Abstract
Unlike parasitic protist groups that are defined by the absence of mitochondria, the Pelobiontida is composed mostly of free-living species. Because of the presence of ultrastructural and cellular features that set them apart from all other eukaryotic organisms, it has been suggested that pelobionts are primitively amitochondriate and may represent the earliest-evolved lineage of extant protists. Analyses of rRNA genes, however, have suggested that the group arose well after the diversification of the earliest-evolved protists. Here we report the sequence of the gene encoding the largest subunit of DNA-dependent RNA polymerase II (RPB1) from the pelobiont Mastigamoeba invertens. Sequences within RPB1 encompass several of the conserved catalytic domains that are common to eubacterial, archaeal, and eukaryotic nuclear-encoded RNA polymerases. In RNA polymerase II, these domains catalyze the transcription of all nuclear pre-mRNAs, as well as the majority of small nuclear RNAs. In contrast with rDNA-based trees, phylogenetic analyses of RPB1 sequences indicate that Mastigamoeba represents an early branch of eukaryotic evolution. Unlike sequences from parasitic amitochondriate protists that were included in our study, there is no indication that Mastigamoeba RPB1 is attracted to the base of the eukaryotic tree artifactually. In addition, the presence of introns and a heptapeptide C-terminal repeat in the Mastigamoeba RPB1 sequence, features that are typically associated with more recently derived eukaryotic groups, raise provocative questions regarding models of protist evolution that depend almost exclusively on rDNA sequence analyses.
The protistan order Pelobiontida is composed predominantly of free-living amoeboflagellates that inhabit microoxic and anoxic environments (1–3). Because they lack mitochondria, Golgi bodies, and most membrane-bound organelles it has been argued that pelobionts, along with other protist groups defined by an absence of mitochondria (Metamonada, Parabasalia, and Microsporidia) branched from the eukaryotic line before the establishment of an endosymbiotic relationship with the ancestor of mitochondria and, therefore, represent the earliest eukaryotic lineages (1). Among these groups, the Pelobiontida exhibit cytologic features that have been interpreted as ancestral even to other amitochondrial protists (2). The presence of a single basal body and flagellum, unique flagellar-root ultrastructure, and a highly reduced endomembrane complement suggest that pelobionts are distinct from all other eukaryotes (2, 3). Based primarily on these cytologic features, amitochondriate amoebae were proposed to be the most ancient of extant eukaryotes (2). Initial molecular analyses have not supported this conclusion.
The amitochondriate amoeboid gut parasite Entamoeba histolytica does not branch near the base of eukaryotes in phylogenetic analyses of small-subunit ribosomal RNA gene sequences (SSU rDNA) (4). In addition, the presence in Entamoeba of two genes apparently of mitochondrial origin suggests that it is secondarily amitochondriate (5). Because no flagellate stage has been observed in Entamoeba, however, corroborative ultrastuctural evidence relating Entamoeba to the Pelobiontida is lacking, and proposed relationships between the two taxa (2) have no support. Therefore, neither the derived position of Entamoeba in phylogenetic analyses, nor indications that its ancestors contained mitochondria, should be construed as evidence against the antiquity and possibly primitive amitochondrial nature of pelobiont amoebae.
Analysis of the SSU rDNA sequence from the pelobiont Mastigamoeba balamuthi (under the synomym Phreatamoeba balamuthi) also does not indicate an ancient origin of the Pelobiontida (6). Although the sequence contains large insertions in variable regions, and is one of the longest SSU rRNA genes yet reported, the M. balamuthi sequence branches at a derived position near or within the so-called “crown” (7) eukaryotes (6, 8). A partial large-subunit rDNA sequence from an unidentified species ascribed to the pelobiont genus Pelomyxa also does not branch near the base of the eukaryotic tree (9). To our knowledge, however, neither this sequence nor a specific identification of the taxon has been published.
The unusually long M. balamuthi rDNA sequence has been the only molecular character from a clearly defined pelobiont amoeba available for comparative evolutionary analysis. Given the cytological and ultrastructural evidence for a possible ancient origin of the Pelobiontida, additional molecular markers are needed to provide an accurate account of their evolutionary position among eukaryotes. Here we report the sequence of the gene encoding the largest subunit of RNA polymerase II (RPB1) from ‘Mastigamoeba invertens’ Klebs. Analyses of this sequence suggest that Mastigamoeba represents an early eukaryotic lineage and raise questions about current models of eukaryotic evolution that depend heavily on phylogenetic analyses of aligned rDNA sequences.
MATERIALS AND METHODS
Culturing.
