Gier et al., 1996; Houben et al., 2000, 2002; Samuelson et al., 2000). Previously, we have shown that nascent Lep already associates with the membrane at a length of 50 amino acids (Houben et al., 2002). In this study, we have investigated the nature and order of molecular contacts of nascent Lep that direct it into this pathway. Using site-directed photo–cross-linking from amino acid 3, we identified interactions of nascent Lep of 9–50 amino acids within the ribosome, cytosol, and membrane. Consecutive interactions with L4, L22, L23, SRP, SecY, and YidC were identified.
Based on atomic structures of the large ribosomal subunit, nascent polypeptides have been proposed to traverse the ribosome via an ∼100-Å-long cavity that runs from the PTC toward an exit site, where protein L23 is located (Nissen et al., 2000; Harms et al., 2001; Fig. 5). The extensive contacts of short nascent Lep species with L4, L22, and L23 (Fig. 1) indeed suggest that Lep exits the ribosome via this main tunnel (Nissen et al., 2000; Fig. 5). Most interesting, the overlap in cross-linking to the ribosomal proteins suggests multiple and flexible conformations of nascent Lep in the ribosome rather than a rigid structure with specific vectorial contacts. When the probe is six amino acids from the PTC, cross-linking to L4 and L22 was already observed, implying that this part of nascent Lep reaches, in extended form, the narrow constriction in the tunnel that is formed by the tips of L4 and L22, which are located ∼30 Å below the PTC. The most extensive contact (from 9 to 50Lep) is observed with L22, which is consistent with L22's major contribution to the tunnel wall between the constriction area and exit site (Nissen et al., 2000). Fully extended nascent Lep would require ∼27 amino acids to reach the ribosomal surface (Fig. 5). This indicates that part of nascent Lep has a nonstretched conformation, most likely past the constriction site. On the other hand, cross-linking to L23 (localized at the exit site) starts at a length of 24 amino acids, suggesting that part of nascent Lep does reach L23 in an extended conformation. Remarkably, cross-linking to L4 is observed by using chains up to 24 residues in length, indicating that nascent Lep can fold back to the constriction site. Consistent with our results, several studies have previously indicated that nascent chains can fold considerably and acquire various conformations inside the ribosome (Choi et al., 1998; Matlack et al., 1998; Tsalkova et al., 1998; Mingarro et al., 2000). In addition, recent cryo-EM maps of translating ribosomes suggest that the region past the constriction area can expand extensively to accommodate large segments of nascent chains that could be partially folded (Gilbert et al., 2004). Our cross-link data suggest that the first TM (H1) in nascent Lep might at least acquire an α-helical conformation in this region of the tunnel. Fluorescence resonance energy transfer analysis of a nascent membrane protein (111p) in eukaryotic ribosomes supports the acquisition of an α-helical structure by TMs located inside the ribosome (Woolhead et al., 2004). The same study showed cross-linking of the TM to ribosomal proteins that have been tentatively identified as L4, L17 (homologue of L22), and L39 (replaced in prokaryotes by a part of L23) based on the molecular mass of the cross-linked products because antibodies against these proteins were not available. Cross-linking to L17 and L39 appeared specific for a TM of a membrane protein and did not occur in the signal peptide of a nascent secretory protein. An earlier study from the same group has indicated that a TM inside the ribosome induces conformational changes in the Sec–translocon (Liao et al., 1997). The combined data led the authors to propose not only that folding of the TM is induced in the ribosome but also that the TM is specifically recognized by L17/L39, which in turn influences downstream processes such as the membrane integration of TM via the Sec–translocon. Evidence for dynamic and responsive properties of the prokaryotic exit tunnel has also been presented (for review see Tenson and Ehrenberg, 2002).
