Abstract
In melanocytes and melanoma cells α-melanocyte stimulating hormone (α-MSH), via the cAMP pathway, elicits a large array of biological responses that control melanocyte differentiation and influence melanoma development or susceptibility. In this work, we show that cAMP transcriptionally activates Hif1a gene in a melanocyte cell–specific manner and increases the expression of a functional hypoxia-inducible factor 1α (HIF1α) protein resulting in a stimulation of Vegf expression. Interestingly, we report that the melanocyte-specific transcription factor, microphthalmia-associated transcription factor (MITF), binds to the Hif1a promoter and strongly stimulates its transcriptional activity. Further, MITF “silencing” abrogates the cAMP effect on Hif1a expression, and overexpression of MITF in human melanoma cells is sufficient to stimulate HIF1A mRNA. Our data demonstrate that Hif1a is a new MITF target gene and that MITF mediates the cAMP stimulation of Hif1a in melanocytes and melanoma cells. Importantly, we provide results demonstrating that HIF1 plays a pro-survival role in this cell system. We therefore conclude that the α-MSH/cAMP pathway, using MITF as a signal transducer and HIF1α as a target, might contribute to melanoma progression.
Introduction
In the skin physiological system, α-melanocyte stimulating hormone (α-MSH) constitutes an important keratinocyte secreted factor in response to the UV radiation of sunlight. Keratinocytes account for the main epidermal cell type, and melanocytes, situated at the basal layer of the epidermis, are responsible for melanin synthesis and skin color. In melanocytes, α-MSH, through the binding to the melanocortin-1 receptor (MC1R) and up-regulation of the cAMP pathway, induces pigmentation by controlling several parameters of melanocyte differentiation such as melanin synthesis (melanogenesis) (Buscà and Ballotti, 2000), dendrite outgrowth (Buscà et al., 1998), and melanosome transport, required for melanin distribution within the skin (Passeron et al., 2004).
The α-MSH/MC1R/cAMP signaling pathway has also been reported to influence melanoma biology, but in contrast to the undisputed role of cAMP in promoting pigmentation, data relating cAMP to melanoma progression currently appear contradictory. α-MSH has been reported to inhibit melanoma growth and adhesion (Robinson and Healy, 2002), suggesting that a compromised cAMP signaling could favor melanoma progression. In agreement with this hypothesis, variants of MC1R with altered cAMP response have been associated with high melanoma susceptibility (Valverde et al., 1995; Robinson and Healy, 2002). In contrast, α-MSH and cAMP increase c-met expression in melanoma cells and could thereby favor the melanoma metastatic potential (Rusciano et al., 1999). Consistently, the CREB/ATF family of transcription factors that function downstream of the cAMP pathway have also been reported to act as survival factors and contribute to the malignant phenotype (for review see Poser and Bosserhoff, 2004).
The cAMP-triggered mechanisms controlling melanocyte pigmentation have been thoroughly investigated. cAMP, through PKA and CREB activation, increases the expression of the melanocyte-specific form of microphthalmia-associated transcription factor (MITF) (Bertolotto et al., 1998a), a basic helix-loop-helix transcription factor that plays a key role not only in melanin synthesis, but also in melanocyte development and survival (Hodgkinson et al., 1993; Hughes et al., 1993).
Noteworthy, in melanocytes and melanoma cells, cAMP regulates several signaling pathways that control multiple biological functions. Indeed, cAMP inhibits the phosphatidylinositol 3-kinase/AKT pathway that influences both differentiation and survival of melanocytes (Buscà et al., 1996; Khaled et al., 2002; Larribere et al., 2004). Further, α-MSH and cAMP stimulate the MAPK pathway (Englaro et al., 1995; Buscà et al., 2000) through B-Raf activation. Indeed, B-Raf has been recently found mutated and activated in a vast majority of melanoma (Davies et al., 2002).
To further investigate the molecular events elicited by α-MSH and cAMP in melanocytes and better understand the implication of its signaling in melanoma biology, we performed a large-scale gene screening by using a DNA microarray (Moreilhon et al., 2004). We used B16 mouse melanoma cells to analyze the overall modification of gene expression after 24 h of treatment with the cAMP-elevating agent forskolin.
Very interestingly, we found that cAMP stimulates the expression of the Hif1a gene whose protein product, hypoxia-inducible factor 1α (HIF1α), constitutes the α subunit of the HIF1, a master regulator of oxygen homeostasis. After dimerization with HIF1β (also named ARNT), HIF1α binds to a consensus sequence 5′-RCGTG-3′ called hypoxia-responsive element (HRE), and controls the expression of several genes involved in many aspects of cancer progression, including metabolic adaptation, apoptosis resistance, angiogenesis, invasion, and metastasis (for review see Semenza, 2002). To date, HIF1α has been shown to be tightly regulated by multiple post-translational modifications that mainly control the protein stability through the ubiquitin–proteasome pathway (Brahimi-Horn et al., 2001; Berra et al., 2003).
In this paper, we show that cAMP transcriptionally regulates Hif1a gene expression because the cAMP-elevating agent forskolin increases Hif1a mRNA and regulates Hif1a promoter activity. cAMP increases the expression of a functional HIF1α protein that results in the induction of Vegf expression. Furthermore, the effect of cAMP on HIF1α is restricted to melanocyte cells. This cell specificity is given by MITF that binds to the Hif1a promoter and strongly stimulates its transcriptional activity. MITF silencing abrogates the effect of cAMP on HIF1α expression, and infection of human melanoma cells with an MITF-encoding adenovirus increases Hif1a mRNA expression, clearly demonstrating that Hif1a is a new MITF target gene. Because we demonstrate that HIF1α plays a pro-survival role in melanoma and we suggest that it is involved in neovascularization, we conclude that the α-MSH/cAMP pathway, through a molecular cascade sequentially involving MITF and HIF1α, might contribute to melanoma development.
Results
Hif1a gene expression is induced by the cAMP pathway in B16 melanoma cells
In an attempt to screen for cAMP-induced genes in melanocyte cells, we used a laboratory-designed DNA microarray (Moreilhon et al., 2004) described in the Materials and methods section. B16 mouse melanoma cells were treated (or not) with the cAMP-elevating agent forskolin for 24 h. Total mRNAs were extracted, labeled, and hybridized with microarray slides.
The induction of several already-known cAMP-regulated genes validated our array approach. The biochip analysis identified several cAMP-regulated genes which protein products have previously been clearly involved in melanogenesis (Table I) such as tyrosinase, tyrosinase-related protein 1, Mitf, or proteins involved in melanosome transport such as Myosin Va and Rab27a. These results were confirmed by real-time quantitative PCR (Table I).
Table I. cAMP induces Hif1a mRNA expression.
| Gene | Microarray | Q-PCR |
|---|---|---|
| Tyr | 6.32 ± 1.0 | 3.32 ± 0.4 |
| Tyrp1 | 1.8 ± 0.4 | 2.8 ± 0.5 |
| MyoVa | 8.70 ± 0.3 | 2.9 ± 0.2 |
| Rab27a | 2.27 ± 1.0 | 3.05 ± 0.2 |
| Mitf | 3.71 ± 0.9 | 4.92 ± 0.9 |
| Hif1a | 3.05 ± 1.0 | 2.85 ± 0.4 |
Microarray analysis performed as described in the Materials and methods section. The results correspond to the fold stimulation of genes stimulated in B16 mouse melanoma cells after 24 h of treatment with the cAMP-elevating agent, forskolin (20 μM). The values are the average of four independent biochip experiments. Real time quantitative PCR analysis (Q-PCR) was performed in the same conditions. The results are represented as the fold stimulation after forskolin treatment and correspond to the average of five independent experiments.