“Mastigamoeba invertens” was obtained from the American Type Culture Collection (ATCC no. 50338; www.atcc.org/atcc.html) and cultured in #1773 Hexamita medium following ATCC instructions. Cultures were maintained in 16 × 122 mm screw-capped tubes at 25°C for 5–9 days to achieve peak cell densities of 1–2 × 106 cells per ml and subsequently either subcultured or harvested. Tubes were sampled every few days and examined under ×320 phase-contrast microscopy to determine cell densities and to monitor for the presence of eukaryotic contaminants.
DNA Extraction.
Cells were pelleted at 2,000 × g and 4°C for 10 min. Pellets were gently resuspended in lysis buffer (2% cetyltrimethylammonium bromide/1.4 M NaCl/20 mM EDTA/100 mM Tris⋅Cl, pH 8.0/10 μg/ml RNase A) and immediately recentrifuged at 12,000 × g for 10 min to pellet unlysed bacteria and cellular debris. The supernatant was then extracted with an equal volume of 1:1 (vol/vol) chloroform/isoamyl alcohol. DNA was precipitated with two-thirds volume of isopropyl alcohol, resuspended in TE (10 mM Tris·HC1, pH7.4/1 mM EDTA) buffer, and column purified (Qiagen, Chatsworth, CA).
Isolation of Mastigamoeba Genes.
Conserved core regions A to G of RBP1 (10) were PCR-amplified by using a combination of universal and specific primers as described previously (11). The 3′ end of the gene was isolated by using suppression PCR genomic walking using Mastigamoeba-specific primers in opposition to linker primers ligated to genomic DNA (12, 13). All PCR fragments were cloned and sequenced in complementary directions.
Sequence and Phylogenetic Analyses.
The inferred Mastigamoeba RPB1 amino acid sequence was aligned with eukaryotic polymerase II, RNA polymerase III (RPC1), and archaeal largest subunits by using Clustal W (14). Sequences were further adjusted by eye and areas with ambiguous gaps were removed from the alignment. The remaining gaps were treated as missing data in phylogenetic analyses using unweighted parsimony (paup 3.1.1 at default settings) (15), distance (phylip 5.372), (16) and maximum-likelihood (puzzle) (17) algorithms. Distance matrices were constructed in protdist by using the George/Baker/Hunt categories model for weighting amino acid changes. Distance trees were produced by neighbor-joining. Maximum-likelihood analyses were performed under the JTT (Jones, Taylor, and Thornton) model of amino acid-substitution probabilities and assuming a gamma distribution, estimated from the data set, for rate variation among sites.
One hundred bootstrap replicates were performed in parsimony and distance analyses. Parsimony values were produced by using 10 random-edition subreplicates per round of bootstrapping. Likelihood support values were obtained from puzzle based on the number of times a given bipartition was supported in 10,000 quartet-puzzling trees (17). Analyses were carried out by using 1,138 aligned positions from 24 RPB1, RPC1, and archaeal sequences. A second set of analyses were performed with a somewhat larger alignment (1,232 positions) of only the 15 RPB1 sequences to better assess their relative branching order in the absence of more-distant outgroups.
Paired-sites tests were used to compare the relative support for different basal sequences as the earliest RPB1 branch as well as support for the early branching position of Mastigamoeba. Alternative trees were constucted using paup by constraining only the relevant branches and finding the remaining topology that was most parsimonious. These alternative topologies were supplied as user trees in Kishino–Hasegawa and Templeton tests (18, 19) by using puzzle and protpars (phylip), respectively.
To examine the tendency of different RPB1 sequences to attract to long branches in phylogenetic reconstructions (20), we produced 100 random sequences based on maximum-likelihood estimates (puzzle) of RPB1 amino acid composition using MacClade 3.01 (21). One hundred independent parsimony analyses were performed with the 1,232-position alignment to determine which branches of the RPB1 tree attracted each random sequence. Five separate searches were used with random addition of taxa to find the most parsimonious point of attachment for each random sequence. Because parsimony may be more prone to long-branch effects (20), we also perfomed maximum-likelihood analyses of 1,000 quartet-puzzling trees with two randomly generated sequences and the substitution parameters described above.
RESULTS
A total of 5,866 bp were sequenced from the Mastigamoeba RPB1 gene, from the first highly conserved region A motif through the stop codon. The sequence is interrupted by five large insertions (Fig. 1), each of which disrupts the inferred amino acid sequence either by introducing a stop codon or by producing a shift in the reading frame. Three of these insertions occur in conserved regions of the RPB1 gene that contain no significant indels among aligned eukaryotic sequences, and a fourth interrupts a motif in region F that is universally conserved in size, even in archaeal and eubacterial homologues (22). In all five cases the assumption of the presence of a spliceosomal intron bounded by GT-AG splice sites restores the RPB1 reading frame.
Figure 1.
Insertion sites relative to conserved domains and terminal sequences for five putative introns found in the Mastigamoeba RPB1 gene. Amino acid residues shown in boldface are conserved and can be aligned among all RPB1 sequences. Dinucleotides coding for putative splice donor and acceptor sites are also in boldface.
