Abstract
In both neurons and muscle fibers, specific mRNAs are concentrated beneath and locally translated at synaptic sites. At the skeletal neuromuscular junction, all synaptic RNAs identified to date encode synaptic components. Using microarrays, we compared RNAs in synapse-rich and -free regions of muscles, thereby identifying transcripts that are enriched near synapses and that encode soluble membrane and nuclear proteins. One gene product, LL5β, binds to both phosphoinositides and a cytoskeletal protein, filamin, one form of which is concentrated at synaptic sites. LL5β is itself associated with the cytoplasmic face of the postsynaptic membrane; its highest levels border regions of highest acetylcholine receptor (AChR) density, which suggests a role in “corraling” AChRs. Consistent with this idea, perturbing LL5β expression in myotubes inhibits AChR aggregation. Thus, a strategy designed to identify novel synaptic components led to identification of a protein required for assembly of the postsynaptic apparatus.
Introduction
A cardinal feature of the chemical synapse is the existence of a highly specialized postsynaptic apparatus whose molecular architecture differs dramatically from that of contiguous, nonsynaptic portions of the cell surface (Sanes and Lichtman, 2001; Sheng and Kim, 2002). To understand synaptic development, function, and plasticity, it is important to identify components of the postsynaptic apparatus, learn how their distribution is regulated, and determine the roles they play.
A key advance in addressing these issues was the discovery that some postsynaptic proteins are synthesized locally rather than (or in addition to) being synthesized at a distant site and then transported to the synapse. In forebrain neurons, for example, most protein synthesis occurs in the cell body but specific subsets of mRNAs are transported to dendritic spines, where they are translated (Steward, 1983; for reviews see Steward and Schuman, 2001; Martin and Kosik, 2002). Likewise, at the skeletal neuromuscular junction (NMJ), RNAs encoding some postsynaptic proteins are concentrated and translated directly beneath the postsynaptic membrane (Merlie and Sanes, 1985; Goldman and Staple, 1989; Jasmin et al., 1993, Moscoso et al., 1995b; Valenzuela et al., 1995; Imaizumi-Scherrer et al., 1996; for reviews see Sanes and Lichtman, 1999, 2001; Chakkalakal and Jasmin, 2003). The mechanism that underlies localized protein synthesis at the NMJ differs from that in neurons: RNA localization in muscle results, at least in part, from transcriptional specialization of the few myonuclei within the multinucleated muscle fiber that lie beneath the postsynaptic membrane (Klarsfeld et al., 1991; Sanes et al., 1991; Simon et al., 1992; for review see Schaeffer et al., 2001). The outcome is similar in neurons and muscles, however, in that specific mRNAs are localized to the postsynaptic apparatus.
Identification of these synaptic RNAs provides a means of identifying proteins likely to be important for synapse formation or function. Moreover, determining which synaptic components are synthesized locally is an important step in understanding how synapse assembly is regulated. In the first part of this paper, we report results of an expression-profiling screen designed to identify synaptically enriched RNAs in muscle. We used the elegant methods of Tietjen et al. (2003) to amplify RNAs from microdissected NMJs and to compare them to RNAs from synapse-free portions of muscle. This comparison led to identification of several new synaptic transcripts, encoding membrane, cytoplasmic, cytoskeletal, and nuclear proteins.
In the second part of the paper, we describe LL5β, a protein encoded by one of the synaptic RNAs we identified. LL5β was previously identified in a database search for proteins containing pleckstrin homology (PH) domains, which bind phosphoinositides on the inner leaflet of the plasma membrane (Dowler et al., 2000). Subsequent analysis showed that this binding is regulated by extracellular signals and that LL5β also binds a cytoskeletal, actin-associated protein, filamin (Paranavitane et al., 2003), one form of which is concentrated at the NMJ (Bloch and Hall, 1983; Shadiack and Nitkin, 1991). We show here that LL5β is associated with the postsynaptic membrane at the NMJ, that it is concentrated at borders of the postsynaptic domain as it develops, and that it regulates acetylcholine receptor (AChR) aggregation in this domain. We propose that LL5β is a regulated link in a multimolecular complex that corrals AChRs in the postsynaptic apparatus.
Results
A screen for RNAs at synaptic sites
Our aim was to identify polyA+ RNAs selectively associated with the postsynaptic apparatus at the NMJ. In previous studies, we and others took advantage of the fact that most synapses in mammalian muscles are confined to a central “endplate band” (Fig. 1 A), so that synapse-rich and -free samples can be generated by cutting the muscle into thirds, perpendicular to the long axis of the fibers (Hall, 1973; Merlie and Sanes, 1985; Velleca et al., 1994). However, this strategy is not useful for global comparisons of gene expression because the NMJ occupies only ∼0.1% of each muscle fiber's surface, so synapses comprise only a small fraction of the nominally synapse-rich third. Transcripts that are only severalfold enriched at synapses or that are present at low abundance are not efficiently detected in this comparison.
Figure 1.
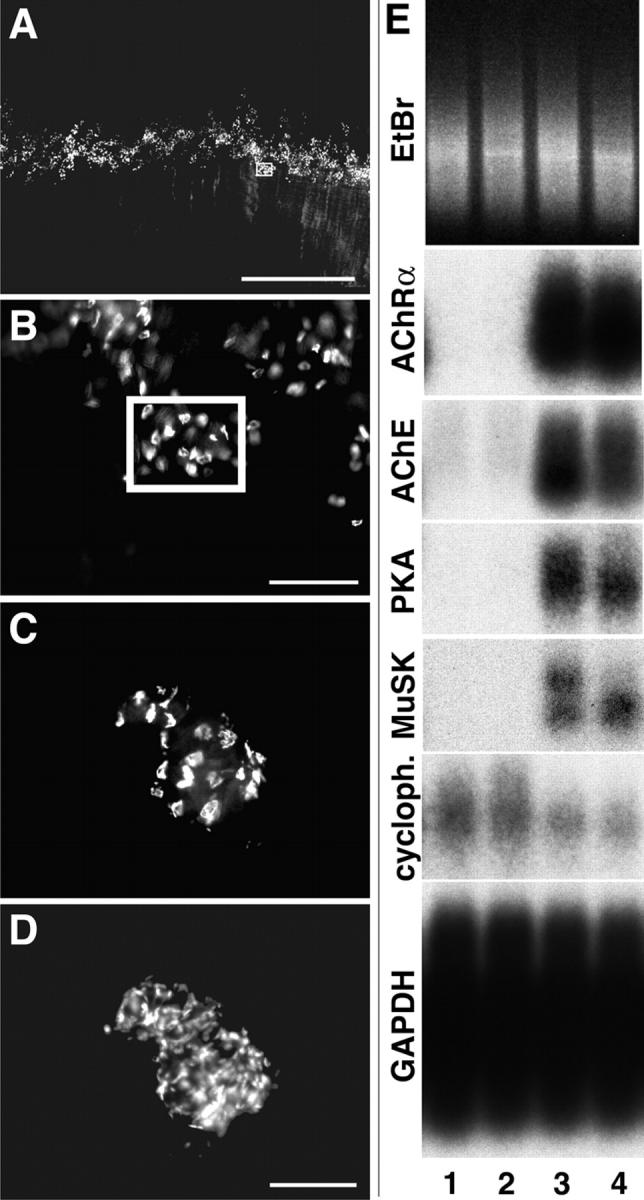
Isolation of synaptic and nonsynaptic samples from muscle. (A) Diaphragm muscle stained with rBTX and viewed under a fluorescence dissecting microscope. Fibers run from top to bottom. NMJs are in a central “endplate band.” (B) Higher-magnification view of the endplate band shows that small groups of NMJs are tightly clustered. Synaptic samples for microarray analysis were obtained by microdissecting groups such as the one boxed in A and B. (C and D) Microdissected synaptic sample, double-stained with rBTX (C) and the nuclear stain DAPI (D). Bars: (A) 1 mm; (B) 200 μm; (C and D) 100 μm. (E) Southern analysis of two nonsynaptic (lanes 1 and 2) and two synaptic (lanes 3 and 4) samples after reverse transcription and PCR amplification. Gel was stained with ethidium bromide (EtBr) to verify equal loading. Blots were probed for transcripts known to be synaptically enriched (AChRα, AChE, PKARIα [PKA], and MuSK) or presumed to be uniformly distributed (cyclophilin [cycloph.] and GAPDH).