In this study, we did not observe sequence-specific contacts in the E. coli ribosome. L4, L22, and L23 were cross-linked to nascent chains of the IMP Lep (irrespective of the presence of a functional TM), the secretory protein PhoE, and the cytoplasmic protein RpoB in approximately the same order. Although the absence of L17/L39 cross-linking to a nascent secretory protein in a eukaryotic translation system (Woolhead et al., 2004) is not conclusive, differences in the specificity of ribosomal contacts might yet be species related. Notably, in eukaryotes, both secretory and membrane proteins are cotranslationally targeted by the SRP to the Sec61 complex in the ER membrane. Therefore, it is conceivable that identification of nascent chains by the ribosome has evolved to adequately distinguish between the two types of nascent chains and prepare the Sec–translocon for the upcoming translocation or integration event. In contrast, in E. coli, the targeting of secretory proteins occurs very differently from the targeting of IMPs. Whereas IMPs are cotranslationally targeted via the SRP pathway, secretory proteins are targeted by the cytosolic chaperone SecB in an essentially posttranslational process. Because cotranslational insertion at the E. coli Sec–translocon occurs exclusively for IMPs, the interaction with the ribosome–nascent chain–SRP complex might be sufficient to prime the Sec–translocon for the membrane integration process. As an alternative mechanism to determine the mode of action (translocation versus insertion), the E. coli Sec–translocon could make use of YidC, which is a Sec-associated IMP that has no homologue in the ER membrane (Luirink et al., 2001). Consistent with a role in the recognition and lateral transfer of TMs in the Sec–translocon, YidC interacts with TMs very early when they have not even fully emerged from the ribosome (see below). In conclusion, the TM of an E. coli IMP might be recognized outside the ribosome by SRP and YidC, precluding the need for an intraribosomal sensing mechanism. In agreement with this notion, the presence of a TM (H1) in the exit tunnel did not induce SRP cross-linking to an arbitrary (nonhydrophobic) upstream sequence exposed outside the ribosome (Fig. 4). In contrast to earlier speculations (Gu et al., 2003; Ullers et al., 2003), this result suggests that the SRP is not recruited at L23 toward the nascent chain before emergence of (part of) the TM.
The uniform intraribosomal cross-link patterns also suggest that all nascent chains, irrespective of their final destination, traverse the “main” ribosomal tunnel and do not exit via alternative channels that appear present in the cryoelectron microscopy reconstruction of the E. coli ribosome (Gabashvili et al., 2001).
Cross-linking to the SRP starts remarkably early during biogenesis of Lep when the nascent chain is 40 amino acids long and the cross-link probe is only 37 amino acids from the PTC. By modeling nascent 40Lep in the crystal structure of the vacant, large ribosomal subunit of Deinococcus radiodurans (Harms et al., 2001), which is presumably very similar to the E. coli structure, it is shown that 27Lep residues are accommodated inside the tunnel in a fully extended conformation (Fig. 5). Thus, no more than half of the TM can be exposed outside the tunnel at this stage (Fig. 5). It is tempting to speculate that the exit region expands once a TM starts to emerge so that at least the M domain of Ffh partly penetrates the exit region to interact with the complete TM. Extensive flexibility of the ribosomal tunnel has also been inferred from cross-link studies at the Wiedmann laboratory that suggest contacts of the nascent polypeptide-associated complex are as near as 17 amino acids from the PTC in wheat germ ribosomes (Wang et al., 1995). However, given the lack of structural data on the SRP–TM or SRP-signal sequence interaction, we cannot formally exclude that part of the TM is sufficient for interaction with the SRP.