In addition to canonical splice-junction dinucleotides, the Mastigamoeba insertions have other features typical of eukaryotic spliceosomal introns. In four of the five insertions the positions immediately following the presumed GT splice donor site match the conserved yeast consensus sequence exactly (ref. 23, Fig. 1). For the most proximal insertion, which occurs in a region of RPB1 that is unconserved and difficult to align, there are several GT-AG boundaries that can restore the ORF; however, none of the possible GT-splice donor sites is followed by the conserved nucleotides present in the four downstream insertions. In contrast to yeast introns, but similar to the corresponding intron regions from plants, vertebrates, and red algae (23), sequences upstream (positions −3 to −15) of the putative 3′-splice acceptor sites are pyrimidine-rich (66% T+C) in the Mastigamoeba insertions (Fig. 1). Although no clearly conserved internal motif is present, there are several potential branch sites in each of the five insertions. Thus, the insertions in Mastigamoeba appear to be best characterized as spliceosomal introns.
The 3′ end of the Mastigamoeba gene encodes 25 tandemly repeated heptads (Fig. 2), similar to those of the C-terminal domain (CTD) of RPB1 sequences from so-called “crown” eukaryotic groups. The CTD has been found in all animals, plants, and fungi, as well as in the slime mold Dictyostelium and in the soil protist Acanthamoeba, but is absent in other, putatively more-ancient protists (13). There are several features that differ, however, between the Mastigamoeba and CTD repeats. The CTD heptads are composed of the consensus sequence YSPTSPS, whereas in Mastigamoeba the C terminus consists of YSPASPA repeats. In addition, in the “crown” sequences there are typically numerous substitutions within individual CTD heptads that cause them to differ from the consensus sequence. For example, in Drosophila melanogaster only 2 perfect heptads are present among 42 repeats. In Saccharomyces, Arabidopsis, and Mus, 65%, 42%, and 40% of repeats, repectively, contain the consensus heptapeptide (24). In contrast, the Mastigamoeba heptads are nearly invariant with only a single amino acid that deviates from the consensus among all 25 repeats (Fig. 2). The Mastigamoeba sequence also exhibits absolute codon bias at sixfold degenerate serine residues; all second-position serine residues are encoded by AGC, and all fifth-position serine residues are encoded by TCN codons (Fig. 2). Likewise, all prolines at position three are encoded by the CCA triplet.
Figure 2.
Codon usage in the 25 tandemly repeated Mastigamoeba heptads. The nonsynonymous substitution is indicated in boldface and underlined, and the resulting amino acid change is shown to the left of the heptad in boldface.
Phylogenetic Position of Mastigamoeba RPB1.
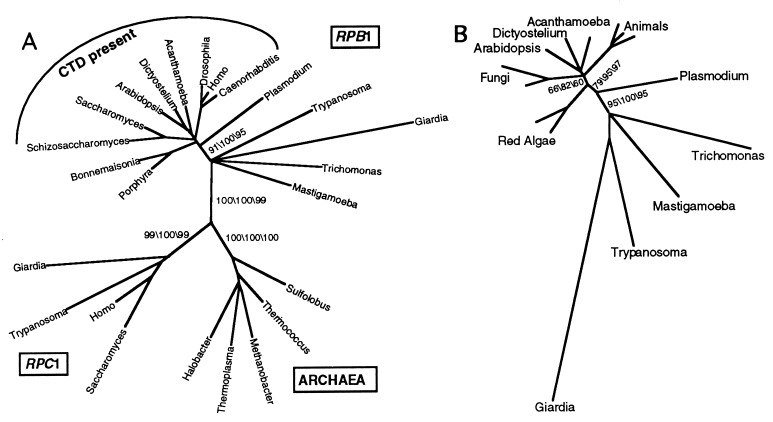
Phylogenetic analyses of aligned RPB1 sequences place Mastigamoeba outside the so-called “crown” of eukaryotic evolution with strong statistical support (Fig. 3A). Parsimony and distance bootstrap values of 91% and 100%, repectively, separate the four most basal taxa, Giardia, Mastigamoeba, Trichomonas, and Trypanosoma from the remaining eukaryotic taxa. Quartet-puzzling maximum likelihood support is 95% for that node. With outgroup sequences removed (Fig. 3B) these respective support values remain robust at bootstrap values of 95%, 100%, and 95% (parsimony, distance, and maximum likellihood). In addition, paired-sites tests using both parsimony (P = 0.0002) and likelihood (P = 0.008) analyses (Table 1) significantly reject grouping Mastigamoeba RPB1 with sequences that contain a canonical CTD.
Figure 3.