We combined two methods to circumvent this problem. First, we labeled live diaphragm muscles with rhodamine-α-bungarotoxin (rBTX) to visualize the AChRs concentrated at synaptic sites and microdissected small groups of closely spaced NMJs (Fig. 1, B and C). In parallel, we dissected regions of equivalent size from completely synapse-free regions of the same muscles. The extent to which the dissection enriched synaptic samples can be estimated from counts of nuclei. In mouse diaphragm, about half of all nuclei are within muscle fibers; the others are in satellite cells, fibroblasts, capillary endothelial cells, and Schwann cells. Each muscle fiber contains ∼500 nuclei, of which approximately five are synapse associated (Merlie and Sanes, 1985). Thus, in the central third of the muscle, ∼1.5% (5/[1/3 × 1000]) of all nuclei are synaptic myonuclei. In the synaptic sample in Fig. 1 (C and D) there were 21 NMJs and 198 nuclei, so ∼50% of nuclei were synaptic, a >30-fold improvement over the previous method.
Second, we used the amplification method of Tietjen et al. (2003) to prepare targets for hybridization to microarrays. After amplification, sample quality was assessed by Southern blotting for four transcripts known to be enriched in synaptic areas (the AChR α-subunit, acetylcholinesterase [AChE], the protein kinase A RIα subunit [PKARIα], and the muscle specific kinase [MuSK]; references in Introduction), as well as two broadly expressed genes (glyceraldehyde-3-phosphate dehydrogenase [GAPDH] and cyclophilin). Pairs of samples were selected in which: (a) the synaptic sample was rich in products amplified from the synaptic genes, (b) the nonsynaptic sample contained little or none of these products, and (c) both samples had nearly equivalent levels of the broadly expressed genes (Fig. 1 E). Of >100 pairs analyzed, the nine that best fulfilled these criteria were hybridized to Affymetrix Murine U74A, B, and C GeneChips, which together contain 36,701 probe sets.
As a first test of the method, we calculated the synaptic enrichment of the six transcripts for which there was the best prior evidence of synaptic enrichment. Four of these (AChRα, AChE, PKARIα, and MuSK) had been used for prescreening by Southern analysis, but the other two (AChRδ and AChRɛ) had not. Enrichment of each transcript was calculated as the ratio of the synaptic to extrasynaptic expression levels (Table S1, available at http://www.jcb.org/cgi/content/full/jcb.200411012/DC1) and as the rank order of the synaptic/nonsynaptic ratios (Table I). All but 1 of the 54 ratios (6 genes × 9 pairs) were >1 (normalization set the mean ratio equal to 1), and all but the corresponding one rank order were above the mean of 18,350 (36,701/2). Both distributions were highly nonrandom (P < 10−14; binomial test). Moreover, the geometric mean of ratios of these six control genes and the arithmetic mean of their rank order (Table S1 and Table I, bottom row) were much higher than expected by chance (P < 0.03 for pair No. 3 and P < 0.0006 for all other pairs; computed using bootstrapping tests). Ratios and rank orders for the transcripts that had been used for prescreening were not better than those that had not.
Table I. Enrichment of known synaptic mRNAs in synaptic samples.
| Gene name | Exp. 1 | Exp. 2 | Exp. 3 | Exp. 4 | Exp. 5 | Exp. 6 | Exp. 7 | Exp. 8 | Exp. 9 | Mean rank | Pooled rank |
|---|---|---|---|---|---|---|---|---|---|---|---|
| AChRα1 | 20 | 1051 | 176 | 4 | 50 | 16 | 10 | 4496 | 66 | 654 | 2 |
| AChRδ | 3631 | 4775 | 8096 | 265 | 565 | 874 | 11596 | 3154 | 83 | 3671 | 21 |
| AChRɛ | 83 | 36 | 80 | 5 | 18 | 88 | 18 | 6 | 119 | 50 | 1 |
| MuSK | 8801 | 3624 | 30131 | 47 | 1807 | 405 | 29 | 1490 | 4271 | 5623 | 20 |
| AChE | 8458 | 14475 | 18126 | 3488 | 1062 | 743 | 64 | 4872 | 6513 | 6422 | 54 |
| PKARIα | 1046 | 8579 | 3298 | 1571 | 329 | 208 | 4957 | 3250 | 968 | 2690 | 18 |
| Mean | 3673 | 5423 | 9985 | 897 | 639 | 389 | 2779 | 2878 | 2003 | 3185 | 19 |
Probe sets for six synaptic genes were ranked each in microarray pairs by calculating ratio of signal in synaptic sample to that in extrasynaptic sample, and then ranking ratios of all 36,701 probe sets. None of the mean rank orders (calculated for each chip or for each gene) are in the top 25, but when signals were pooled and then ranked (see text), five out of six genes ranked in the top 25.
Although known synaptically enriched transcripts ranked high in each GeneChip pair, only a small fraction of the transcripts with high ratios or rank orders were likely to be synaptically enriched, owing to method-derived noise and natural variation among samples. When we combined results from multiple pairs, however, a dramatically different picture emerged. The last column of Table I shows combined rankings for all nine pairs for the six control genes (calculated as S1/E1 × S2/E2…S9/E9). The improved quality of the pooled data can be seen by comparing it with the mean rank orders of genes for the nine pairs taken individually. For example, the mean rankings for the AChRɛ and α subunits are 50 and 654, respectively, whereas those for the pooled ranking are 1 and 2. We also pooled results by dividing the sum of all synaptic scores by the sum of all nonsynaptic scores (S1 + S2…S9/E1 + E2…E9) and by using the significance analysis of microarrays (SAM) method (Tusher et al., 2001), but these methods gave slightly lower rank orders than the multiplicative method (unpublished data) and were not used further. Our method of ranking genes by geometric means of their ratio is equivalent to that independently proposed by Breitling et al. (2004), who also found that nonparametric ranking could outperform the SAM method.
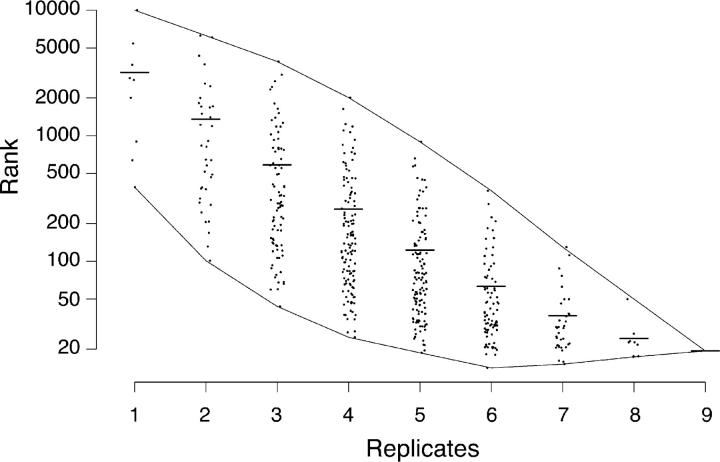
The availability of multiple chip pairs and validated control transcripts afforded the opportunity to ask how many replicates are needed to obtain reliable rank orders. To this end, we calculated the mean rank order for the six control transcripts from each combination of the 36 (9 × 8/2) possible set of two pairs, each of the 84 (9 × 8 × 7/3 × 2) possible sets of three pairs, and so on. Results are shown in Fig. 2. The two lines enveloping the individual points show the best and worst rank orders that could be obtained; the central line shows the mean rank order (Fig. 2). Three features of the graph are noteworthy. First, as expected, the overall reliability of the rankings increases with the number of replicates (Fig. 2, middle line). Second, even though all pairs provide useful data, there is such great variability among them that the reliability of rankings varies manyfold depending on which particular pairs are considered (Fig. 2, difference between top and bottom lines). Third, if one can prospectively identify control genes, it is possible to obtain more reliable data from a subset of the pairs than from all of them (Fig. 2, bottom line). For example, the mean rank order of the six control genes is 19 for all nine pairs, but 14 for the best six pairs.
Figure 2.
Extent to which replicate sampling enhances reliable detection of synaptic transcripts. Rank orders for six transcripts known to be synaptically enriched (AChRα, AChRδ, AChRɛ, AChE, PKARIα, and MuSK) were calculated for each of nine independent experiments, with the most synaptically enriched equal to 1 and the least equal to 36,701 (see Table I). The mean rank order for all six transcripts was calculated for each combination of the 36 (9 × 8/2) possible sets of two pairs, each of the 84 (9 × 8 × 7/3 × 2) possible sets of three pairs, and so on. The two lines enveloping the individual points show the best and worst rank orders that could be obtained from a given number of pairs; central bars shows means.