Strikingly, the earliest cross-linking of nascent Lep to SRP coincides with cross-linking to SecY when membranes are added during translation. This suggests that nascent Lep of ∼40 amino acids can be handed over from the SRP to SecY in the membrane. Analogous to the SRP, SecY is likely to have access to the ribosome interior for full contact with the TM at this stage. Upon elongation of Lep, the cross-linking shifts from SecY toward YidC, resulting in an almost exclusive YidC cross-linking at 50 amino acids. Notably, the YidC contact is clearly visible from 44Lep, suggesting that YidC also contacts the Lep TM before it is fully emerged from the ribosome. At present, it is not clear how YidC senses the identity of the TM at this early stage. In any case, the cross-link data suggest an early and intimate contact between the exit portal of the ribosome and the Sec–YidC complex during membrane protein integration. Similarly, Sec61 subunits and translocating chain-associated membrane protein in the ER have been cross-linked to nascent membrane proteins early in the insertion process when the cross-linking amino acid in the TM was only 25–27 residues from the PTC (Laird and High, 1997; Woolhead et al., 2004). Structural analysis of a defined early insertion intermediate will be required to reveal the precise architecture of the ribosome–translocon complex “in action” at this stage.
Materials and methods
Reagents and sera
Restriction enzymes and the long template Expand High Fidelity PCR-System were obtained from Roche Diagnostics GmbH. T4 DNA ligase was purchased from Epicenter Technologies. The MEGAshortscript T7 transcription kit was obtained from Ambion. [35S]methionine and protein A–Sepharose beads were obtained from Amersham Biosciences. All other chemicals were supplied by Sigma-Aldrich. Antisera against L23, Ffh, TF, YidC, and SecY have been described previously (Scotti et al., 2000; Houben et al., 2002; Ullers et al., 2003). Antisera against L4 and L22 were gifts from R. Brimacombe (Max-Planck-Institut für Molekulare Genetik, Berlin, Germany).
Strains and plasmid constructs
Strain Top10F′ was used for the maintenance of plasmid constructs. Strain MRE600 was used to prepare translation lysate for the suppression of TAG stop codons in the presence of (Tmd)Phe-tRNAsup. Strain MC4100 was used to obtain IMVs, which were prepared as described previously (de Vrije et al., 1987). Plasmids pC4Meth9-46LepTAG3 were constructed by PCR using pC4Meth50LepTAG3 as a template (Houben et al., 2002). 7, 9, 14, and 19Leu in H1 of pC4Meth15-24-34-44LepTAG3 were replaced by Glu, Lys, Pro, and Arg, respectively, using site-directed mutagenesis obtaining pC4Meth15-24-34-44LepKOTAG3. Plasmids pC4Meth15/44pre-PhoETAG3 and pC4Meth15/44RpoBTAG3 were constructed by nested PCR using pC4Meth150pre-PhoE and C4Meth150RpoB, respectively, as templates (Ullers et al., 2004). The 18 NH2-terminal amino acids of Pf3 with a stop codon at position 7 (TAG) were fused to 30Lep and 30LepKO by nested PCR and were cloned in the pC4Meth vector, resulting in plasmids pC4Meth48Pf3LepTAG7 and pC4Meth48Pf3LepKOTAG7.
In vitro transcription, translation, targeting, and cross-linking
Truncated mRNA was prepared as described previously (Scotti et al., 2000) from HindIII-linearized Lep plasmids. In vitro translation, cross-linking, and ribosome–nascent chain complex purification through a high salt sucrose cushion of nascent Lep derivatives carrying the photo-activatable amino acid l[3-(trifluoromethyl)-3-diazirin-3H-yl] phenylalanine ([Tmd]Phe) were performed as described previously (Ullers et al., 2003). The Tmd was a gift from J. Brunner (Institut für Biochemie der Eidgenössischen Technischen Hochschule, Zürich, Switzerland). Targeting to IMVs and carbonate extraction have been described previously (Scotti et al., 2000; Urbanus et al., 2001). High salt sucrose pellet fractions and carbonate-insoluble fractions were either analyzed directly by SDS-PAGE and phosphoimaging or were first immunoprecipitated by using two-to fourfold the amount used for direct analysis.