Consensus trees from parsimony, neighbor-joining, and maximum-likelihood phylogenetic analyses. Branch lengths are from maximum-likelihood analyses. (A) Tree based on an alignment of 24 RNA polymerase largest-subunit homologues. Bootstrap support values (parsimony, neighbor-joining, and maximum-likelihood) are shown at nodes relevant to the branching position of Mastigamoeba. (B) The consensus tree based on RPB1 sequences produced in the absence of outgroups.
Table 1.
Paired-sites analyses with maximum-likelihood/parsimony of alternative tree topologies with each of the four earliest-branching sequences constrained as the most-basal branch
| Tree tested | δ 1nL/steps | SD-ML/SD-MP | P |
|---|---|---|---|
| Most basal RPB1 branch | |||
| Giardia | best/15 | —/25 | —/0.54 |
| Trichomonas | 18.4/best | 26.1/— | 0.48/— |
| Trypanosoma | 44.5/31 | 24.7/18.3 | 0.07/0.09 |
| Mastigamoeba | 9.1/28 | 13.5/24.3 | 0.50/0.25 |
| M.i. + CTD taxa | 97.3/102 | 36.9/26.8 | 0.008/0.0002 |
Also tested is the placement of Mastigamoeba(M.i.) with the CTD-containing taxa. δ/nL, change in natural logarithm of likelihood; SD, standard deviation; ML, maximum likelihood; MP, Maximum parsimony.
The relative branching order among the four basal taxa is not well-resolved. Bootstrap values are low in all analyses, and branching topologies differ among parsimony, distance, and maximum-likelihood trees. Paired-sites analyses (Table 1) indicate that placing any of these four taxa as the earliest eukaryotic branch does not represent a significantly worse topology, although the tree with Trypanosoma at the base is rejected at P = 0.09 and P = 0.07 in parsimony and likelihood analyses, respectively. There appears to be no significant or even consistent preference for Giardia, Trichomonas, or Mastigamoeba as the most-basal RPB1 sequence.
Reliability of Basal Branches?
Several methods were used to assess the possibility that any of the four sequences occupying a deep branching position may do so because of an increased rate of amino acid substitution with corresponding long-branch effects (20). We examined the frequency of individual substitutions at the most-highly conserved positions in our alignment. Unique autapomorphic changes at otherwise invariable positions were counted, as well as nonconservative amino acid substitutions (George/Baker/Hunt categories) (16) at otherwise conservative positions. Because some increase in such changes is expected in more ancient eukaryotes, we also scored the reverse condition, that is, positions at which each basal taxon matched an invariable or conserved amino acid in the archaebacterial sequences that did not occur in other RPB1 sequences.
There is an overabundance of substitutions at highly conserved positions primarily in Giardia but also in Trichomonas (Fig. 4A). Mastigamoeba RPB1 has the fewest of these unique substitutions of the four basal sequences. The Giardia sequence has several amino acid substitutions at positions that are invariant in RPB1 and archaebacterial sequences and in nearly all largest-subunit homologues (ref. 25, Fig. 5). For example, there are differences in the Giardia sequence at positions in the D and G conserved motifs that are otherwise strongly conserved and are known to participate in forming the catalytic center of DNA-dependent RNA polymerases (25). Furthermore, the only other sequence in our full alignment that has a substitution anywhere in the highly conserved D motif is the largest subunit of Giardia RNA polymerase III (Fig. 5).
Figure 4.
Indicators for long branch attraction among RPB1 sequences. (A) The number of unique changes in each sequence at otherwise universally conserved sites as well as sites with a conserved ancestral character shared only with archaebacterial outgroups. G, Giardia; Tr, Trichomonas; Ty, Trypanosoma; M, Mastigamoeba; P, Plasmodium; R, red algae; C, other crown taxa. Averages are shown for red algae and for other crown eukaryotes. (B) The number of times randomly generated sequences were attracted to each taxon in 100 independent parsimony analyses.
Figure 5.
Substitutions (boldface and underlined) in the most highly conserved polymerase largest-subunit motifs. Ac, Acanthamoeba; At, Arabidopsis; Dd, Dictyostelium; Gl, Giardia; Hh, Halobacterium; Hs, Homo; Mi, Mastigamoeba; Mt, Methanobacterium; Sa, Sulfolobus; Sc, Saccharomyces; Tb, Trypanosoma; Tv, Trichomonas; B1, RPB1; C1, RPC1.
Given the great number of substitutions at the most-conserved RPB1 positions, it stands to reason that the Giardia sequence would have evolved even more rapidly at less-conserved sites. This increase in substitution rate appears to be reflected in the extremely long branch length of Giardia RPB1 in phylogenetic analyses (Fig. 3). Moreover, the Giardia sequence is the only one of the four basal sequences that deviates significantly (χ2 test) from a maximum-likelihood estimate of RPB1 amino acid composition for all eukaryotes.