Identification of novel synaptic transcripts
We ranked the 36,701 probe sets from the “best” eight pairs of samples (Table I; Exp. 3 was omitted based on the low ranking of the six control genes) and asked how many of the top 25 represented transcripts enriched at synaptic sites. In some cases, relevant data have been reported previously; for the others, we assayed localization with in situ hybridization and/or quantitative RT-PCR (Fig. 3). As summarized in Table II, genes fell into the following five categories.
Figure 3.
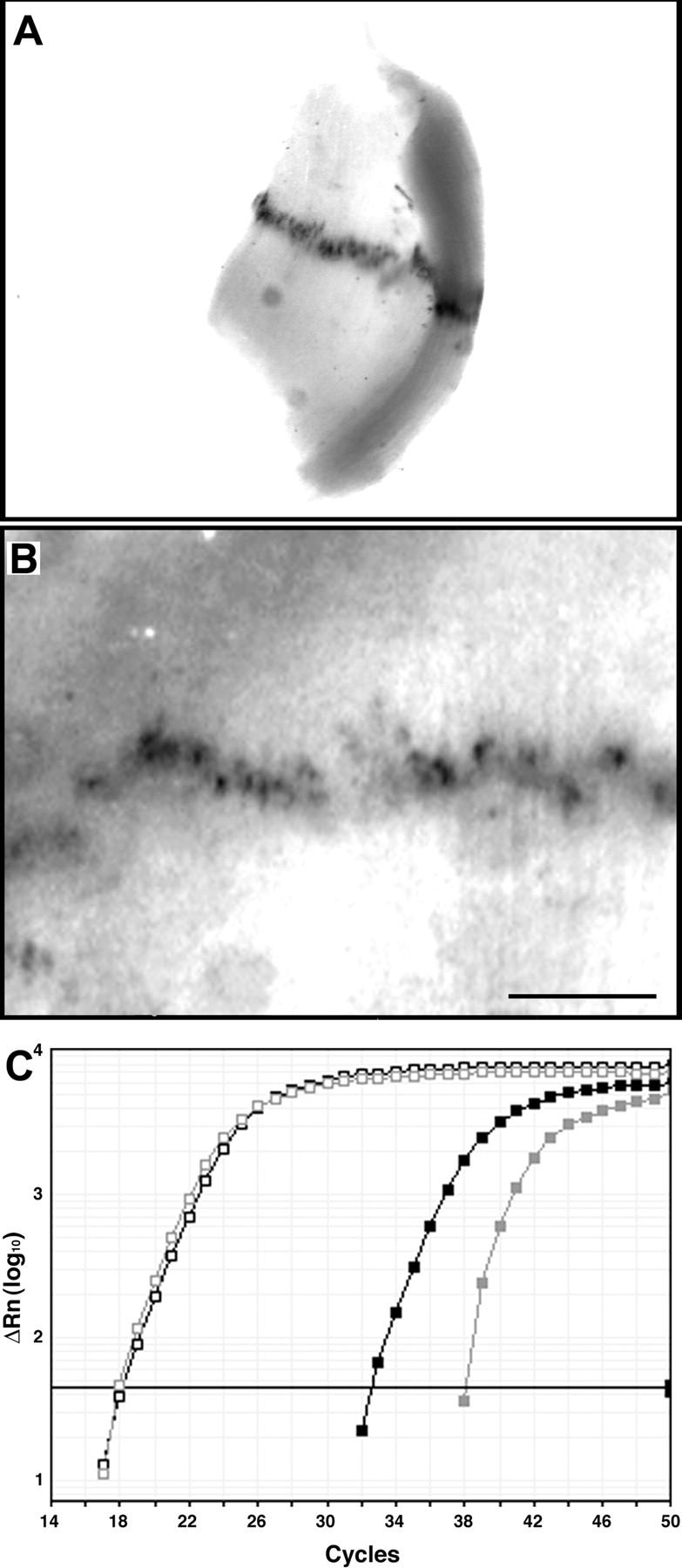
Verification of novel synaptic transcripts by whole mount in situ hybridization and quantitative RT-PCR. (A and B) Neonatal diaphragm was hybridized with probes for CD24 (A) or LL5β (B). RNA is concentrated at discrete sites identified as NMJs based on their number and position and by histochemical staining of adjacent muscle pieces with rBTX or a histochemical stain for AChE. Bar, 0.5 mm. (C) Quantitative RT-PCR of synaptic and extrasynaptic samples. A broadly distributed transcript (GAPDH, open squares) is present at similar levels in both samples, whereas dynein intermediate chain (Table II, No. 3; filled squares) is greater than sixfold enriched in the synaptic sample. Black and gray lines show data from synaptic and extrasynaptic samples, respectively. Thick line is the threshold value, used to calculate Ct (threshold cycle), from which differences between samples were determined.
Table II. Transcripts with highest enrichment in synaptic compared with extrasynaptic samples.
| Rank | Ratio | UniGene (Mm.) | Gene Name | Validity |
|---|---|---|---|---|
| 1 | 83.64 | 4980 | AChRɛ | K-R |
| 2 | 56.78 | 4583 | AChRα1 | K-R |
| 3 | 30.28 | 79127 | Dynein int. chain 1 | SY-? |
| 4 | 20.76 | 291463 | Trifunctional enz.-β | – |
| 5 | 18.93 | 276739 | SOX10 | SC |
| 6 | 16.39 | 29742 | CD24 | Sy-M |
| 7 | 16.28 | 37644 | rep seq | – |
| 8 | 15.22 | 66293 | SDR2 | Sy-? |
| 9 | 14.91 | 33268 | Ssb4 | Sy-? |
| 10 | 13.32 | 5025 | PEA3 | SC |
| 11 | 12.86 | 225050 | Unc53H3 | Sy-M |
| 12 | 11.63 | 16148 | MuSK | K-R |
| 13 | 11.40 | 155708 | ERM | K-R |
| 14 | 10.88 | 23742 | Nestin | K-P |
| 15 | 10.49 | 172931 | rep seq | – |
| 16 | 10.40 | 211477 | LL5β | Sy-M |
| 17 | 10.14 | 317642 | Serf2 | – |
| 18 | 9.69 | 124666 | NTA | Sy-M |
| 19 | 8.88 | 4980 | AChRɛ | K-R |
| 20 | 8.63 | 32648 | unnamed | Sy-? |
| 21 | 8.46 | 30039 | PKARIα | K-R |
| 22 | 8.40 | 2811 | AChRδ | K-R |
| 23 | 7.67 | 9986 | MPZ | SC |
| 24 | 7.58 | 295263 | Cpg16 | Sy-? |
| 25 | 7.52 | 335452 | noncoding RNA | – |
Rankings and ratios, calculated as in Table I and Table S1, respectively, are shown along with UniGene number (http://www.ncbi.nlm.nih.gov) and gene name. Last column classifies genes: K-R, known to be synaptic at RNA level; K-P, known to be synaptic at protein level; SC, known to be selectively expressed by Schwann cells; Sy-M, synaptic mRNA shown by in situ hybridization to be expressed by muscle in this study; Sy-?, mRNA shown to synaptic by quantitative PCR, but cell of origin unknown; –, no evidence for synaptic enrichment.
Known synaptically enriched transcripts, n = 6.
Of the six transcripts for which evidence of synaptic localization is strongest (AChRα, AChRδ, AChRɛ, MuSK, PKARIα, and AChE; see Introduction), all but AChE ranked in the top 25. A second probe set for AChRɛ was also within the top 25.
Known synaptically-enriched proteins, n = 1.
Nestin, an intermediate filament protein originally identified in stem cells, is concentrated at the NMJ (Carlsson et al., 1999; Vaittinen et al., 1999). We find that its RNA is also enriched at synaptic sites.
Genes expressed by Schwann cells, n = 3.
Motor nerve terminals are ensheathed by Schwann cells, the glia of the peripheral nervous system. Therefore, synapse-rich samples used for expression profiling contained Schwann cells, whereas extrasynaptic samples, isolated from nerve-free regions, were Schwann cell-free. Therefore, we expected that genes expressed by Schwann cells would be enriched in the synaptic samples. Consistent with this expectation, three genes known to be selectively expressed by Schwann cells were within the top 25 transcripts: the transcriptional regulators PEA-3 and SOX-10 and the myelin protein zero (Jessen and Mirsky, 2002).