Surprisingly, cAMP clearly induced the expression of Hif1a gene (threefold stimulation; Table I). Due to the pivotal role of HIF1 transcription factor in cancer progression, we decided to thoroughly investigate the molecular mechanisms governing Hif1α gene regulation by cAMP in melanoma cells.
cAMP induces a functional HIF1α protein
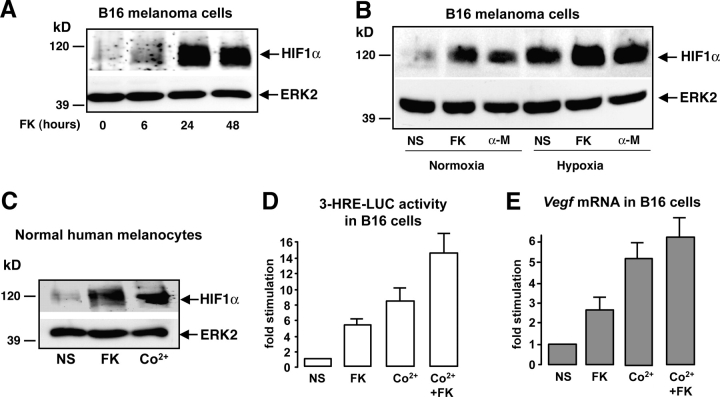
Western blot experiments using a specific anti HIF1α antibody showed that HIF1α protein slightly increased at 6 h and reached its maximal levels upon 24 h of forskolin treatment. After 48 h, HIF1α protein expression remained stable (Fig. 1 A). α-MSH, a physiological activator of the cAMP pathway in melanocytes, also increased HIF1α protein levels similarly to hypoxia (24 h), which was used as a positive control of HIF1α protein induction. In addition, cAMP plus hypoxia appeared to have additive effects (Fig. 1 B). The same results were obtained using normal human melanocytes (NHM). In this case we used Co2+, to mimic hypoxia (Berra et al., 2003), as a control (Fig. 1 C). Therefore, HIF1α protein increased upon cAMP up-regulation in B16 melanoma cells and NHM independently of the oxygen context.
Figure 1.
cAMP increases the expression of a functional HIF1α protein in melanocyte cells. (A) B16 cells were stimulated for 6, 24, and 48 h with forskolin (FK) and cell extracts were subjected to Western blot analysis to detect HIF1α protein levels. A control of the protein loading was performed by detecting ERK2. (B) The same experiment was performed by stimulating B16 cells either with forskolin (FK) or α-MSH (α-M) for 24 h. Cells were incubated, either in normal oxygen conditions (Normoxia, 20% O2) or maintained under hypoxia (1–2% O2). (C) A Western blot to analyze HIF1α protein expression was performed using extracts from normal human melanocytes. Cells were starved and treated with forskolin (FK) for 24 h or with cobalt (Co2+) to mimic hypoxia as a positive control. (D) B16 cells were transfected with the 3-HRE-LUC reporter construct and treated (or not) (NS) with forskolin (FK) for at least 36 h, with cobalt (Co2+) for 12 h, or with both (Co2+ + FK). Luciferase activity was normalized by the β-galactosidase activity and data are expressed in fold stimulation of the basal 3-HRE-LUC activity. Data are means ± SE of five experiments performed in triplicate. (E) Real-time quantitative PCR to detect Vegf mRNA levels on total RNA extracts from B16 cells treated as described in D.
To evaluate whether the cAMP-induced HIF1α formed a functional transcriptional complex, we measured the activity of the 3-HRE-LUC reporter gene (Berra et al., 2003).
The results obtained showed that the 3-HRE-LUC construct was responsive (by a five- to sixfold stimulation) to cAMP in B16 melanoma cells. Co2+ stimulation was used as a positive control and again the effect of Co2+ and cAMP appeared additive (Fig. 1 D).
Among the multiple HIF1 target genes containing an HRE core sequence in their promoter region, Vegf has been largely studied because it is required for tumor angiogenesis. We therefore analyzed the effects of cAMP in endogenous Vegf expression in B16 cells. Real-time quantitative PCR experiments showed that forskolin increased vascular endothelial growth factor (VEGF) mRNA in these melanocyte cells. As expected, Co2+ highly induced Vegf and enhanced the cAMP response (Fig. 1 E). Together, these results demonstrate that cAMP induced a functional HIF1α protein.
cAMP up-regulation if Hif1a gene transcription is cell specific
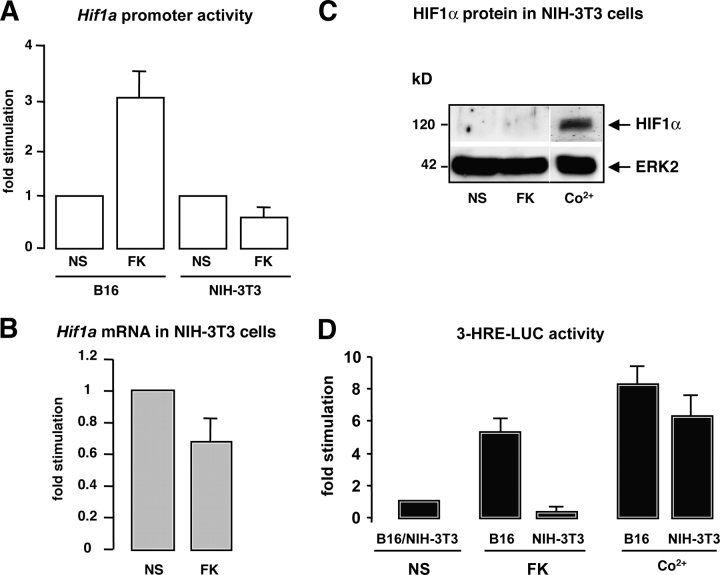
To further elucidate the mechanisms by which cAMP regulates HIF1α expression, we performed Hif1a promoter activity assays. An Hif1a promoter fragment cloned upstream of the luciferase reporter gene (Wenger et al., 1998) was transfected in B16 melanoma cells. The promoter activity, monitored by the luciferase levels, increased upon cAMP treatment (Fig. 2 A). The same experiment was performed in NIH-3T3 fibroblasts, and we observed that forskolin had no impact on Hif1a promoter activity (Fig. 2 A). Furthermore, in these cells cAMP decreased Hif1a mRNA amount (Fig. 2 B). At the protein level, cAMP treatment did not affect HIF1α expression in NIH-3T3 fibroblasts (Fig. 2 C), whereas the control Co2+ induced the protein expression. A comparison of the functional HIF1α protein levels between B16 melanocyte cells and NIH-3T3 nonmelanocyte cells was performed using the 3-HRE-LUC reporter system (Fig. 2 D). We observed that although forskolin significantly increased the reporter transactivation in B16 cells, this did not occur in NIH-3T3 fibroblasts. In contrast, Co2+ treatment increased luciferase expression in both cell lines. These results indicate that cAMP increases Hif1α gene transcription in a melanocyte cell–specific manner.
Figure 2.
cAMP induces HIF1α expression in a cell-specific manner. (A) B16 cells and NIH-3T3 cells were transfected with a fragment of the Hif1a gene promoter cloned upstream of the luciferase reporter gene, and were then stimulated (or not) (NS) with forskolin (FK). Luciferase activity was normalized by the β-galactosidase activity and results are represented as the fold stimulation of the cAMP-activated promoter compared with the basal value. (B) Real-time quantitative PCR to detect Hif1a mRNA levels was performed on total RNAs from NIH-3T3 fibroblasts nonstimulated (NS) or treated for 24 h with forskolin (FK). (C) NIH-3T3 extracts were subjected to Western blot analysis to detect HIF1α protein levels after cell stimulation with forskolin (FK) for 24 h, or cobalt (Co2+) for 12 h. ERK2 levels show a control of the gel protein loading. White lines indicate that intervening lanes have been spliced out. (D) 3-HRE-LUC reporter assay on B16 and NIH-3T3 cells nonstimulated (NS) of stimulated with forskolin (FK) or cobalt (Co2+).