A possible explanation for the high level of autapomorphic substitutions in Giardia or Trichomonas may be that they represent groups that arose long before the emergence of other protists and, therefore, that they had far more time to accumulate unique differences in their RPB1 sequences. In that event we may also expect to see an excess of unique conserved residues shared between Giardia or Trichomonas and archaebacterial outgroups. This is because equally numerous but different autapomorphic substitutions also should have accumulated in the long branch leading to all remaining eukaryotic groups. The number of such putatively ancestral sites, however, is approximately equal in the sequences from each of the four most basal branches of the RPB1 tree (Fig. 4A).
Random Rooting of RPB1 Trees.
To further explore the tendency of long branches to attract each of the four basal taxa, we rooted RPB1 trees with 100 randomly generated sequences (26). Of the 202 most parsimonious trees produced in these analyses, the random sequence was attracted to the Giardia branch 88% of the time (Fig. 4B). In contrast, Mastigamoeba was attracted to the random sequence only once among all 202 most-parsimonious trees. When Giardia was removed from the alignment and the analyses repeated, the Mastigamoeba sequence still failed to attract the random sequence (only 6%) compared with Trichomonas (53%) and Trypanosoma (41%). In analyses performed using maximum likelihood, the random sequences were attracted to Giardia in 96%, Giardia+Trichomonas in 19%, and Trichomonas alone in 3% of quartet-puzzling bipartitions. Although these data suggest that the positions of Giardia and possibly Trichomonas near the base of the RPB1 clade should be viewed with caution, there is no indication that the position of Mastigamoeba is artifactual.
DISCUSSION
Phylogenetic analyses based on conserved domains of RPB1 provide strong evidence that Mastigamoeba belongs to an early branch of the eukaryotic tree. Unlike other branches near the base of the RPB1 clade, there is no indication that the Mastigamoeba sequence is attracted to outgroups because of an increased rate of substitution or a biased amino acid composition. An early evolutionary origin of the Pelobiontida is consistent with the proposal that the group diverged before the endosymbiotic origin of mitochondria (2, 3, 27) and with features of cellular ultrastructure that suggest they may be the most ancient surviving eukaryotic lineage (2, 3).
C-Terminal Repeats and Introns in an Ancient Eukaryote?
The presence of introns and a heptad repeat in the RPB1 gene of an early eukaryote raise intriguing questions. The CTD, consisting of tandemly repeated heptapeptides with the consensus sequence YSPTSPS, appears to be a synapomorphic feature that unites certain “crown” eukaryotes including green plants, animals, fungi, and several related protist lineages (12, 28). Heptads with the CTD consensus are not present in RPB1 genes isolated from any eukaryote outside of these taxa (Fig. 3) including red algae, which appear to have emerged just before the radiation of CTD-containing lineages (13, Fig. 3). Mastigamoeba RPB1, however, is not the only gene from a eukaryote outside of this group that has tandem heptads with a non-CTD consensus. The apicomplexan parasite Plasmodium falciparum also contains an RPB1 C-terminal heptapeptide with its own regularly repeated difference from the CTD consensus: lysine in place of serine at position seven (29). The Plasmodium heptads also show codon bias and, based on phylogenetic analyses, are believed to be the result of a recent amplification of this YSPTSPK heptad only in the P. falciparum lineage (30).
Likewise, the unique characteristics of the tandem repeats in Mastigamoeba are most easily explained if they resulted from the amplification of a YSPASPA heptad that occurred independently of the initial amplification of the YSPTSPS repeats present in certain “crown” eukaryotes. Small tracts with partial sequence similarity to CTD heptads are present in virtually all eukaryotic RPB1 C termini. Although no tandem repeats occur in other putatively ancient protists, the RPB1 C termini of Trichomonas, Giardia, and Trypanosomids all are enriched in amino acids that make up the CTD, and each contains fragmentary sequences that are identical to portions of the CTD consensus heptapeptide. Presumably the ancestral RPB1 gene had similar sequences that provided the raw material for subsequent but independent heptad multiplications in different lineages. The hypothesis that distinct repeats were amplified independently in Mastigamoeba and in the “crown” group is supported by their statistically robust separation in all RPB1-based phylogenetic analyses (Fig. 3, Table 1), combined with the lack of a canonical CTD in groups that branch between the Mastigamoeba and the CTD-containing taxa.
It is also possible that the common ancestor of Mastigamoeba and “crown” eukaryotes contained a CTD, but that the consensus sequence diverged between the two lineages. These ancestral repeats would then have been lost independently at least from red algae and from the Plasmodium lineage and then regained in Plasmodium falciparum. In this case, small heptad-like segments in other protists also may represent the remains of degenerate CTD regions. Given the centrality of the CTD to the entire mRNA transcription cycle (ref. 31, and see below) such wholesale loss in so many different lineages seems unlikely; however, the functional role of tandem repeats has been investigated only in plants, animals, and fungi (31). Repeated heptads might have been under different selective constraints during the evolutionary history of other eukaryotic lineages.