False positives, n = 5.
We obtained no evidence that transcripts expressed by five of the probe sets (Table II, Nos. 4, 7, 15, 17, and 25) were enriched at synaptic sites.
Novel transcripts enriched in synaptic regions, n = 10.
Quantitative RT-PCR and/or in situ hybridization confirmed synaptic enrichment of 10 gene products that had not previously been localized to the NMJ. As noted above, these transcripts might be expressed by muscle fibers, Schwann cells, or perhaps other synapse-associated cells (e.g., a specialized perisynaptic population of fibroblasts; Gatchalian et al., 1989). For five of the mRNAs, in situ hybridization signals were too weak or diffuse to allow distinction among these sites (Table II, Nos. 3, 8, 9, 20, and 24). For the other five, however, in situ hybridization revealed a punctate pattern of expression associated with individual muscle fibers (Fig. 3, A and B); this finding was most consistent with localization in the postsynaptic apparatus (Moscoso et al., 1995a,b), although association with Schwann cells cannot be ruled out. One of these gene products, LL5β (No. 16; Fig. 3 B), is described in detail below (see Figs. 4–6). The other four are described in the following paragraphs.
Figure 4.
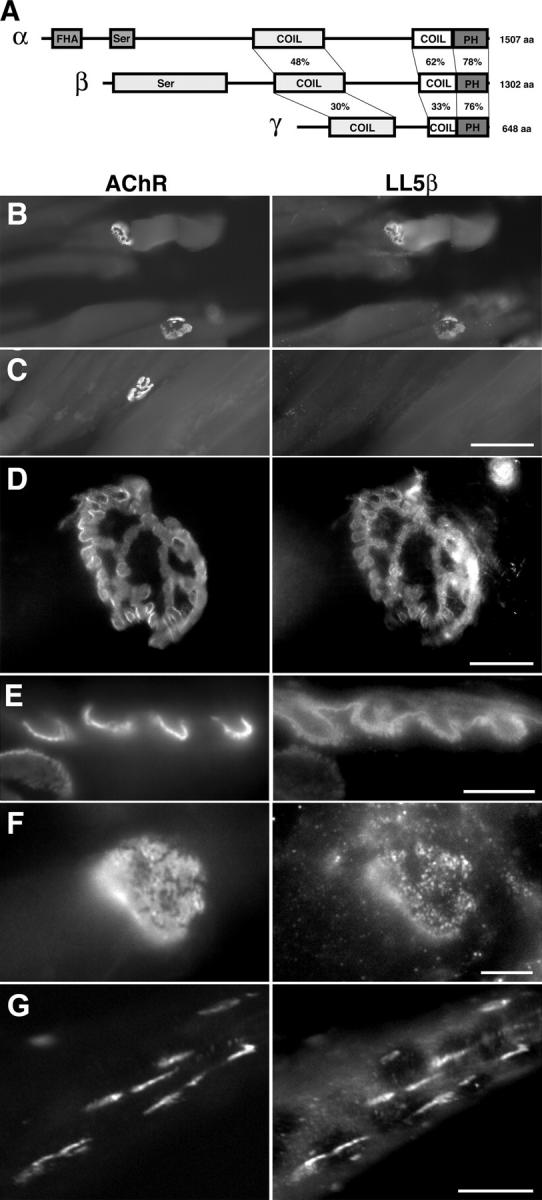
Structure and synaptic localization of LL5β. (A) Structures of LL5β and two homologues, LL5α and LL5γ. The percentage of amino acid identity of conserved domains is shown; amino termini of the three gene products are dissimilar. FHA, forkhead-associated domain; Ser, serine-rich region; COIL, coil-coil domain; PH, pleckstrin homology domain. (B–D) Longitudinal sections of adult muscle stained with mAb to LL5β (low magnification, B; high magnification, D) or control antibody of the same isotype (C) plus rBTX. LL5β is colocalized with AChRs. (E) Cross section of an adult rat NMJ showing LL5β concentrated in the depths of junctional folds and between AChR-rich areas at the crests of folds. (F) Synaptic concentration and colocalization were evident by prenatal day 4. (G) C2 myotubes were treated with agrin to induce AChR clusters, and then stained with anti-LL5β and rBTX. LL5β is concentrated at AChR-rich clusters. Bars: (B and C) 100 μm; (D) 20 μm; (E and F) 5 μm; (G) 10 μm.
Figure 6.
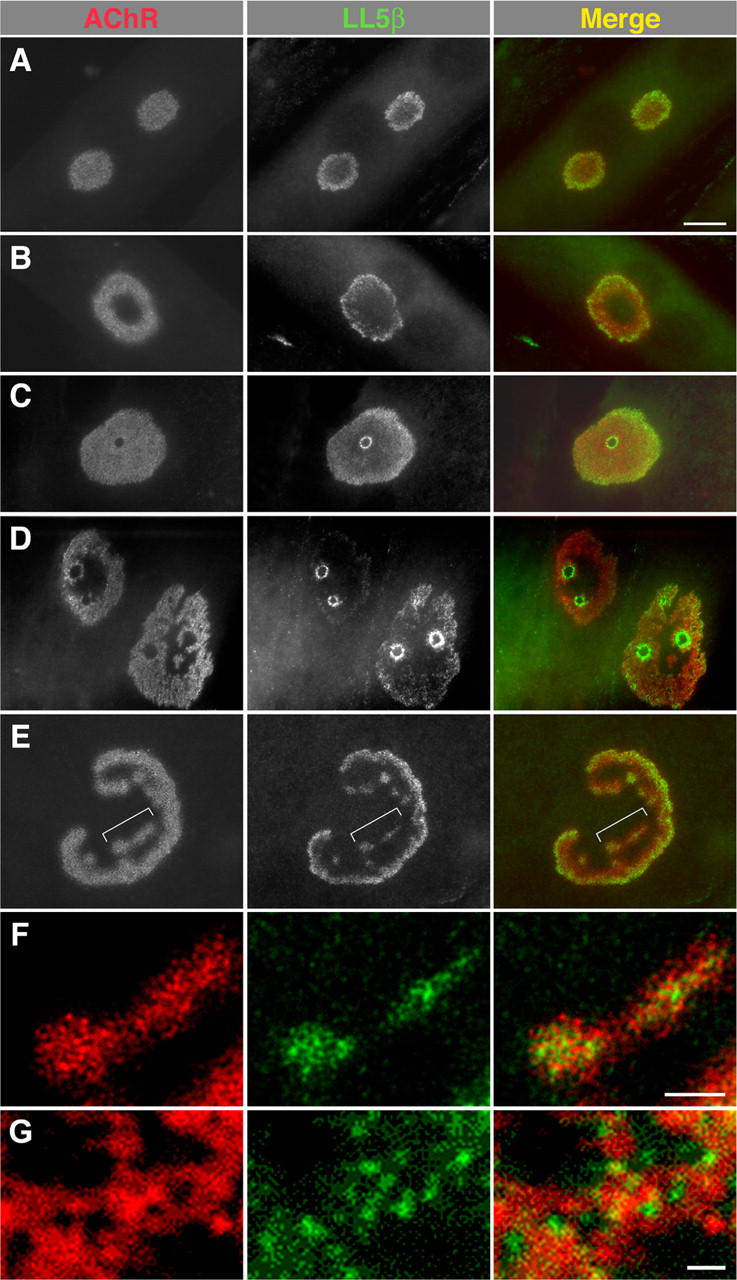
LL5β is associated with borders of developing AChR clusters. C2 myotubes were cultured on a laminin substrate to induce development of complex, branched AChR aggregates, and then double-stained with anti-LL5β and rBTX. (A) LL5β is colocalized with AChRs in simple plaques but concentrated at the circumference of the clusters. (B and E) Peripheral distribution of LL5β becomes more marked as clusters become perforated and then branched. Bracketed region in E is shown at higher magnification in F. (C and D) In some cases, LL5β is most concentrated at the borders of the perforations that transform plaques into annuli and then into branched aggregates. (F) Within aggregates, AChRs and LL5β have complementary distributions. (G) High magnification view of an NMJ from a P7 mouse (Fig. 4 F) showing similar complementarity in vivo. Bars: (A–E) 10 μm; (F and G) 1 μm.