Structure modeling
The image of the ribosome–nascent chain complex (Fig. 5) was performed with PyMol. Poly-Arg (40 aa in length) peptide was modeled on the SecM extended polypeptide (Berisio et al., 2003) by using the program O (Jones et al., 1991). The poly-Arg peptide was then geometrically minimized in order to avoid clashes with the large ribosomal subunit of D. radiodurans (D50S; protein data bank ID, 1NKW). The P-site tRNA (1GIY) was superimposed on the D50S structure as described previously (Harms et al., 2001) and marked the COOH-terminal position of the polypeptide. Poly-Arg was used in order not to be influenced by the amino acids' type and shape and to ensure the free path of the peptide in the ribosome–nascent chain tunnel.
Acknowledgments
We thank C. ten Hagen-Jongman and G. Koningstein for technical assistance, A. Bashan for preliminary modeling work, and J.-W. de Gier, N. Harms, D.-J. Scheffers, and A. Yonath for critical reading of the manuscript.
This work was supported by the Dutch Earth and Life Sciences Foundation, which is subsidized by the Netherlands Organisation for Scientific Research.
Abbreviations used in this paper: IMP, inner membrane protein; IMV, inner membrane vesicle; Lep, leader peptidase; PTC, peptidyl transferase center; SR, SRP receptor; SRP, signal recognition particle; TF, trigger factor; TM, transmembrane domain.
References
- Beck, K., L.F. Wu, J. Brunner, and M. Müller. 2000. Discrimination between SRP- and SecA/SecB-dependent substrates involves selective recognition of nascent chains by SRP and trigger factor. EMBO J. 19:134–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berisio, R., F. Schluenzen, J. Harms, A. Bashan, T. Auerbach, D. Baram, and A. Yonath. 2003. Structural insight into the role of the ribosomal tunnel in cellular regulation. Nat. Struct. Biol. 10:366–370. [DOI] [PubMed] [Google Scholar]
- Bernstein, H.D., and J.B. Hyndman. 2001. Physiological basis for conservation of the signal recognition particle targeting pathway in Escherichia coli. J. Bacteriol. 183:2187–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, K.M., J.F. Atkins, R.F. Gesteland, and R. Brimacombe. 1998. Flexibility of the nascent polypeptide chain within the ribosome–contacts from the peptide N-terminus to a specific region of the 30S subunit. Eur. J. Biochem. 255:409–413. [DOI] [PubMed] [Google Scholar]
- de Gier, J.-W.L., P. Mansournia, Q.A. Valent, G.J. Phillips, J. Luirink, and G. von Heijne. 1996. Assembly of a cytoplasmic membrane protein in Escherichia coli is dependent on the signal recognition particle. FEBS Lett. 399:307–309. [DOI] [PubMed] [Google Scholar]
- de Vrije, T., J. Tommassen, and B. de Kruijff. 1987. Optimal posttranslational translocation of the precursor of PhoE protein across Escherichia coli membrane vescicles requires both ATP and the proton motive force. Biochim. Biophys. Acta. 900:63–72. [DOI] [PubMed] [Google Scholar]
- Ferbitz, L., T. Maier, H. Patzelt, B. Bukau, E. Deuerling, and N. Ban. 2004. Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature. 431:590–596. [DOI] [PubMed] [Google Scholar]
- Flanagan, J.J., J.C. Chen, Y. Miao, Y. Shao, J. Lin, P.E. Bock, and A.E. Johnson. 2003. Signal recognition particle binds to ribosome-bound signal sequences with fluorescence-detected subnanomolar affinity that does not diminish as the nascent chain lengthens. J. Biol. Chem. 278:18628–18637. [DOI] [PubMed] [Google Scholar]