The functional significance of the Mastigamoeba heptads also is unclear. The CTD in animals, plants, and fungi acts as a coordinating center for much of the mRNA transcription cycle (31, 32). In an unphosphorylated state, the CTD plays a key role in the assembly of polymerase II holoenzyme components needed to establish the transcription initiation complex. Subsequent phosphorylation of the CTD allows RNA polymerase II to release the promoter and undergo processive elongation. When phosphorylated, the CTD also interacts with a myriad of protein cofactors that mediate most aspects of pre-mRNA processing (31). The coupling of these various stages of mRNA synthesis afforded by the RNA polymerase II CTD is essential to the complex development and tissue differentiation that occurs in multicellular plants and animals. Which if any of these functions are performed by noncanonical C-terminal heptads in a pelobiont amoeba is unknown; however, the presence of introns in Mastigamoeba suggests at least one role for these tandem repeats.
One of the key functions of the CTD is to act as an organizational platform for bringing together the elongating RNA polymerase II, spliceosomes, and related splicing factors for the efficient cotranscriptional excision of large numbers of introns that are present in many genes (32). The presence of both introns and a tandemly repeated RPB1 heptad in Mastigamoeba, therefore, is probably not coincidental. Spliceosomal introns and C-terminal repeats are absent from other putatively ancient protist lineages (13, 33). That they are both present in Mastigamoeba RPB1 suggests that its heptad repeat may act to recruit spliceosomes and that the repeat arose concurrently with the spread of introns through the Mastigamoeba genome.
The presence of introns in Mastigamoeba RPB1 raises an additional question regarding the placement of pelobionts among the earliest eukaryotic lineages. The observed distribution of introns among eukaryotic genomes has led to the idea that their presence is restricted to more recently evolved groups (33). The relatively derived position of Mastigamoeba in rDNA trees and the presence of introns in Mastigamoeba RPB1 appear at first glance to provide two independent pieces of evidence that the Pelobiontida is not an ancient eukaryotic lineage. However, the argument that introns first appeared in late-evolving eukaryotes is based on correlations between intron presence or absence along with position on the rDNA tree (33). Therefore, both pieces of evidence absolutely depend on the ability of rDNA phylogenies to recover the earliest events in eukaryotic evolution accurately.
Early Eukaryotic Evolution and rDNA.
It has been suggested that parasitic organisms cluster at the base of the rDNA tree because of increased rates of sequence evolution or variant base composition (34–36). A growing body of cytologic, biochemical, and now molecular evidence (34) suggests that such an artifactual misplacement in rDNA trees is true for at least one parasitic amitiochondriate group, the Microsporidia. Phylogenetic analyses of both mitochondrial-type HSP70 (34) and tubulin (37) sequences indicate a close relationship between microsporidia and fungi. Although phylogenies based on EF-1α place a microsporidian as the most basal eukaryotic taxon, the presence of a shared insertion is consistent with a close relationship between microsporidia and fungi and suggests that the deep branching position is artifactual, because of the extremely long branch length associated with this microsporidian sequence (38). Despite the evolutionary connection to fungi indicated by these various independent characters, microsporidia branch consistently near the root of the eukaryotic tree in rDNA-based phylogenetic analyses, even when corrections are made for variations in substitution rates and base composition among sequences (39).
We have provided evidence that Giardia and probably Trichomonas RPB1 sequences are unusually divergent and attract long branches. This suggests that their deep branching position in RPB1 phylogenies may be unreliable, perhaps because of more rapid rates of sequence evolution in these organisms. Data from microsporidia suggest that such increased substitution rates can affect multiple genes (34, 38). Given the deep, robust, but apparently incorrect position of microsporidia in rDNA phylogenies, it seems reasonable to ask whether similar effects may underlie the early branching position of other protists as well.
Fossils and Molecular Clocks.
The extremely long internodes on rDNA trees between archaebacterial outgroups and the first branches of the eukaryotic “crown” radiation (40) also are inconsistent with the fossil record and with dates for the origin of eukaryotes from protein sequence-based molecular clock estimates. Fossils of multicellular algae that are considered part of the rDNA crown radiation indicate that several of these groups already had achieved broad diversity by 1 billion years ago (Bya) (41). Large, blade-like algae are found from at least 1.7 Bya (42), whereas multicellular eukaryotes of unknown affiliation appear as long ago as 2.1 Bya (43). Molecular clock calibrations based on numerous protein-coding genes place the origin of all eukaryotes at just over 2 Bya (44). Independent clock estimates from rDNA sequences suggest an origin for all ciliated protists, a lineage that emerges near the base of the “crown” radiation, at over 2 Bya (45).