MITF binds and transactivates the Hif1a promoter
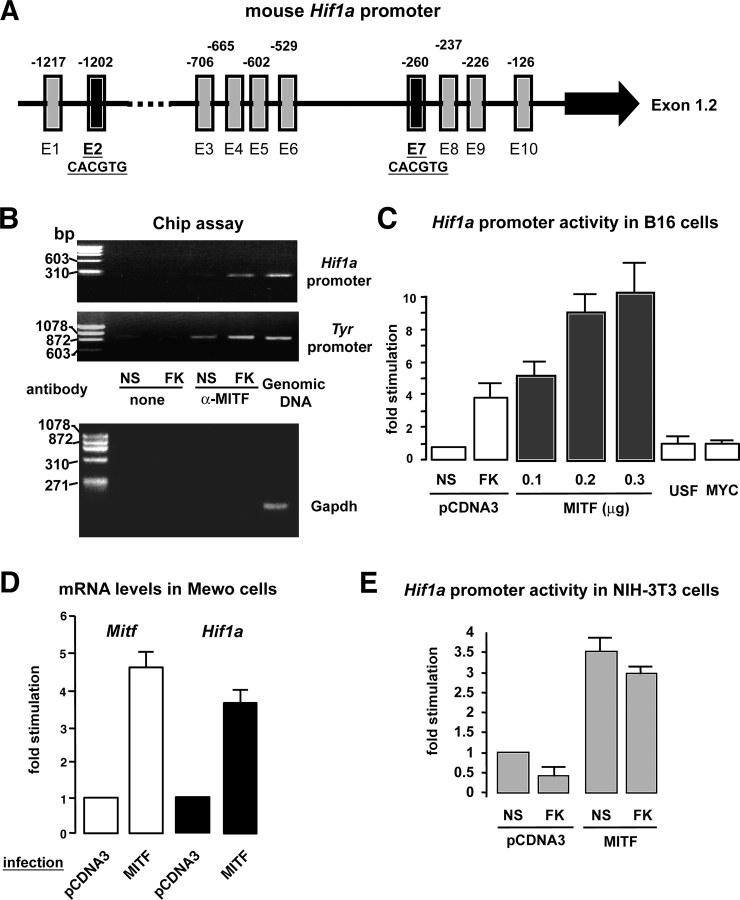
The cell specificity of HIF1α induction by cAMP prompted us to hypothesize the existence of specific molecular mechanisms responsible for HIF1α regulation in melanocytes and melanoma cells. Sequence analysis of the Hif1a promoter (Wenger et al., 1998) revealed 10 core E-box consensus sequences composed by the CANNTG motif (Fig. 3 A). Noteworthy, among these 10 E-boxes, 2 perfectly match with the motif of MITF binding (CACGTG) (Aksan and Goding, 1998). We therefore investigated whether MITF could account for the cell-specific cAMP regulation of HIF1α expression. To evaluate the MITF binding to the Hif1a promoter region in the endogenous chromatin context, we performed chromatin immunoprecipitation assays (Fig. 3 B). B16 cells were stimulated with the cAMP-elevating agent forskolin for 5 h to maximally increase MITF protein expression (Bertolotto et al., 1998a). Chromatin complexes were immunoprecipitated with an anti-MITF antibody and a PCR was performed using specific primers to the mouse Hif1a promoter region. As shown in Fig. 3 B, we detected a specific amplification of the Hif1a promoter region that increased after forskolin treatment. As a positive control, we used specific primers spanning the tyrosinase promoter region, which is well known to bind MITF. As expected, the tyrosinase promoter amplification also increased upon cAMP exposure. A specific band corresponding to the Hif1a or tyrosinase promoter was detected when using mouse genomic DNA. In the absence of antibody, no specific amplification was observed. Another negative control was performed by applying 40 cycles of PCR to the MITF immunoprecipitates using a primer pair to the mouse GAPDH. As expected for a sequence that does not present MITF binding sites, no GAPDH amplification was observed while a clear PCR band was detected using the genomic DNA as a template (Fig. 3 B, bottom panel). We therefore conclude that MITF binds to the Hif1a promoter in the chromatin context of B16 melanoma cells.
Figure 3.
MITF binds to the Hif1a promoter and induces Hif1 α gene expression. (A) Schematic representation of the Hif1a promoter. Black arrow denotes the ubiquitously expressed 1.2 exon (Wenger et al., 1998). Vertical rectangles and the associated numbers represent the several E-boxes found in this promoter fragment and their localization within the sequence. Note that E2 and E7 match perfectly with the previously described MITF binding sequence (CACGTG) (Weilbaecher et al., 1998). (B) Chromatin immunoprecipitations were performed on extracts from nonstimulated (NS) and 5-h forskolin-stimulated (FK) B16 melanoma cells as described in Materials and methods. Immunoprecipitation was performed using a specific anti-MITF antibody and primers spanning the Hif1a or the tyrosinase promoter region were used for the PCR amplification. A control of PCR amplification was performed using mouse genomic DNA, which showed a 300-bp and a 900-bp band corresponding to the amplification of the Hif1a and tyrosinase promoter regions, respectively. Another control was performed using a primer pair to the mouse GAPDH (150-bp amplicon). (C) Hif1a promoter activity after cotransfection of B16 cells with the Hif1a promoter fragment together with an empty plasmid (pcDNA3), a vector encoding MITF transcription factor, and two other constructs encoding USF and MYC. As a control of the promoter activation, cells were stimulated with forskolin (FK). Results are shown as the fold stimulation compared with the basal nonstimulated promoter activity (NS). (D) Real-time quantitative PCR analysis to detect Mitf and Hif1a mRNA levels after infection of Mewo (human melanoma cells) with either an empty adenovirus (Adeno-pCDNA3) or an adenovirus encoding MITF. (E) Hif1a promoter activity after cotransfection of NIH-3T3 fibroblasts with an empty vector (pCDNA3) or a construct encoding MITF. Cells were next stimulated (or not) (NS) with forskolin (FK) for 24 h.
Next, we evaluated whether the MITF binding transactivated Hif1a promoter. Thus, we cotransfected the reporter construct encoding the Hif1a promoter fragment, together with different amounts of a plasmid encoding the MITF transcription factor. We clearly observed that MITF highly transactivated the Hif1a promoter activity in a dose-dependent manner (Fig. 3 C), whereas two other transcription factors of the same basic helix-loop-helix family, like USF and myc, did not. The functionality of the constructs encoding USF and myc was verified by their capacity to transactivate specific target promoters (unpublished data).
Next, we analyzed whether MITF could stimulate the expression of the endogenous Hif1a gene. In this aim we infected Mewo human melanoma cells either with an empty adenovirus (pCDNA3) or an adenovirus encoding MITF. 48 h after infection, total mRNAs were extracted and subjected to real-time quantitative PCR to control MITF overexpression and evaluate Hif1a expression. As shown in Fig. 3 D, MITF mRNA levels increased after MITF infection (∼4.6-fold) as expected, and Hif1a mRNA expression was significantly induced by MITF overexpression (∼3.7-fold).
To elucidate whether MITF could also transactivate the Hif1a promoter in a nonmelanocyte cell context, we cotransfected NIH-3T3 with this promoter together with the plasmid encoding MITF. The results obtained revealed that MITF significantly increased the Hif1a promoter activity (∼3.5-fold) (Fig. 3 E).
Together, these results strongly indicate that MITF binds to the proximal region of the Hif1a promoter and stimulates its transactivation leading to increased expression of Hif1a mRNA in melanoma cells. Moreover, MITF seems to be sufficient to transactivate the Hif1a promoter in nonmelanocyte cells.
MITF mediates the effect of cAMP on Hif1a promoter activity and protein expression
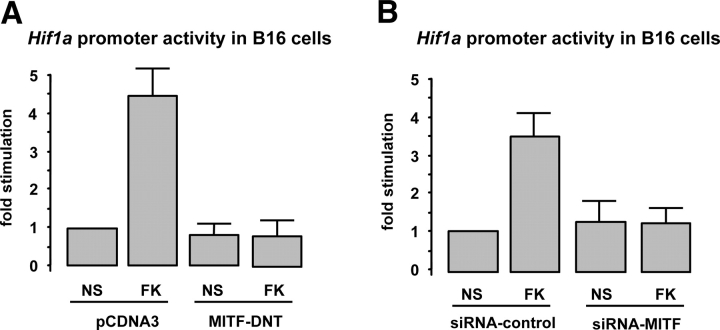
To evaluate whether MITF was required for the cAMP induction of the Hif1a gene expression, B16 cells were cotransfected with the Hif1a promoter fragment together with a dominant-negative mutant of MITF (MITF DNT) lacking the NH2-terminal transactivation domain, and cells were then stimulated with forskolin. Interestingly, we observed that MITF DNT completely abolished the cAMP stimulation of the Hif1a promoter activity (Fig. 4 A). To further strengthen the MITF specificity of such response, we performed the same experiment by using specific small interference RNAs (siRNAs) targeting MITF. As a control, cells were cotransfected with the Hif1a promoter together with a nonrelevant siRNA (control siRNA). Fig. 4 B shows that MITF silencing clearly inhibited the cAMP-induced Hif1a promoter transactivation, thereby indicating that MITF mediates the cAMP effects on Hif1a gene expression.
Figure 4.