- Gabashvili, I.S., S.T. Gregory, M. Valle, R. Grassucci, M. Worbs, M.C. Wahl, A.E. Dahlberg, and J. Frank. 2001. The polypeptide tunnel system in the ribosome and its gating in erythromycin resistance mutants of L4 and L22. Mol. Cell. 8:181–188. [DOI] [PubMed] [Google Scholar]
- Gilbert, R.J., P. Fucini, S. Connell, S.D. Fuller, K.H. Nierhaus, C.V. Robinson, C.M. Dobson, and D.I. Stuart. 2004. Three-dimensional structures of translating ribosomes by Cryo-EM. Mol. Cell. 14:57–66. [DOI] [PubMed] [Google Scholar]
- Gu, S.Q., F. Peske, H.J. Wieden, M.V. Rodnina, and W. Wintermeyer. 2003. The signal recognition particle binds to protein L23 at the peptide exit of the Escherichia coli ribosome. RNA. 9:566–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic, M., T. Becker, M.R. Pool, C.M. Spahn, R.A. Grassucci, J. Frank, and R. Beckmann. 2004. Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature. 427:808–814. [DOI] [PubMed] [Google Scholar]
- Harms, J., F. Schluenzen, R. Zarivach, A. Bashan, S. Gat, I. Agmon, H. Bartels, F. Franceschi, and A. Yonath. 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 107:679–688. [DOI] [PubMed] [Google Scholar]
- Hesterkamp, T., S. Hauser, H. Lütcke, and B. Bukau. 1996. Escherichia coli trigger factor is a prolyl isomerase that associates with nascent polypeptide chains. Proc. Natl. Acad. Sci. USA. 93:4437–4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben, E.N.G., P.A. Scotti, Q.A. Valent, J. Brunner, J.-W.L. de Gier, B. Oudega, and J. Luirink. 2000. Nascent Lep inserts into the Escherichia coli inner membrane in the vicinity of YidC, SecY and SecA. FEBS Lett. 476:229–233. [DOI] [PubMed] [Google Scholar]
- Houben, E.N.G., M.L. Urbanus, M. van der Laan, C.M. ten Hagen-Jongman, A.J. Driessen, J. Brunner, B. Oudega, and J. Luirink. 2002. YidC and SecY mediate membrane insertion of a Type I transmembrane domain. J. Biol. Chem. 277:35880–35886. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., J.Y. Zou, S.W. Cowan, and Kjeldgaard. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 47:110–119. [DOI] [PubMed] [Google Scholar]
- Koch, H.G., M. Moser, and M. Müller. 2003. Signal recognition particle-dependent protein targeting, universal to all kingdoms of life. Rev. Physiol. Biochem. Pharmacol. 146:55–94. [DOI] [PubMed] [Google Scholar]
- Kramer, G., T. Rauch, W. Rist, S. Vorderwülbecke, H. Patzelt, A. Schulze-Specking, N. Ban, E. Deuerling, and B. Bukau. 2002. L23 protein functions as a chaperone docking site on the ribosome. Nature. 419:171–174. [DOI] [PubMed] [Google Scholar]
- Kusters, R., T. de Vrije, E. Breukink, and B. de Kruijff. 1989. SecB protein stabilizes a translocation-competent state of purified prePhoE protein. J. Biol. Chem. 264:20827–20830. [PubMed] [Google Scholar]
- Laird, V., and S. High. 1997. Discrete cross-linking products identified during membrane protein biosynthesis. J. Biol. Chem. 272:1983–1989. [DOI] [PubMed] [Google Scholar]
- Lee, J.I., A. Kuhn, and R.E. Dalbey. 1992. Distinct domains of an oligotopic membrane protein are Sec-dependent and Sec-independent for membrane insertion. J. Biol. Chem. 267:938–943. [PubMed] [Google Scholar]
- Liao, S., J. Lin, H. Do, and A.E. Johnson. 1997. Both lumenal and cytosolic gating of the aqueous ER translocon pore are regulated from inside the ribosome during membrane protein integration. Cell. 90:31–41. [DOI] [PubMed] [Google Scholar]
- Luirink, J., T. Samuelsson, and J.W. de Gier. 2001. YidC/Oxa1p/Alb3: evolutionarily conserved mediators of membrane protein assembly. FEBS Lett. 501:1–5. [DOI] [PubMed] [Google Scholar]