The first gene product, CD24 (No. 6), is a small, highly glycosylated, membrane-associated protein. It has been studied most intensively in the immune system but is also expressed in the nervous system, where it has been implicated in the control of neurogenesis and neurite outgrowth (Calaora et al., 1996; Shewan et al., 1996; Kleene et al., 2001; Belvindrah et al., 2002).
The second gene product, Unc53H3/neuron navigator 3/ POMFIL1 (No. 11; Coy et al., 2002; Maes et al., 2002; Merrill et al., 2002), is predicted, on the basis of its sequence, to be a cytoplasmic, actin-binding adaptor protein. It is one of three mammalian orthologues of Caenorhabditis elegans unc-53. Unc 53 mutants exhibit defects in axon branching and muscle formation (Hekimi and Kershaw, 1993; Stringham et al., 2002), and unc53H2-deficient mice show defects in sensory acuity and optic nerve growth (Peeters et al., 2004).
The third gene product, ERM (No. 13), is a member of a large family of “Ets-domain” transcriptional regulators (Sharrocks, 2001). Ets-domain proteins are believed to promote selective transcription of AChR subunit genes by synaptic nuclei (Schaeffer et al., 2001). Attention has centered on the Ets-related proteins GABP-α and -β, but both RNAs are present in extrasynaptic as well as synaptic regions (Fromm and Burden, 1998; Schaeffer et al., 1998), suggesting that additional factors are involved in limiting GABP-mediated transcription to specific sites. That Ets-domain factors function as heterodimers makes it tempting to speculate that ERM plays such a role. Consistent with this possibility, Hippenmeyer et al. (2002) recently published micrographs suggesting selective localization of ERM RNA in synaptic regions of embryonic muscle.
Finally, NTA (No. 18) RNA encodes a 217–amino acid protein conserved in several mammalian species (mouse, rat, and human; UniGene identifiers are Mm 21836, Rn. 13571, and Hs. 157779, respectively). This RNA begins just 200 bp 3′ to the last known exon of the AChE gene (Wilson et al., 2001), so we have given its locus the working name “NTA” for “next to AChE.” It is unclear whether it represents an alternative transcriptional product of the AChE locus or a distinct gene. Our data are consistent with both possibilities: Northern analysis using an NTA-specific probe revealed transcripts of 1.2 and 3.8 kb in muscle, the former excluding and the latter including AChE-derived sequences (unpublished data). The differential regulation of NTA and AChE RNAs by innervation (see the following section) also implies the existence of distinct mRNAs.
In summary, of the top 25 genes from our microarray analysis, the synaptic enrichment of 20 was validated by a second method (RT-PCR, in situ hybridization, or published immunohistochemical localization). Of these 20, at least five encode RNAs that are concentrated very near the NMJ but have not previously been studied in the context of the NMJ. Remarkably, all five are plausible candidates for synaptic roles, based on their chromosomal localization (NTA), roles in other neural systems (Unc5h3 and CD24), known properties of synapse-specific transcription (ERM), or our own data (LL5β; see Figs. 7 and 8).
Figure 7.
LL5β is required for AChR clustering. (A) COS cells were cotransfected with GFP-LL5β plus control duplex VIII siRNA (left), LL5β-RNAi#1 (center), or LL5β-RNAi#2 (right), and then examined for GFP fluoresence. RNAi#1 and #2 suppressed GFP-LL5β expression. (B and C) Myotubes cultured on laminin (Fig. 6) were transfected with the indicated RNAi vectors, or control RNAi vector, fused to form myotubes, and then stained with rBTX. Both LL5β RNAi's decreased the number of AChR aggregates, whereas control RNAi (a sequence that effectively suppressed GFP) had no effect. (C) Examples are shown in B and data from one experiment are graphed (C; 15–10× microscope fields were counted per culture). The difference between LL5β RNAi and either control is significant at P < 0.001 by t test. Similar results were obtained in two additional experiments. Western blots of myotube lysates, using anti-LL5β antibody 7E3, are shown in C (bottom). Densitometry showed that RNAi#1 and #2 decreased LL5β protein levels by 40–50% compared with mock-transfected and control RNAi-transfected cultures. Error bars show mean ± SEM. (D) GFP-transfected myotubes were stained with anti-GFP to show high transfection efficiency. Only the brightest myotubes would appear positive for endogenous GFP fluorescence. Inset shows mock-transfected myotubes stained with anti-GFP. Bar, 100 μm.
Figure 8.
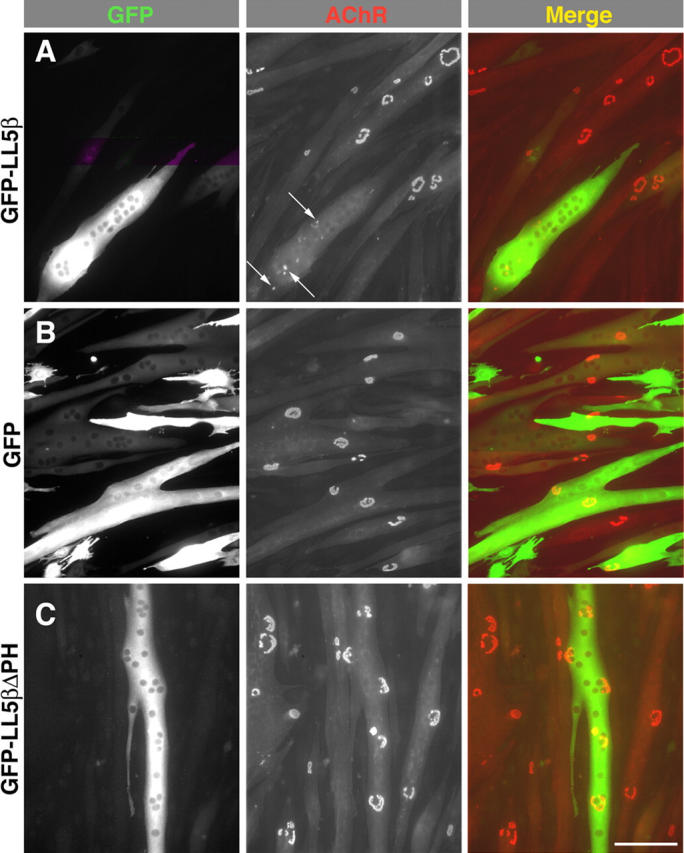
Overexpression of LL5β perturbs AChR clustering. Myoblasts cultured on laminin (Fig. 6) were transfected with the indicated constructs, fused to form myotubes, and then stained with rBTX. (A) Few large AChR clusters are present in myotubes that overexpress GFP-LL5β; instead, receptors are present diffusely and in small patches (arrows). (B and C) Forced expression of GFP (B) or a GFP-LL5β fusion protein lacking the PH domain (GFP-LL5β-ΔPH; C) has no effect on the frequency of aggregation. Bar, 100 μm. (D) Percent of transfected (strongly fluorescent) or untransfected myotubes with complex AChR aggregates. 80 myotubes were analyzed per condition: 40 in each of two separate experiments. Difference between GFP-LL5β and each other condition is significant at P < 0.001. Error bars show mean ± SEM.
The 25 probe sets that we analyzed gave signals that were >7.5-fold higher in synaptic than in extrasynaptic regions of muscle. Of the 36,701 probe sets queried, a total of 42, 70, 139, and 424 showed greater than six-, five-, four-, and threefold synaptic enrichment, respectively (Table S2, available at http://www.jcb.org/cgi/content/full/jcb.200411012/DC1). Almost surely, the incidence of false positives will increase as the enrichment decreases. Moreover, some of the true positives may encode proteins expressed by nonmuscle cells, such as Schwann cells or perisynaptic fibroblasts. Nonetheless, it seems likely that the list in Table S2 includes many additional novel synaptic components.
Coregulation of synaptically enriched transcripts
Several RNAs previously shown to be enriched at synaptic sites—notably the AChRα, β, and δ subunits, N-CAM, and MuSK—share two additional features: their abundance in muscle decreases as development proceeds and increases after denervation (Merlie et al., 1984; Covault et al., 1986; Goldman et al., 1988; Valenzuela et al., 1995). We asked whether or not RNAs encoding other components of the postsynaptic apparatus are regulated similarly.