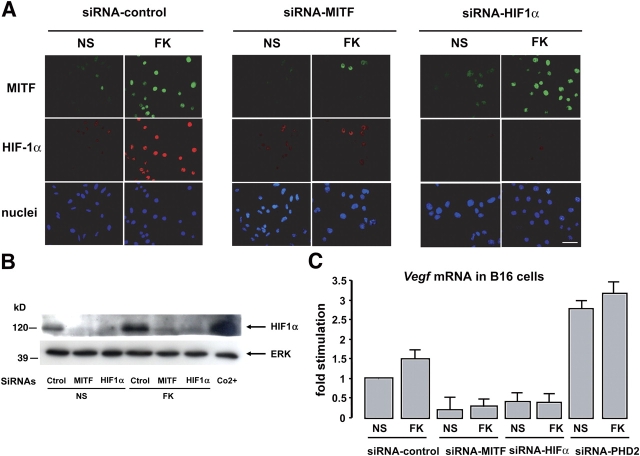
MITF is required for cAMP-dependent Hif1a expression. (A) Hif1a promoter activity assays on B16 cells cotransfected with either an empty vector (pCDNA3) or a construct encoding a dominant-negative mutant of MITF (MITF-DNT) and treated (or not) (NS) with the cAMP-elevating agent forskolin (FK). Results are expressed as the fold stimulation of the luciferase activity compared with the pcDNA3-transfected nonstimulated condition. (B) The same experiment was performed by cotransfecting the Hif1a promoter with either an siRNA-targeting MITF (siRNA-MITF) or a nonrelevant siRNA (siRNA-control).
To study the impact of MITF on the endogenous HIF1α protein, the same siRNA approach was undertaken. The expression of MITF and HIF1α proteins was then monitored by immunofluorescence using specific antibodies to MITF (green fluorescein labeling) and HIF1α (Texas red labeling). After 24 h of stimulation with the cAMP-elevating agent forskolin we observed an increased nuclear labeling corresponding to MITF expression, as well as a significant increase in HIF1α protein (Fig. 5 A). As expected, MITF silencing led to an important inhibition of cAMP-dependent MITF induction as well as the cAMP-induced HIF1α expression. In the presence of an siRNA to silence HIF1α expression, although cAMP elevating agents expectedly increased MITF expression, HIF1α expression was inhibited in both basal and cAMP conditions. A Western blot analysis of samples treated in parallel (Fig. 5 B) confirmed the same results in a more quantitative way. We therefore demonstrate that MITF controls HIF1α transactivation and constitutes a critical step in the cAMP up-regulation of HIF1α expression.
Figure 5.
MITF mediates the cAMP increase of HIF1α protein. (A) Immunofluorescence study of B16 cells transfected with a nonrelevant siRNA (siRNA control) (left), an siRNA-targeting MITF (middle), and an siRNA to HIF1α (right). Cells were either left nonstimulated (NS) or were stimulated for 24 h with the cAMP-elevating agent forskolin (FK). MITF protein expression was detected with the C5 monoclonal primary antibody and a secondary fluorescein-conjugated antibody (green), HIF1α was labeled with a primary pAb and a secondary Texas red–conjugated antibody (red), and nuclei were stained with bisbenzidine (blue). Bar, 20 μM. (B) Western blot analysis of HIF1α protein from B16 cell extracts treated as in A. Cells were stimulated with Co2+ for 12 h as a positive control. ERK2 detection showed the equal protein loading of each gel lane. (C) Real-time quantitative PCR to detect Vegf mRNA levels on total RNA extracts from B16 cells transfected and treated as described in A. A transfection condition using an siRNA to silence PHD2 (to increase HIF1α protein) was added.
MITF and HIF1α are responsible for the cAMP-dependent Vegf mRNA induction
Vegf being a well-known target gene of HIF1α involved in angiogenesis and tumor growth, we analyzed whether MITF and HIF1α could account for the cAMP-induced Vegf expression in B16 melanoma cells. Quantitative PCR experiments were performed on mRNAs extracted from B16 cells transfected with a control siRNA or siRNAs targeting MITF or HIF1α to deplete cells from these transcription factors, as well an siRNA to silence HIF prolyl-hydroxylase 2 (PHD2), which is specifically responsible for HIF1α protein degradation (Berra et al., 2003). We observed that MITF or HIF1α silencing inhibited the basal as well as the forskolin-induced Vegf expression, whereas PHD2 cell depletion, which increased HIF1α protein levels (unpublished data), exerted the opposite effect by significantly increasing the Vegf mRNA levels.
These data are consistent with Vegf being a MITF-HIF1α target gene in our melanoma model, and show that the cAMP-induced Vegf expression is mediated by the MITF-HIF1α cascade.
HIF1α plays a pro-survival role in melanoma
To investigate the physiological relevance of HIF1α induction in melanocyte cells, and taking into account the role attributed to HIF1 factor in cell survival, we evaluated the putative implication of HIF1α in this feature of melanoma cell physiology.
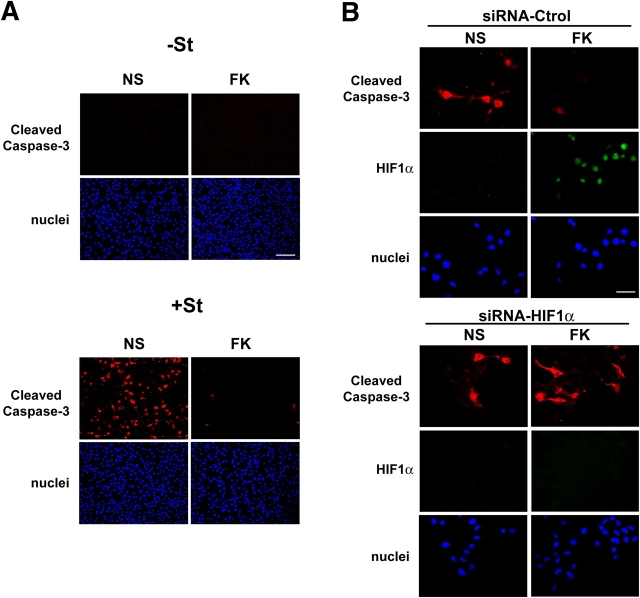
For that we treated B16 melanoma cells with staurosporin, considered a strong cell death inducing agent, and we used an immunofluorescence analysis to detect the presence of cleaved caspase-3 as a marker of cell death. We observed that treatment with staurosporin for 6 h greatly induced caspase-3 cleavage in our cell system (Fig. 6 A). We next pretreated B16 cells for 24 h with forskolin to increase HIF1α. We interestingly observed that cAMP strongly reduced the cleaved caspase-3 labeling thereby indicating that cAMP inhibits the staurosporin-induced cell death (Fig. 6 A).
Figure 6.
HIF1α plays a pro-survival role in B16 melanoma cells. (A) Immunofluorescence study to detect the presence of cleaved caspase-3 in B16 cells nonstimulated (NS) or stimulated for 24 with forskolin (FK), and nontreated (−St) or treated with staurosporin (+St) for 6 h. Bar, 40 μM. (B) Immunofluorescence analysis of B16 cells transfected with a nonrelevant siRNA (siRNA-Ctrol) or an siRNA to invalidate HIF1α. Cleaved caspase-3 was labeled with a secondary Texas red–conjugated antibody (red) and HIF1α protein was detected by an mAb and a secondary FITC-conjugated secondary one (green). Nuclei were labeled with bisbenzidine (blue). Bar, 20 μM.
Then we used an siRNA approach to dissect the contribution of HIF1α in this cell death protection. B16 cells were transfected with a nonrelevant siRNA or an siRNA to invalidate Hif1a expression. Cells were next treated (or not) with forskolin for 24 h, and they were finally exposed to 6 h of staurosporin. Fig. 6 B shows that in control siRNA–transfected cells, forskolin increased HIF1α protein expression and decreased caspase-3 cleavage, whereas in siRNA-Hif1α–transfected cells, the forskolin-dependent decrease of caspase-3 cleavage was nearly abolished. This result indicates that HIF1α protects cells from the staurosporin-induced cell death, thereby suggesting a pro-survival role of HIF1 in the melanoma cell model.
Discussion
In the present work, we have identified Hif1a as a new cAMP-induced gene in melanoma cells and we have characterized the molecular mechanisms underlying this regulation.