- Matlack, K.E.S., W. Mothes, and T.A. Rapoport. 1998. Protein translocation: tunnel vision. Cell. 92:381–390. [DOI] [PubMed] [Google Scholar]
- Mingarro, I., I. Nilsson, P. Whitley, and G. von Heijne. 2000. Different conformations of nascent polypeptides during translocation across the ER membrane. BMC Cell Biol. 1:10.1186/1471-2121-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa, H., and K. Ito. 2002. The ribosomal exit tunnel functions as a discriminating gate. Cell. 108:629–636. [DOI] [PubMed] [Google Scholar]
- Nissen, P., J. Hansen, N. Ban, P.B. Moore, and T.A. Steitz. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science. 289:920–930. [DOI] [PubMed] [Google Scholar]
- Pool, M.R., J. Stumm, T.A. Fulga, I. Sinning, and B. Dobberstein. 2002. Distinct modes of signal recognition particle interaction with the ribosome. Science. 297:1345–1348. [DOI] [PubMed] [Google Scholar]
- Raine, A., R. Ullers, M. Pavlov, J. Luirink, J.E. Wikberg, and M. Ehrenberg. 2003. Targeting and insertion of heterologous membrane proteins in E. coli. Biochimie. 85:659–668. [DOI] [PubMed] [Google Scholar]
- Samuelson, J.C., M. Chen, F. Jiang, I. Möller, M. Wiedmann, A. Kuhn, G.J. Phillips, and R.E. Dalbey. 2000. YidC mediates membrane protein insertion in bacteria. Nature. 406:637–641. [DOI] [PubMed] [Google Scholar]
- Scotti, P.A., M.L. Urbanus, J. Brunner, J.-W.L. de Gier, G. von Heijne, C. van der Does, A.J.M. Driessen, B. Oudega, and J. Luirink. 2000. YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 19:542–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenson, T., and M. Ehrenberg. 2002. Regulatory nascent peptides in the ribosomal tunnel. Cell. 108:591–594. [DOI] [PubMed] [Google Scholar]
- Tsalkova, T., O.W. Odom, G. Kramer, and B. Hardesty. 1998. Different conformations of nascent peptides on ribosomes. J. Mol. Biol. 278:713–723. [DOI] [PubMed] [Google Scholar]
- Ullers, R.S., E.N.G. Houben, A. Raine, C.M. Ten Hagen-Jongman, M. Ehrenberg, J. Brunner, B. Oudega, N. Harms, and J. Luirink. 2003. Interplay of signal recognition particle and trigger factor at L23 near the nascent chain exit site on the Escherichia coli ribosome. J. Cell Biol. 161:679–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullers, R.S., J. Luirink, N. Harms, F. Schwager, C. Georgopoulos, and P. Genevaux. 2004. SecB is a bona fide generalized chaperone in Escherichia coli. Proc. Natl. Acad. Sci. USA. 101:7583–7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanus, M.L., P.A. Scotti, L. Fröderberg, A. Sääf, J.-W.L. de Gier, J. Brunner, J.C. Samuelson, R.E. Dalbey, B. Oudega, and J. Luirink. 2001. Sec-dependent membrane protein insertion: sequential interaction of nascent FtsQ with SecY and YidC. EMBO Rep. 2:524–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valent, Q.A., D.A. Kendall, S. High, R. Kusters, B. Oudega, and J. Luirink. 1995. Early events in preprotein recognition in E. coli: interaction of SRP and trigger factor with nascent polypeptides. EMBO J. 14:5494–5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S., H. Sakai, and M. Wiedmann. 1995. NAC covers ribosome-associated nascent chains thereby forming a protective environment for regions of nascent chains just emerging from the peptidyl transferase center. J. Cell Biol. 130:519–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolhead, C.A., P.J. McCormick, and A.E. Johnson. 2004. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell. 116:725–736. [DOI] [PubMed] [Google Scholar]