Taken together, these mutually consistent data imply that the earliest branching “crown” taxa began to radiate shortly after the common origin of all extant eukaryotes. Yet the vast majority of the total length of the rDNA eukaryotic tree occurs in the segments leading to the radiation of multicellular organisms (40). Indeed, the assertion that “crown taxa” evolved late in eukaryotic evolution is based largely on the extremely long branches at the base of the rDNA tree (7). If the fossil record and protein-clock data are more accurate reflections of the evolutionary record, most of this length in rDNA trees must be artifactual.
Statistical analysis of over 200 eukaryotic rDNA sequences, accounting for biases in base composition and rate variation, as well as nonindependence of substitutions across sites, indicates that the basal branches of the eukaryotic tree are not well-resolved in rDNA phylogenies (39). If, as suggested by these authors, the base of the rDNA tree is reduced to a polytomy, the conflict between protein and rDNA-based phylogenetic analyses of pelobionts is eliminated. It is partly because of historical accident that rDNA was the first gene to be sampled widely, thus providing the first molecular framework for early eukaryotic evolution. An objective understanding of the complex historical clues contained in modern genomes must leave room for a broad interpretation of accumulating molecular data from the protist world, and not be restricted by phylogenetic topologies based on rDNA or by any other single gene sequence.
The RPB1 sequence from Mastigamoeba raises many questions about eukaryotic evolution. Additional RPB1 genes should be sampled from diverse eukaryotes to provide a larger data set for phylogenetic analyses, a better understanding of the evolutionary history of C-terminal repeats, their relationship to the presence of introns, and the evolution of mRNA transcription in general. In particular, RPB1 sequences from other pelobionts will help to determine whether the heptad repeats and introns in ‘Mastigamoeba invertens’ are representative of the Pelobiontida as a whole. Molecular analyses also are needed to determine whether there is vestigial evidence, as has been found in all parasitic protist groups (34, 40), that pelobionts once contained mitochondria. Their simple intracellular structure, ultrastructural features, and lack of parasitism, all combined in organisms that occur sympatrically with many mitochondriate protists (3), argue for a further exploration of the intriguing hypothesis that pelobiont amoebae may be a true lineage of primitively amitochondriate eukaryotes.
ABBREVIATIONS
- CTD
C-terminal domain of RNA polymerase II
- RPB1
RNA polymerase II largest subunit
- RPC1
RNA polymerase III largest subunit
- SSU rDNA
small-subunit ribosomal RNA
- Bya
billion years ago
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF083338).
References
- 1. Cavalier-Smith T. In: Endocytobiology II. Schwemmler W, Schenk H E A, editors. Berlin: de Gruyter; 1983. pp. 1027–1034. [Google Scholar]
- 2.Cavalier-Smith T. Biosystems. 1991;25:25–38. doi: 10.1016/0303-2647(91)90010-i. [DOI] [PubMed] [Google Scholar]
- 3.Simpson A G B, Bernard C, Fenchel T, Patterson D J. Eur J Protistol. 1997;33:87–98. [Google Scholar]
- 4.Sogin M L. Curr Opin Genet Dev. 1991;1:457–463. doi: 10.1016/s0959-437x(05)80192-3. [DOI] [PubMed] [Google Scholar]
- 5.Clark C G, Roger A J. Proc Natl Acad Sci USA. 1995;92:6518–6521. doi: 10.1073/pnas.92.14.6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinkle G, Leipe D D, Nerad T A, Sogin M L. Nucleic Acids Res. 1994;22:465–469. doi: 10.1093/nar/22.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knoll A H. Science. 1992;256:622–627. doi: 10.1126/science.1585174. [DOI] [PubMed] [Google Scholar]
- 8.Cavalier-Smith T, Chao E E. J Mol Evol. 1996;43:551–562. doi: 10.1007/BF02202103. [DOI] [PubMed] [Google Scholar]
- 9.Morin L, Mignot J-P. Eur J Protistol. 1995;31:402. [Google Scholar]
- 10.Jokerst R S, Weeks J R, Zehring W A, Greenleaf A L. Mol Gen Genet. 1989;215:266–275. doi: 10.1007/BF00339727. [DOI] [PubMed] [Google Scholar]
- 11.Stiller J W, Hall B D. Proc Natl Acad Sci USA. 1997;94:4520–4525. doi: 10.1073/pnas.94.9.4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siebert P D, Chenchik A, Kellogg D E, Lukyanov K A, Lukyanov S A. Nucleic Acids Res. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stiller, J. W. & Hall, B. D. (1998) J. Phycol.34, in press.