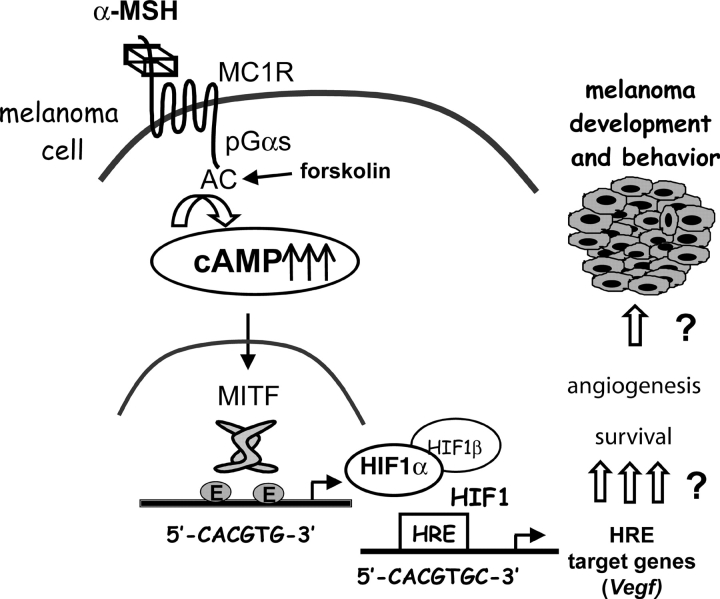
We demonstrate for the first time that cAMP regulates Hif1a at a transcriptional level, leading to increased expression of HIF1α mRNA and protein. HIF1α forms a HIF1 complex that is functional and able to transactivate target genes such as Vegf. Very interestingly, HIF1α induction by cAMP is restricted to melanocyte cells, prompting us to propose a cell-specific regulation through the melanocyte-specific transcription factor MITF. We demonstrate that MITF binds and transactivates the Hif1a promoter, and the overexpression of MITF is sufficient to increase Hif1a mRNA levels. Furthermore, by silencing MITF or Hif1α expression, we show that cAMP-induced HIF1α expression is mediated by MITF, and that the MITF/HIF1α pathway mediates the cAMP induction of Vegf expression (a model illustrating these conclusions is shown in Fig. 7). We therefore define Hif1a as a new MITF target gene and we provide data revealing the pro-survival role of the MITF-HIF1 cascade in melanoma cells.
Figure 7.
Model for the mechanisms of cAMP induction of HIF1α expression. α-MSH, by up-regulating the cAMP pathway, increases MITF expression. MITF binds and transactivates the Hif1a promoter, thereby increasing the expression of Hif1a gene and protein in a melanocyte cell–specific manner. HIF1α dimerizes with HIF1β to constitute a functional HIF1 transcription factor that is capable of activating target genes such as Vegf and up-regulate other genes involved in melanocyte cell survival and angiogenesis to participate in melanoma development and behavior.
HIF1α activation triggered by hypoxia is mainly governed by post-translational modifications involving hydroxylation of specific amino acid residues in the HIF1α subunit (Berra et al., 2003). Indeed, O2-dependent prolyl hydroxylation regulates proteasome-mediated destruction of HIF1α, whereas asparaginyl hydroxylation blocks the interaction with the p300/CBP coactivators (for review see Lando et al., 2002). In addition to hypoxia, growth factors, vaso-active hormones, cytokines, and tumor suppressor mutations also stimulate HIF1α synthesis via activation of the phosphatidylinositol 3-kinase or MAPK pathways (Zhong et al., 2000; Zundel et al., 2000; Fukuda et al., 2002).
As a novelty, our study clearly demonstrates that cAMP regulates HIF1α at a transcriptional level independently of the oxygen context. To date, very few works have described a regulation of Hif1a mRNA (Jiang et al., 1997; Blouin et al., 2004; Liu et al., 2004). Nevertheless, no studies have so far dissected or identified a transcriptional mechanism accounting for HIF1α gene expression. In our work, we go further in the characterization of the molecular mechanisms that regulate Hif1a gene transcription because we identify for the first time a transcription factor (i.e., MITF) that binds and functionally transactivates the Hif1a promoter and controls this gene expression independently of the oxygen conditions.
We also show that cAMP and hypoxia have additive effects on HIF1α protein levels. Both stimuli via transcriptional and post-translational mechanisms, respectively, cooperate to highly induce HIF1α protein. It is tempting to propose that, independently of the oxygen availability, MITF acts as a fine modulator of the final HIF1α protein levels to modify the biological fate of melanocytes and melanoma cells.
In addition to regulating Hif1a transcription, we could imagine that cAMP, like hypoxia, might act on HIF1α protein stabilization by modulating its hydroxylation or enhancing its translation. This does not seem to be the case because MITF inhibition by a dominant-negative mutant or a specific siRNA completely abolishes the cAMP-induced HIF1α protein expression. However, we cannot definitively rule out the possibility that cAMP might also regulate HIF1 DNA binding or transactivation potential by controlling other HIF1α post-translational modifications such as phosphorylation. In support of that, we have previously described that cAMP specifically activates the MAPK pathway in melanocytes (Englaro et al., 1995; Buscà et al., 2000) and the transcriptional activity of HIF1 has been reported to be stimulated upon phosphorylation by MAPK (Richard et al., 1999; Suzuki et al., 2001; Lee et al., 2002).
HIF1α has a broad impact on gene expression (>60 direct target genes have been identified), making it a crucial element in the regulation of many cell features going from cell proliferation to motility and apoptosis. Furthermore, HIF1α has a special impact on cancer progression because of its effects on angiogenesis and glucose metabolism (Semenza, 2003). Further, HIF1 mediates tumor cell adaptive response to hypoxia by regulating genes implicated in cell survival (Semenza 2001). In nontumor cells, a recent work by Xie et al. (2005) also involves HIF1 as a survival factor that protects neurons from NGF deprivation-induced cell death. In contrast, in other contexts HIF1α has been shown to participate in hypoxia-induced cell death via interaction with p53 (Halterman and Federoff, 1999) or modulation of several pro-apoptotic targets such as NIP3 (Bruick, 2000), Noxa (Kim et al., 2004), BNIP3 and NIX (Sowter et al., 2001), RTP801 (Shoshani et al., 2002), and the recently described HGTD-P (Lee et al., 2004). In this case, MITF, by regulating HIF1α expression, would rather favor cell death. In our melanoma cell system, we find that HIF1 plays a pro-survival role, as observed by the inhibition of the staurosporin-induced caspase-3 cleavage. We can therefore imagine that the cAMP/MITF/HIF1 pathway might contribute to higher cell viability and resistance to any putative physiological or environmental stresses such as UV exposure, for example.
The fact that cAMP increases Vegf mRNA in our melanoma cells also leads us to hypothesize an angiogenic role of the cAMP/MITF/HIF1 cascade. In agreement with this hypothesis, UV radiation has been reported to increase cutaneous angiogenesis (Bielenberg et al., 1998) and nevus growth (Menzies et al., 2004). Because α-MSH production in skin is stimulated upon UV radiation, we could imagine that α-MSH, by up-regulating the cAMP-MITF-HIF1 pathway could participate to the angiogenic and growth-promoting effects of UV radiation.
Thus, together, these observations suggest that the stimulation of HIF1α expression by cAMP and MITF might facilitate both melanoma survival, neo-vascularization, and metabolic adaptation, thereby favoring the development of these tumors.
Although the role of MITF in survival and development of melanocyte precursors has been clearly documented, the growth and survival functions of MITF in mature melanocytes and melanoma cells remain a matter of debate. Indeed, it has been demonstrated that overexpression of MITF into a melanoma cell line results in slower xenograft tumor growth (Selzer et al., 2002), and MITF was recently shown to inhibit cell growth by activating the cell cycle inhibitor INK4A (Loercher et al., 2005) as well p21CIP1 expression (Carreira et al., 2005). However, recent reports have shown that MITF is a melanoma survival factor presumably through the up-regulation of the anti-apoptotic gene BCL2 (McGill et al., 2002) and promotes melanoma cell growth by stimulating CDK2 expression (Du et al., 2004).
All of these data suggest that MITF, as well HIF1α, can elicit opposite cellular responses. The decision between pro-survival and growth inhibition might be dictated by the cellular context, the extracellular signals, the microenvironment, or the mutational status of melanoma. In our case, cAMP constitutes a signal that clearly tilts the balance toward survival and presumably angiogenesis and melanoma growth.