First, we determined the synaptic enrichment of transcripts encoding basal lamina, plasma membrane, and cytoskeletal proteins concentrated in the postsynaptic apparatus (Peters et al., 1994; Miner and Sanes, 1994; Altiok et al., 1995; Moscoso et al., 1995a,b; Zhu et al., 1995; Patton et al., 1997; Sanes and Lichtman, 1999). In addition to those transcripts listed in Tables I and II, RNAs encoding AChRβ, AChRγ, rapsyn, ColQ, erbB3, utrophin, N-CAM, laminins α5, and collagen α3(IV) were enriched in the synaptic samples (Table III). In contrast, RNAs encoding the basal lamina protein laminin β2, the membrane protein erbB4, and the cytoskeletal protein syntrophin β2 were not appreciably enriched in synaptic mRNA (unpublished data), even though their protein products are concentrated at synaptic sites.
Table III. Regulation of synaptic genes during development and after denervation.
| Rank | Gene name | Synaptic/ extrasynaptic |
Denervation/ adult |
Embryonic/ adult |
|---|---|---|---|---|
| 1 | AChRɛ | 83.64 | 0.28 | 0.04 |
| 2 | AChRα1 | 56.78 | 25.35 | 3.58 |
| 6 | CD24 | 16.39 | 1.93 | 0.34 |
| 11 | Unc53H3 | 12.86 | 1.35 | 3.17 |
| 12 | MuSK | 11.63 | 4.89 | 7.58 |
| 13 | ERM | 11.40 | 0.93 | 0.47 |
| 14 | nestin | 10.88 | 0.41 | 4.03 |
| 16 | LL5β | 10.40 | 1.95 | 4.04 |
| 18 | NTA | 9.69 | 1.49 | 2.57 |
| 21 | PKARIα | 8.46 | 1.66 | 1.08 |
| 22 | AChRδ | 8.40 | 2.42 | 7.12 |
| 47 | AChE | 5.78 | 0.09 | 1.95 |
| 79 | collagen α3(VI) | 4.82 | 1.08 | 1.58 |
| 122 | ColQ | 4.14 | 1.15 | 1.91 |
| 307 | utrophin | 3.28 | 4.76 | 2.75 |
| 455 | AChRγ | 2.95 | 1.10 | 19.36 |
| 1728 | AChRβ1 | 2.13 | 6.16 | 1.09 |
| 2227 | NCAM | 1.99 | 1.77 | 0.95 |
| 2845 | laminin α5 | 1.88 | 1.35 | 3.40 |
| 4925 | rapsyn | 1.61 | 0.50 | 1.57 |
| 7374 | ErbB3 | 1.44 | 2.83 | 8.69 |
Synaptic enrichment of transcripts previously known to encode synaptic proteins was determined as in Table S1; those with rankings in the top 20% are listed, along with new genes identified in this study. Changes in abundance during development and after denervation were assessed with duplicate microarrays.
Second, we asked which synaptically enriched transcripts are present at higher levels in embryonic (embryonic day 15) or denervated muscles than in normal adult muscle. For this purpose, we generated new targets by conventional methods that did not involve amplification. 76% of the synaptically enriched transcripts queried (16/21) were up-regulated after denervation, and 81% (17/21) were down-regulated during development (Table III). Three genes identified in this study, NTA, unc53H3, and LL5β, share this pattern.
Synaptic genes are not, however, completely coregulated. Some variations from the common pattern have been noted previously, for example the postnatal up-regulation of AChRɛ, and the down-regulation of AChE after denervation (Sanes and Lichtman, 1999). Another pattern is that of ERM RNA, which changes little during development or after denervation. Synaptic nuclei acquire specialized properties during development and maintain them in denervated muscle. Although AChR subunit genes are selectively transcribed by these nuclei, they are also transcribed by extrasynaptic nuclei during development and after denervation. Expression of a gene involved in controlling synapse-specific transcription might be expected to remain more confined to synaptic nuclei than expression of the genes it regulates. Our results are consistent with the idea that ERM is one such gene.
Synaptic localization of LL5β
The rationale for our screen was that RNAs concentrated in synaptic areas encode proteins concentrated at synaptic sites. This idea was plausible but unproven, in that all synaptically enriched RNAs described to date were identified because their protein products were already known to be synaptic components. Therefore, we asked whether or not a novel gene identified in our screen, LL5β, encoded a synaptic component.
LL5β is an ∼160-kD protein containing two predicted coiled-coil domains, a serine-rich region predicted to contain multiple protein kinase C phosphorylation sites, and a phosphoinositide-binding module called a PH domain. Many PH domains selectively bind 3-phosphorylated phosphoinositides that are generated in the inner leaflet of the plasma membrane by phosphatidylinositol 3-kinase (PI3K); therefore, they serve to recruit signaling proteins to the membrane in response to PI3K activation (Lemmon and Ferguson, 2000). Paranavitane et al. (2003) showed that LL5β binds both phosphatidylinositol (3,4,5)-triphosphate and a cytoskeletal adaptor protein, γ-filamin, and that activation of PI3K regulates its association with the membrane. Two partial homologues of LL5β are present in the mouse genome: LL5α, which binds phosphoinositides (Dowler et al., 2000) but has not been studied further, and a novel sequence that we found by database searching and call LL5γ. The three members of the LL5 family all have two coiled-coiled domains and a PH domain, but strong homology is restricted to their carboxy termini; their amino termini are divergent in both sequence and domain structure (Fig. 4 A). To date, no physiological role has been reported for any member of the LL5β family.
To ask if LL5β is synaptically localized, we raised monoclonal antibodies to a region with low homology to LL5α and γ (see Materials and methods) and used them to stain longitudinal sections of adult mouse and rat muscles. NMJs were selectively stained (Fig. 4, B–D), although nerve bundles and fibroblasts were also LL5β-positive (not depicted). Similar results were obtained with two other antibodies to LL5β, whereas control antibodies of the same isotype did not stain NMJs. High-power views of cross sections suggested that the distribution of AChRs and LL5β was not identical within NMJs: AChRs are concentrated at the crests of junctional folds (Sanes and Lichtman, 2001), whereas levels of LL5β were highest in the depths of junctional folds and between AChR-rich regions (Fig. 4 E). Substantial colocalization of LL5β and AChRs was evident by P4 (Fig. 4 F). LL5β was also colocalized with AChR-rich patches in cultured myotubes (Fig. 4 G).
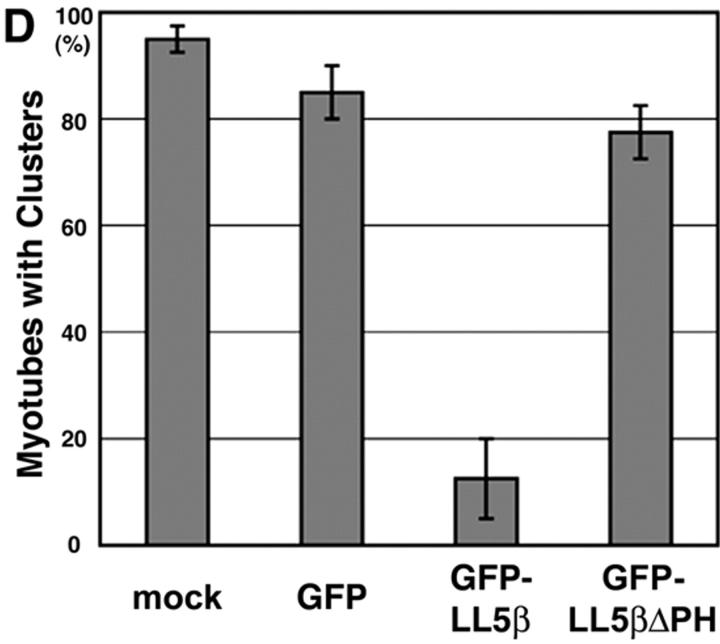
These results demonstrate an association of LL5β with the postsynaptic membrane of the NMJ. To ask if the PH domain mediates this association, we fused GFP to LL5β or to a mutant LL5β from which the PH domain had been deleted (LL5βΔPH). Expression plasmids encoding GFP-LL5β, GFP-LL5βΔPH, or GFP alone were electroporated into mouse anterior tibialis muscles. By 4 d after electroporation, GFP was colocalized with AChRs in muscle fibers expressing GFP-LL5β (Fig. 5 A; n = 10). In contrast, GFP immunofluorescence was diffuse and cytoplasmic in fibers from muscles that had been transfected with GFP-LL5βΔPH or GFP alone (Fig. 5, B and C; n = 10 each). Likewise, GFP-LL5β but not GFP-LL5βΔPH localized to AChR-rich clusters in cultured myotubes (unpublished data). Thus, its PH domain is essential for targeting LL5β to the NMJ. In contrast, fusion of GFP to isolated PH domains did not localize to AChR clusters in myotubes (unpublished data), indicating that although the PH domain is necessary for localization it is not sufficient.