- 14.Thompson J D, Higgens D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swofford D L. paup. Champaign, IL: Illinois Natural History Survey; 1993. , version 3.1.1. [Google Scholar]
- 16.Felsenstein J. Cladistics. 1989;5:164–165. [Google Scholar]
- 17.Strimmer K, von Haeseler A. Mol Biol Evol. 1996;13:964–969. [Google Scholar]
- 18.Kishino H, Hasagawa M. J Mol Biol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 19.Felsenstein J. Syst Zool. 1985;34:152–161. [Google Scholar]
- 20.Hendy M D, Penny D. Syst Zool. 1989;38:297–309. [Google Scholar]
- 21.Maddison W P, Maddison D R. MacClade. Sunderland, MA: Sinauer; 1992. , version 3.0. [Google Scholar]
- 22.Pühler G, Leffers H, Gropp F, Palm P, Klenk H-P, Lottspeich F, Garrett R A, Zillig W. Proc Natl Acad Sci USA. 1989;86:4569–4573. doi: 10.1073/pnas.86.12.4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liaud M-F, Brandt U, Cerff R. Plant Mol Biol. 1995;28:313–325. doi: 10.1007/BF00020250. [DOI] [PubMed] [Google Scholar]
- 24.Corden J L. Trends Biochem Sci. 1990;15:383–387. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- 25.Mustaev A, Kozlov M, Markovtsov V, Zaychikov E, Denissova L, Goldfarb A. Proc Natl Acad Sci USA. 1997;94:6641–6645. doi: 10.1073/pnas.94.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graham S. Ph.D. Dissertation. Canada: University of Toronto; 1997. [Google Scholar]
- 27.Brugerolle G. Protoplasma. 1991;164:70–90. [Google Scholar]
- 28.Klenk H-P, Zillig W, Lanzendörfer M, Brampp B, Palm P. Arch Protistenkd. 1995;145:221–230. [Google Scholar]
- 29.Li W-B, Bzik D J, Gu H, Tanaka M, Fox B A, Inselburg J. Nucleic Acids Res. 1989;17:9621–9636. doi: 10.1093/nar/17.23.9621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giesecke H, Barale J-C, Langsley G, Cornelissen W C A. Biochem Biophys Res Commun. 1991;180:1350–1355. doi: 10.1016/s0006-291x(05)81344-0. [DOI] [PubMed] [Google Scholar]
- 31.Steinmetz E J. Cell. 1997;89:491–494. doi: 10.1016/s0092-8674(00)80230-5. [DOI] [PubMed] [Google Scholar]
- 32.Corden J L, Patturajan M. Trends Biochem Sci. 1997;22:413–416. doi: 10.1016/s0968-0004(97)01125-0. [DOI] [PubMed] [Google Scholar]
- 33.Palmer J D, Logsdon J M., Jr Curr Opin Genet Dev. 1991;1:470–477. doi: 10.1016/s0959-437x(05)80194-7. [DOI] [PubMed] [Google Scholar]
- 34.Germot A, Philippe H, Le Guyader H. Mol Biochem Parasitol. 1997;87:159–168. doi: 10.1016/s0166-6851(97)00064-9. [DOI] [PubMed] [Google Scholar]
- 35.Siddall M E, Hong H, Desser S S. J Protozool. 1992;39:361–367. doi: 10.1111/j.1550-7408.1992.tb01465.x. [DOI] [PubMed] [Google Scholar]
- 36.Hasegawa M, Hashimoto T. Nature (London) 1993;361:23. doi: 10.1038/361023b0. [DOI] [PubMed] [Google Scholar]
- 37.Keeling P J, Doolittle W F. Mol Biol Evol. 1996;13:1297–1305. doi: 10.1093/oxfordjournals.molbev.a025576. [DOI] [PubMed] [Google Scholar]
- 38.Baldauf S L, Doolittle W F. Proc Natl Acad Sci USA. 1997;94:12007–12012. doi: 10.1073/pnas.94.22.12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar S, Rzhetsky A. J Mol Evol. 1996;42:183–193. doi: 10.1007/BF02198844. [DOI] [PubMed] [Google Scholar]
- 40.Sogin M L. Curr Opin Genet Dev. 1997;7:792–799. doi: 10.1016/s0959-437x(97)80042-1. [DOI] [PubMed] [Google Scholar]
- 41.Xiao S, Zhang Y, Knoll A H. Nature (London) 1998;391:553–558. [Google Scholar]
- 42.Shixing Z, Huineng C. Science. 1995;270:620–622. [Google Scholar]
- 43.Han T-M, Runnegar B. Science. 1992;257:232–235. doi: 10.1126/science.1631544. [DOI] [PubMed] [Google Scholar]
- 44.Feng D-F, Cho G, Doolittle R F. Proc Natl Acad Sci USA. 1997;94:13028–13033. doi: 10.1073/pnas.94.24.13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wright A D G, Lynn D H. Arch Protistenkd. 1997;148:329–341. [Google Scholar]