In conclusion, here we show for the first time a transcriptional up-regulation of HIF1α by the melanocyte-specific transcription factor MITF, thereby establishing a possible link between MITF and melanoma survival and vascularization. Taking into account the functional plasticity of both MITF and HIF1α, further studies are underway to elucidate the precise physiological relevance of the MITF-HIF1α cascade in melanoma development. Modulation of such regulatory mechanisms could provide novel therapeutic approaches to this aggressive and metastatic cancer.
Materials and methods
Cell culture
B16/F10 (mouse melanoma cells), Mewo (human melanoma cells), and NIH 3T3 (mouse fibroblasts) were cultured in DME (GIBCO BRL) supplemented with 7% FBS (Hyclone Laboratories) and penicillin/streptomycin (100 IU/50 μg/ml; Sigma-Aldrich) in a humidified atmosphere containing 5% CO2 in air at 37°C. Primary human melanocytes (NHM) were obtained from neonatal foreskins of Caucasian individuals, and grown in MCDB medium (GIBCO BRL) according to a modification of the method of Eisinger and Marko (1982) and Aberdam et al. (1993). Before cAMP treatment, NHM were depleted in forskolin for at least 24 h.
When indicated, cells were treated with forskolin (20 μM), α-MSH (1 μM), or cobalt (200 μM) to mimic hypoxia. To induce caspase-3 cleavage, cells were treated with 1 μM staurosporin (Calbiochem) for 6 h. Hypoxic conditions were produced by incubation of cells in a sealed “Bug-Box” anaerobic workstation (Ruskin Technologies). The oxygen in this workstation was maintained at 1–2%, with the residual gas mixture being 93–94% nitrogen and 5% CO2.
Plasmids, adenovirus, siRNAs, and PCR oligonucleotide sequences
The plasmid encoding the mouse Hif1a promoter fragment upstream of the luciferase reporter gene was provided by Dr. R. Wenger (University of Zürich, Zürich, Switzerland). The 3-HRE-LUC reporter vector contains three copies of the HRE from the erythropoietin promoter cloned upstream of a minimal thymidine kinase promoter and the luciferase reporter gene (Berra et al., 2003). The plasmids encoding the mouse MITF and MITFDNT are described in Bertolotto et al. (1998a). The adenovirus encoding MITF is reported in Gaggioli et al. (2003).
The DNA sequence corresponding to the siRNA-targeting Mitf is: 5′-GGTGAATCGGATCATCAAG-3′; Hif1a: 5′-CCTATATCCCAATGGATGATG-3′. As nonrelevant siRNAs (control siRNAs) we used the siLUC: 5′-CGTACGCGGAATACTTCGA-3′ and the Drosophila melanogaster Hif1a (SIMA): 5′-CCTCATCCCGATCGATGATG-3′ described in Berra et al. (2003). The mouse siRNA sequence targeting PHD2 was: 5′-GAACTCAAGCCCAATTCAGTT-3′.
The oligonucleotides to amplify the Hif1a and tyrosinase promoter regions were: 5′-CACGGGCACGAAGTGTTCCTTTG-3′ (forward) and 5′-GCTCAGCCGCACCCGTCACT-3′ (reverse) for Hif1a and 5′-GATCTCTAAATACAACAGGCT-3′ (forward) and 5′-TCATACAAAATCTGCACCAA-3′ (reverse) for tyrosinase, and to amplify GAPDH we used: 5′-TGGTATGACAACGAATTTG-3′ (forward) and 5′-TCTACATGGCAACTGT-GAGG-3′ (reverse).
The oligonucleotide sequences corresponding to mouse Hif1a, Mitf, tyrosinase, tyrosinase-related protein 1, MyoVa and Rab27a, and human MITF and HIF1A used for real-time quantitative PCR are available upon request.
Antibodies
The mAb to MITF (C5) was obtained from Dr. D. Fisher (Dana Farber Cancer Institute, Boston, MA; Weilbaecher et al., 1998). The polyclonal anti-MITF antibody used for the chromatin immunoprecipitation assays is described in Bertolotto et al. (1996). The rabbit anti-HIF1α (antiserum 2087) is described in Richard et al. (1999) and the monoclonal anti-HIF1α antibody used for the HIF1α/caspase-3 double labeling was from Abcam. The pAb to cleaved caspase-3 was from Cell Signaling. The pAb anti-extracellular signal-regulated kinase 2 (ERK2) is from Santa Cruz Biotechnologies, Inc. The anti–mouse and anti–rabbit peroxidase-conjugated secondary antibodies as well as the FITC and Texas red–conjugated anti–mouse and anti–rabbit antibodies were from Dakopatts.
Biochips screening
The laboratory-designed microarray containing 4,200 genes was previously described (Moreilhon et al., 2004). The list of probes spotted on the microarray is available at http://medlab.ipmc.cnrs.fr/gene_List.html. Total RNAs (10 μg) were reverse transcribed using the CyScribe First-Strand cDNA labeling kit (RPN 6200; Amersham Biosciences). Microarrays were hybridized with the Cy3/Cy5 mixed sample overnight at 42°C, and were washed at RT in 1× SSC/0.03% SDS for 5 min, in 0.2× SSC for 5 min, and in 0.05× SSC for 5 min. Slides were scanned using a confocal scanner (Scan Array 5000; GSI Lumonics). Images were analyzed by the Quant Array software.
Quantitative real-time one-step RT-PCR and Western blot
RNAs were extracted using TRIzol reagent (Invitrogen) and Rneasy protect mini kit (QIAGEN). mRNA expression levels were quantified by real-time one-step RT-PCR using SYBR-Green PCR Master Mix (Applied Biosystems). The results are the average of three separate experiments.
For the Western blot analysis, subconfluent cells were lysed in Laemmli buffer. The protein concentration was determined using the Lowry assay and 50 μg total cell extracts was resolved by SDS-PAGE (7.5% for HIF1α detection) and electrophoretically transferred onto a PVDF membrane (Millipore). Immunoreactive bands were visualized using the ECL system (Amersham Biosciences).
Chromatin immunoprecipitation assay
B16 cells were stimulated (or not) with forskolin for 5 h and were then treated with 1% formaldehyde for 5 min at 37°C and for 30 min at 4°C. Next, cells were harvested and suspended in the lysis buffer (10 mM Hepes, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.1% NP-40, and protease inhibitors). After 10 min incubation on ice, cells were centrifuged (5,000 rpm, 5 min at 4°C) to pellet the nuclei. Nuclei were then suspended in the nuclei lysis buffer (50 mM Tris-Cl, pH 8.1, 10 mM EDTA, 1% SDS, and protease inhibitors) and incubated on ice for 10 min. The sheared chromatin was then immunoprecipitated using (or not) a specific polyclonal anti-MITF antibody (Bertolotto et al., 1998b). After immunoprecipitation, the cross-link was reverted by heat treatment (67°C overnight and proteinase K digestion). The genomic captured fragments were then recovered by phenol-chloroform extraction. Identification of the captured Hif1a or tyrosinase promoter fragments was performed by PCR analysis using the promoter primers. 30 cycles of PCR were performed and the amplified products were analyzed on a 2% agarose gel.
Transfections and luciferase assays
B16 melanoma cells and NIH-3T3 fibroblasts were seeded in 24-well dishes, and transient transfections were performed the following day using 2 μl Lipofectamine (GIBCO BRL) and 0.5 μg total plasmid DNA or 20 nM annealed siRNAs, in a 200-μl final volume. pCMVβGal was cotransfected with the test plasmids to control the variability of transfection efficiency in the reporter assays. 24 h after transfection, soluble extracts were harvested in 50 μl of lysis buffer and assayed for luciferase and β-galactosidase activities. All transfections were repeated several times using different plasmid preparations.
Immunofluorescence microscopy
siRNA-transfected B16 melanoma cells cultured on glass coverslips were washed three times in PBS, fixed for 20 min in 3% PFA, washed again, and incubated for 10 min in 50 mM NH4Cl. After three more washes, cells were incubated with the primary antibody diluted in PBS containing 0.5% BSA and 0.02% saponin for 1 h, washed abundantly in PBS, and incubated for another hour with the appropriate conjugated secondary antibody. After washing, the coverslips were incubated with bisbenzidine (0.5 μg/ml) to stain the nuclei, they were mounted on glass slides, and viewed with an Axiophot fluorescent microscope (Carl Zeiss MicroImaging, Inc.), using a 40×/0.75 Plan Neofluar objective lens. To acquire the images, a COHU (High performance CCD camera) and the Q-Fish (Leica) informatic software was used.