Figure 5.
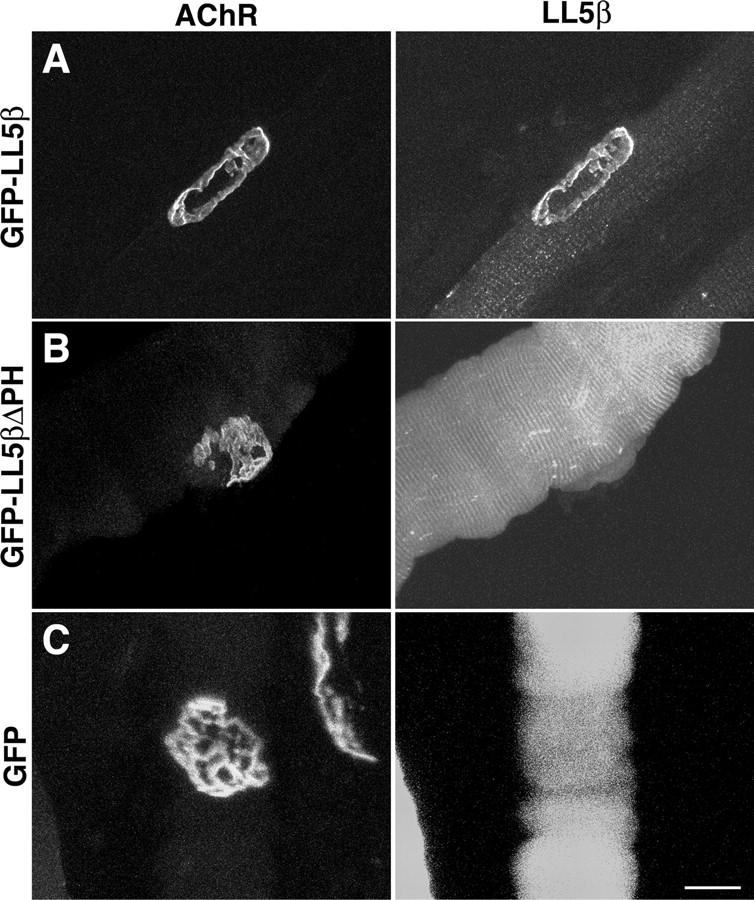
Synaptic localization of LL5β requires its PH domain. Muscles were electroporated with the indicated constructs. 4 d later, fibers were fixed, teased, and stained with rBTX. (A) GFP-LL5β colocalizes with AChRs at the NMJ. (B and C) A fusion protein lacking the carboxy-terminal PH domain (GFP-LL5β-ΔPH [B]) and GFP alone (C) are diffusely distributed. Bar, 20 μm.
Role of LL5β in AChR clustering
For further analysis of LL5β localization and function, we used a culture system in which myotubes form elaborate, branched AChR aggregates that resemble adult NMJs in several respects. The aggregates form on the substrate-facing surface, and can thus be imaged in a single plane; and they form from simple plaques in a series of steps resembling those that transform the embryonic NMJ into the adult structure in vivo (Kummer et al., 2004). They are therefore useful for following developmental changes in LL5β distribution that could not be discerned in vivo.
LL5β was associated with AChR-rich plaques in this system throughout their evolution, but its distribution differed systematically from that of AChRs. In simple plaques, AChRs were uniformly distributed, whereas LL5β was concentrated around the circumference (Fig. 6 A). Subsequently, as plaques became perforated at their centers, the peripheral distribution of LL5β became more marked (Fig. 6 B). In some cases, LL5β also rimmed the outer circumference of the perforations that transform plaques into annuli and then into branched structures (Fig. 6, C and D). The tendency of LL5β to concentrate at the periphery persisted in branched aggregates but was less striking (Fig. 6 E). Within aggregates, LL5β and AChRs were often distributed in complementary patterns (Fig. 6 F); the distribution of LL5β is reminiscent of that of vinculin and dystrophin (Bloch and Pumplin, 1988; Dmytrenko et al., 1993). Likewise, LL5β and AChRs appeared to have complementary distributions at developing NMJs in vivo (Fig. 6 G), as well as at adult NMJs (Fig. 4 E), although the more favorable geometry of myotube cultures made details of the localization more readily appreciated in vitro.
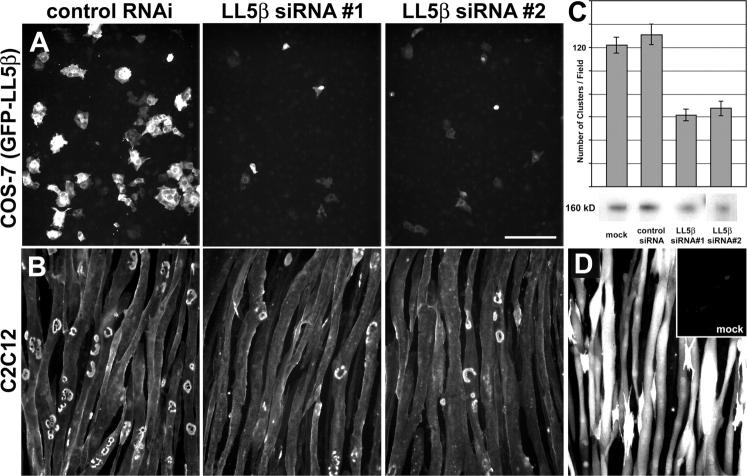
To find whether or not LL5β plays a role in AChR clustering, we used RNAi to suppress its expression in myotubes. Two of four sequences tested effectively suppressed LL5β expression in heterologous cells (Fig. 7 A and see Materials and methods). These sequences were used to generate expression vectors, which were introduced into muscle cells. Both vectors decreased the level of endogenous LL5β by 40–50% and the frequency of AChR clustering by ∼50% compared with untreated cultures, whereas control vectors had no significant effect (Fig. 7, B and C; and not depicted). None of the RNAi vectors had any apparent effect on cluster size or shape.
Given the low transfection efficiency that we and others have noted for muscle cells, we were surprised that LL5β RNAi suppressed AChR clustering in so many myotubes. Therefore, we assayed transfection efficiency with GFP. Endogenous GFP fluorescence was detectable in only ∼10% of myotubes, but immunostaining with anti-GFP revealed low levels of expression in an additional ∼50% of myotubes (Fig. 7 D). We suggest that RNAi vectors are also expressed at low levels in most myotubes, and that these levels are sufficient to suppress LL5β function because RNAi acts catalytically. Consistent with this idea, the 40–50% decrease in total LL5β levels in the myotube cultures implies that levels are decreased by 65–80% in the transfected myotubes.
As a second approach to assessing the function of LL5β, we overexpressed GFP-LL5β in myotubes. The fusion protein was associated with AChR aggregates when it was expressed at modest levels (unpublished data; levels assessed by staining with anti-LL5β, and comparing to endogenous levels in controls). In contrast, GFP-LL5β was diffusely distributed in myotubes that expressed high levels of fusion protein, and few AChR aggregates were present in these myotubes (Fig. 8 A). Instead, AChRs in these myotubes were distributed diffusely and in small patches (Fig. 8 A). No perturbations were observed in myotubes expressing GFP alone, the GFP-LL5βΔPH fusion protein, or the GFP-PH fusions (Fig. 8, B–D; and not depicted), indicating that the effects of the LL5β were specific. However, lacking another PH domain–containing protein that localizes to AChR clusters, we cannot rule out the possibility that the effects of overexpression result from a nonphysiological perturbation of the AChR-rich domain.
Discussion
Although much is known about the molecular architecture of the synapse, it is clear that additional components remain to be discovered. To identify some of them, we exploited the finding that all of ∼10 mRNAs previously reported to be concentrated in synaptic regions of muscle fibers encode components of the postsynaptic apparatus at the NMJ (Merlie and Sanes, 1985; Goldman and Staple, 1989; Jasmin et al., 1993, Moscoso et al., 1995b; Valenzuela et al., 1995; Imaizumi-Scherrer et al., 1996; for reviews see Sanes and Lichtman, 1999, 2001; Chakkalakal and Jasmin, 2003). Therefore, we used microarray-based expression profiling to seek additional synaptic RNAs. By combining microdissection, controlled amplification, sample prescreening, and multiple replicates, we reduced the false positive rate to ∼20% for the top tier of 25 genes, validated 10 novel RNAs enriched in synaptic areas, and provided >200 additional candidates.