Acknowledgments
We are grateful to the facilities of the Nice-Sophia Antipolis Transcriptome Platform of the Marseille-Nice Genopole in which the microarray experiments were carried out. We thank Dr. R. Wenger for providing the Hif1a promoter construct and Dr. D. Fisher for the C5 anti-MITF antibody. We also thank Dr. P. Pognonec for the plasmids encoding USF and myc. We are very grateful to Dr. P. Lenormand for all his help and unconditional support in this work.
Abbreviations used in this paper: α-MSH, α-melanocyte stimulating hormone; ERK2, extracellular signal-regulated kinase 2; HIF1α, hypoxia-inducible factor 1α; HRE, hypoxia-responsive element; MC1R, melanocortin-1 receptor; MITF, microphthalmia-associated transcription factor; NHM, normal human melanocytes; PHD2, prolyl-hydroxylase 2; siRNA, small interference RNA; VEGF, vascular endothelial growth factor.
References
- Aberdam, E., C. Roméro, and J.P. Ortonne. 1993. Repeated UVB irradiations do not have the same potential to promote stimulation of melanogenesis in cultured normal human melanocytes. J. Cell Sci. 106:1015–1022. [DOI] [PubMed] [Google Scholar]
- Aksan, I., and C.R. Goding. 1998. Targeting the microphthalmia basic helix-loop-helix-leucine zipper transcription factor to a subset of E-box elements in vitro and in vivo. Mol. Cell. Biol. 18:6930–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berra, E., E. Benizri, A. Ginouves, V. Volmat, D. Roux, and J. Pouyssegur. 2003. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1α in normoxia. EMBO J. 22:4082–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotto, C., K. Bille, J.-P. Ortonne, and R. Ballotti. 1996. Regulation of tyrosinase gene expression by cAMP in B16 melanoma cells involves two CATGTG motifs surrounding the TATA box: implication of the microphthalmia gene product. J. Cell Biol. 134:747–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotto, C., P. Abbe, T.J. Hemesath, K. Bille, D.E. Fisher, J.-P. Ortonne, and R. Ballotti. 1998. a. Microphthalmia gene product as a signal transducer in cAMP-induced differentiation of melanocytes. J. Cell Biol. 142:827–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolotto, C., R. Buscà, P. Abbe, K. Bille, E. Aberdam, J.-P. Ortonne, and R. Ballotti. 1998. b. Different cis-acting elements are involved in the regulation of TRP1 and TRP2 promoter activities by cyclic AMP: pivotal role of M boxes (GTCATGTGCT) and of microphthalmia. Mol. Cell. Biol. 18:694–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielenberg, D.R., C.D. Bucana, R. Sanchez, C.K. Donawho, M.L. Kripke, and I.J. Fidler. 1998. Molecular regulation of UVB-induced cutaneous angiogenesis. J. Invest. Dermatol. 111:864–872. [DOI] [PubMed] [Google Scholar]
- Blouin, C.C., E.L. Page, G.M. Soucy, and D.E. Richard. 2004. Hypoxic gene activation by lipopolysaccharide in macrophages: implication of hypoxia-inducible factor 1α. Blood. 103:1124–1130. [DOI] [PubMed] [Google Scholar]
- Brahimi-Horn, C., E. Berra, and J. Pouyssegur. 2001. Hypoxia: the tumor's gateway to progression along the angiogenic pathway. Trends Cell Biol. 11:S32–S36. [DOI] [PubMed] [Google Scholar]
- Bruick, R.K. 2000. Expression of the gene encoding the proapoptotic Nip3 protein is induced by hypoxia. Proc. Natl. Acad. Sci. USA. 97:9082–9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscà, R., and R. Ballotti. 2000. Cyclic AMP a key messenger in the regulation of skin pigmentation. Pigment Cell Res. 13:60–69. [DOI] [PubMed] [Google Scholar]
- Buscà, R., C. Bertolotto, J.P. Ortonne, and R. Ballotti. 1996. Inhibition of the phosphatidylinositol 3-kinase/p70(S6)-kinase pathway induces B16 melanoma cell differentiation. J. Biol. Chem. 271:31824–31830. [DOI] [PubMed] [Google Scholar]
- Buscà, R., C. Bertolotto, P. Abbe, W. Englaro, T. Ishizaki, S. Narumiya, P. Boquet, J.P. Ortonne, and R. Ballotti. 1998. Inhibition of Rho is required for cAMP-induced melanoma cell differentiation. Mol. Biol. Cell. 9:1367–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buscà, R., P. Abbe, F. Mantoux, E. Aberdam, C. Peyssonnaux, A. Eychene, J.P. Ortonne, and R. Ballotti. 2000. Ras mediates the cAMP-dependent activation of extracellular signal-regulated kinases (ERKs) in melanocytes. EMBO J. 19:2900–2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreira, S., J. Goodall, I. Aksan, S.A. La Rocca, M.D. Galibert, L. Denat, L. Larue, and C.R. Goding. 2005. Mitf cooperates with Rb1 and activates p21Cip1 expression to regulate cell cycle progression. Nature. 433:764–769. [DOI] [PubMed] [Google Scholar]
- Davies, H., G.R. Bignell, C. Cox, P. Stephens, S. Edkins, S. Clegg, J. Teague, H. Woffendin, M.J. Garnett, W. Bottomley, et al. 2002. Mutations of the BRAF gene in human cancer. Nature. 417:949–954. [DOI] [PubMed] [Google Scholar]
- Du, J., H.R. Widlund, M.A. Horstmann, S. Ramaswamy, K. Ross, W.E. Huber, E.K. Nishimura, T.R. Golub, and D.E. Fisher. 2004. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell. 6:565–576. [DOI] [PubMed] [Google Scholar]
- Eisinger, M., and O. Marko. 1982. Selective proliferation of normal human melanocytes in vitro in presence of phorbol ester and cholera toxin. Proc. Natl. Acad. Sci. USA. 79:2018–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englaro, W., R. Rezzonico, M. Durand-Clément, D. Lallemand, J.-P. Ortonne, and R. Ballotti. 1995. Mitogen-activated protein kinase pathway and AP-1 are activated during cAMP-induced melanogenesis in B-16 melanoma cells. J. Biol. Chem. 270:24315–24320. [DOI] [PubMed] [Google Scholar]
- Fukuda, R., K. Hirota, F. Fan, Y.D. Jung, L.M. Ellis, and G.L. Semenza. 2002. Insulin-like growth factor 1 induces hypoxia-inducible factor 1-mediated vascular endothelial growth factor expression, which is dependent on MAP kinase and phosphatidylinositol 3-kinase signaling in colon cancer cells. J. Biol. Chem. 277:38205–38211. [DOI] [PubMed] [Google Scholar]
- Gaggioli, C., R. Buscà, P. Abbe, J.P. Ortonne, and R. Ballotti. 2003. Microphthalmia-associated transcription factor (MITF) is required but is not sufficient to induce the expression of melanogenic genes. Pigment Cell Res. 16:374–382. [DOI] [PubMed] [Google Scholar]
- Halterman, M.W., and H.J. Federoff. 1999. HIF-1α and p53 promote hypoxia-induced delayed neuronal death in models of CNS ischemia. Exp. Neurol. 159:65–72. [DOI] [PubMed] [Google Scholar]
- Hodgkinson, C.A., K.J. Moore, A. Nakayama, E. Steingrímsson, N.G. Copeland, N.A. Jenkins, and H. Arnheiter. 1993. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 74:395–404. [DOI] [PubMed] [Google Scholar]
- Hughes, M.J., J.B. Lingrel, J.M. Krakowski, and K.P. Anderson. 1993. A helix-loop-helix transcription factor-like gene is located at the mi locus. J. Biol. Chem. 268:20687–20690. [PubMed] [Google Scholar]
- Jiang, B.H., F. Agani, A. Passaniti, and G.L. Semenza. 1997. V-SRC induces expression of hypoxia-inducible factor 1 (HIF-1) and transcription of genes encoding vascular endothelial growth factor and enolase 1: involvement of HIF-1 in tumor progression. Cancer Res. 57:5328–5335. [PubMed] [Google Scholar]
- Khaled, M., L. Larribere, K. Bille, E. Aberdam, J.P. Ortonne, R. Ballotti, and C. Bertolotto. 2002. Glycogen synthase kinase 3β is activated by cAMP and plays an active role in the regulation of melanogenesis. J. Biol. Chem. 277:33690–33697. [DOI] [PubMed] [Google Scholar]
- Kim, J.Y., H.J. Ahn, J.H. Ryu, K. Suk, and J.H. Park. 2004. BH3-only protein Noxa is a mediator of hypoxic cell death induced by hypoxia-inducible factor 1α. J. Exp. Med. 199:113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lando, D., D.J. Peet, J.J. Gorman, D.A. Whelan, M.L. Whitelaw, and R.K. Bruick. 2002. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 16:1466–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larribere, L., M. Khaled, S. Tartare-Deckert, R. Buscà, F. Luciano, K. Bille, G. Valony, A. Eychene, P. Auberger, J.P. Ortonne, et al. 2004. PI3K mediates protection against TRAIL-induced apoptosis in primary human melanocytes. Cell Death Differ. 11:1084–1091. [DOI] [PubMed] [Google Scholar]
- Lee, E., S. Yim, S.K. Lee, and H. Park. 2002. Two transactivation domains of hypoxia-inducible factor-1α regulated by the MEK-1/p42/p44 MAPK pathway. Mol. Cells. 14:9–15. [PubMed] [Google Scholar]
- Lee, M.J., J.Y. Kim, K. Suk, and J.H. Park. 2004. Identification of the hypoxia-inducible factor 1α-responsive HGTD-P gene as a mediator in the mitochondrial apoptotic pathway. Mol. Cell. Biol. 24:3918–3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q., U. Moller, D. Flugel, and T. Kietzmann. 2004. Induction of plasminogen activator inhibitor I gene expression by intracellular calcium via hypoxia-inducible factor-1. Blood. 104:3993–4001. [DOI] [PubMed] [Google Scholar]
- Loercher, A.E., E.M. Tank, R.B. Delston, and J.W. Harbour. 2005. MITF links differentiation with cell cycle arrest in melanocytes by transcriptional activation of INK4A. J. Cell Biol. 168:35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill, G.G., M. Horstmann, H.R. Widlund, J. Du, G. Motyckova, E.K. Nishimura, Y.L. Lin, S. Ramaswamy, W. Avery, H.F. Ding, et al. 2002. Bcl2 regulation by the melanocyte master regulator Mitf modulates lineage survival and melanoma cell viability. Cell. 109:707–718. [DOI] [PubMed] [Google Scholar]
- Menzies, S.W., G.E. Greenoak, C.M. Abeywardana, K.A. Crotty, and M.E. O'Neill. 2004. UV light from 290 to 325 nm, but not broad-band UVA or visible light, augments the formation of melanocytic nevi in a guinea-pig model for human nevi. J. Invest. Dermatol. 123:354–360. [DOI] [PubMed] [Google Scholar]
- Moreilhon, C., D. Gras, C. Hologne, O. Bajolet, F. Cottrez, V. Magnone, M. Merten, H. Groux, E. Puchelle, and P. Barbry. 2004. Live Staphylococcus aureus and bacterial soluble factors induce different transcriptional responses in human airway cells. Physiol. Genomics. 20:244–255. [DOI] [PubMed] [Google Scholar]
- Passeron, T., P. Bahadoran, C. Bertolotto, C. Chiaverini, R. Buscà, G. Valony, K. Bille, J.P. Ortonne, and R. Ballotti. 2004. Cyclic AMP promotes a peripheral distribution of melanosomes and stimulates melanophilin/Slac2-a and actin association. FASEB J. 18:989–991. [DOI] [PubMed] [Google Scholar]
- Poser, I., and A.K. Bosserhoff. 2004. Transcription factors involved in development and progression of malignant melanoma. Histol. Histopathol. 19:173–188. [DOI] [PubMed] [Google Scholar]
- Richard, D.E., E. Berra, E. Gothie, D. Roux, and J. Pouyssegur. 1999. p42/p44 mitogen-activated protein kinases phosphorylate hypoxia-inducible factor 1α (HIF-1α) and enhance the transcriptional activity of HIF-1. J. Biol. Chem. 274:32631–32637. [DOI] [PubMed] [Google Scholar]
- Robinson, S.J., and E. Healy. 2002. Human melanocortin 1 receptor (MC1R) gene variants alter melanoma cell growth and adhesion to extracellular matrix. Oncogene. 21:8037–8046. [DOI] [PubMed] [Google Scholar]
- Rusciano, D., P. Lorenzoni, and M.M. Burger. 1999. Regulation of c-met expression in B16 murine melanoma cells by melanocyte stimulating hormone. J. Cell Sci. 112:623–630. [DOI] [PubMed] [Google Scholar]
- Selzer, E., V. Wacheck, T. Lucas, E. Heere-Ress, M. Wu, K.N. Weilbaecher, W. Schlegel, P. Valent, F. Wrba, H. Pehamberger, et al. 2002. The melanocyte-specific isoform of the microphthalmia transcription factor affects the phenotype of human melanoma. Cancer Res. 62:2098–2103. [PubMed] [Google Scholar]
- Semenza, G.L. 2001. Hypoxia-inducible factor 1: oxygen homeostasis and disease pathophysiology. Trends Mol. Med. 7:345–350. [DOI] [PubMed] [Google Scholar]
- Semenza, G.L. 2002. Involvement of hypoxia-inducible factor 1 in human cancer. Intern. Med. 41:79–83. [DOI] [PubMed] [Google Scholar]
- Semenza, G.L. 2003. Targeting HIF-1 for cancer therapy. Nat. Rev. Cancer. 3:721–732. [DOI] [PubMed] [Google Scholar]
- Shoshani, T., A. Faerman, I. Mett, E. Zelin, T. Tenne, S. Gorodin, Y. Moshel, S. Elbaz, A. Budanov, A. Chajut, et al. 2002. Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, involved in apoptosis. Mol. Cell. Biol. 22:2283–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowter, H.M., P.J. Ratcliffe, P. Watson, A.H. Greenberg, and A.L. Harris. 2001. HIF-1-dependent regulation of hypoxic induction of the cell death factors BNIP3 and NIX in human tumors. Cancer Res. 61:6669–6673. [PubMed] [Google Scholar]
- Suzuki, H., A. Tomida, and T. Tsuruo. 2001. Dephosphorylated hypoxia-inducible factor 1α as a mediator of p53-dependent apoptosis during hypoxia. Oncogene. 20:5779–5788. [DOI] [PubMed] [Google Scholar]
- Valverde, P., E. Healy, I. Jackson, J. Rees, and A. Thody. 1995. Variants of the melanocyte-stimulating hormone receptor gene are associated with red hair and fair skin in humans. Nat. Genet. 11:328–330. [DOI] [PubMed] [Google Scholar]
- Weilbaecher, K.N., C.L. Hershey, C.M. Takemoto, M.A. Horstmann, T.J. Hemesath, A.H. Tashjian, and D.E. Fisher. 1998. Age-resolving osteopetrosis: a rat model implicating microphthalmia and the related transcription factor TFE3. J. Exp. Med. 187:775–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger, R.H., A. Rolfs, P. Spielmann, D.R. Zimmermann, and M. Gassmann. 1998. Mouse hypoxia-inducible factor-1α is encoded by two different mRNA isoforms: expression from a tissue-specific and a housekeeping-type promoter. Blood. 91:3471–3480. [PubMed] [Google Scholar]
- Xie, L., R. Johnson, and R. Freeman. 2005. Inhibition of NFG deprivation-induced death by low oxygen involves suppression of BIMEL and activation of HIF-1. J. Cell Biol. 168:911–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, H., K. Chiles, D. Feldser, E. Laughner, C. Hanrahan, M.M. Georgescu, J.W. Simons, and G.L. Semenza. 2000. Modulation of hypoxia-inducible factor 1α expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: implications for tumor angiogenesis and therapeutics. Cancer Res. 60:1541–1545. [PubMed] [Google Scholar]
- Zundel, W., C. Schindler, D. Haas-Kogan, A. Koong, F. Kaper, E. Chen, A.R. Gottschalk, H.E. Ryan, R.S. Johnson, A.B. Jefferson, et al. 2000. Loss of PTEN facilitates HIF-1-mediated gene expression. Genes Dev. 14:391–396. [PMC free article] [PubMed] [Google Scholar]