Our analysis demonstrates the power of the Tietjen et al. (2003) method for expression profiling of tiny samples and provides some guidelines for its use. In particular, our data directly demonstrate that the amplification method generates high amplitude noise, which obscures enrichment of transcripts. In fact, most exceptionally high and low values obtained from any single GeneChip represent noise. Because this noise is more or less random, however, it is efficiently diluted when data sets are pooled, facilitating detection of genes that are consistently enriched, even if the degree of enrichment is modest.
We also used conventional expression profiling to show that greater than three fourths of the known and novel synaptic RNAs are coregulated during development and after denervation, suggesting the existence of a common regulatory program. One cis-element critical for synaptic expression is the N-box, a site that binds Ets-domain transcription factors; such sites are already known to play roles in the synaptic transcription of genes encoding AChR subunits, AChE, and utrophin (Koike et al., 1995; Schaeffer et al., 2001). Cis-elements called E-boxes, which bind basic helix-loop-helix factors, have been implicated in down-regulation during development and up-regulation after denervation. Many genes selectively transcribed in synaptic nuclei may share a set of such cis-elements, as envisioned for members of “synexpression groups” (Niehrs and Pollet, 1999). However, synaptic genes are not completely coregulated. Reasons for the exceptions are unknown, but we speculate that expression of genes involved in regulating synapse-specific transcription might be more confined to synaptic nuclei during development and after denervation than expression of the genes they regulate. ERM may be one such gene.
All five of the novel RNAs that we showed to be enriched at or near synaptic sites by in situ hybridization (NTA, Unc5h3, CD24, ERM, and LL5β) are reasonable candidates for synaptic roles. Here, we focused exclusively on one of them, LL5β. LL5β had previously been shown to bind phosphoinositides and filamin (Dowler et al., 2000; Paranavitane et al., 2003), but its localization and physiological function were unknown. We found that LL5β is a component of the postsynaptic apparatus, thereby validating our overall strategy. Moreover, detailed analysis of localization as well as loss- and gain-of-function analyses showed that LL5β is part of the machinery required for aggregation of AChRs.
The mechanism by which LL5β affects AChR aggregation remains to be explored, but some clues exist: (a) LL5β is most highly concentrated in AChR-poor regions directly adjacent to AChR-rich regions (edges of clusters and interstices between microaggregates, in vitro and in vivo: Fig. 4 E; and Fig. 6, F and G). (b) Its association with the postsynaptic membrane is mediated by its phosphoinositide-binding PH domain (Fig. 5). (c) It binds to a cytoskeletal component, filamin (Paranavitane et al., 2003), one form of which is itself colocalized with clustered AChRs in myotubes (Bloch and Hall, 1983; Shadiack and Nitkin, 1991). (d) It can reorganize the cytoskeleton in response to extracellular signals that regulate its association with the membrane (Paranavitane et al., 2003). (e) AChR-rich and -poor regions within AChR aggregates differ in their lipid composition (Pumplin and Bloch, 1983; Scher and Bloch, 1991; Barrantes, 2002). Together, these data suggest a model in which LL5β “corrals” AChRs by forming a dynamic link between specific lipids and the AChR-associated cytoskeleton; the link would remodel as AChR clusters grow or change in shape, and might even help direct shape changes.
In this model, corrals would be unable to form when LL5β is absent, impeding clustering, whereas overexpression of LL5β might inhibit AChR aggregation by preventing microaggregates from coalescing into larger clusters. Alternatively, when LL5β is present in excess, some molecules might bind the cytoskeleton and others the membrane, disrupting the corraling complex and leading to a phenotype similar to that seen when LL5β levels are decreased. The latter explanation is similar to that postulated for rapsyn, another protein associated with the cytoplasmic face of the postsynaptic membrane: no AChR clusters form in the absence of rapsyn (Gautam et al., 1995), but overexpression also inhibits clustering (Yoshihara and Hall, 1993; Han et al., 1999). One way to elucidate LL5β's mechanism of action will be to identify the signals that affect its localization or binding capacity and to ask if it interacts with intracellular pathways already implicated in AChR clustering (Jones and Werle, 2000; Weston et al., 2000; Luo et al., 2002, 2003; Finn et al., 2003).
Materials and methods
Microarray analysis
To isolate NMJs, diaphragms of C57B1 6J mice were incubated in ice-cold PBS containing 10 μg/ml rBTX (Molecular Probes) for 5 min to stain NMJs, and then washed with PBS. Clusters of ∼20 closely spaced NMJs were located with a fluorescent dissecting stereomicroscope (model SZX12; Olympus) and cut out using forceps and a micro-knife. Synapse-free samples of equivalent size were obtained from each muscle and processed in parallel. These samples were used for cDNA synthesis and PCR amplification as described by Dulac and Axel (1995). The cDNAs were assayed by Southern blotting using the following probes: GAPDH (GenBank/EMBL/DDBJ accession no. M32599; nt 529–1228), cyclophilin A (GenBank/EMBL/DDBJ accession no. X52803; 22–708), AChRα (GenBank/EMBL/DDBJ accession no. M17640; 1148–1825), AChE (GenBank/EMBL/DDBJ accession no. X56518; 1372–2062), MuSK (GenBank/EMBL/DDBJ accession no. U37709; 2626–3299), PKARIα (GenBank/EMBL/DDBJ accession no. BC003461; 2591–3279). Based on results of Southern analysis, probes were prepared from nine pairs of samples (six from adult and three from P4 animals) by minor modifications of the methods of Tietjen et al. (2003). In brief, PCR products were reamplified, purified, digested with EcoRI, purified twice on CHROMA SPIN-200 Columns (CLONTECH Laboratories, Inc.), fragmented with RQ1 DNase (Promega) for 13 min at 37°C, and biotinylated with Biotin-N6-ddATP (NEN Life Science Products) and terminal transferase (Invitrogen) for 90 min at 37°C. The targets were hybridized to GeneChip arrays (Murine U74A, B, and C; Affymetrix, Inc.), which were then processed as described by Lockhart et al. (1996).
For analysis of gene regulation, we compared lower leg muscles from day 15 embryos, lower leg muscles from adults 4 d after unilateral section of the sciatic nerve, and contralateral innervated muscles from the same mice. Total RNA was isolated with Trizol Reagent (Invitrogen) and RNeasy Mini Kit (QIAGEN). RNA was converted to double-stranded cDNA with an oligo(dT) primer containing a T7 promotor and used for synthesis of biotinylated cRNA using a Bioarray HighYield Kit (Enzo). The cRNA targets were fragmentated as described previously (Lipshutz et al., 1999), and then hybridized to GeneChips as described for cDNA. Results from duplicate samples were averaged.
Data were analyzed with Affymetrix Microarray Suite 5.0 software package. The numerical signal intensity for each probe set was used for further calculation in Microsoft Excel and R (www.r-project.org). The SAM program (Tusher et al., 2001) was used at the threshold value of 0.05.
Quantitative RT-PCR
Mouse diaphragms were stained with rBTX as in Microarray analysis and divided into synapse-rich and -free segments (Merlie and Sanes, 1985). Total RNA was extracted, and 1-μg aliquots were used for cDNA synthesis. Products were used for quantitative RT-PCR using the TaqMan 7700 Sequence Detection System (PerkinElmer) and SYBER-GREEN dye (Applied Biosystems) as described previously (Wittwer et al., 1997; Morrison et al., 1998). Levels of GAPDH RNA were used for normalization. Assays were repeated on two independently prepared samples, and each sample was assayed in duplicate.
Antibody production
A fragment of LL5β corresponding to aa 418–760 was fused to a polyhistidine epitope tag, expressed in Escherichia coli, purified with Ni-NTA Spin Kit (QIAGEN), and used to immunize BalbC mice. Hybridomas were produced by standard methods, and their products were assayed by ELISA, using immobilized immunogen, and by immunostaining of COS cells transfected with a GFP-LL5β fusion. Three mAbs (1H12, 6B9, and 7E3) were subcloned.
Evidence that these antibodies specifically recognize LL5β is as follows: (a) the antibodies stain COS cells transfected with LL5β but not untransfected cells (Fig. S1 B, available at http://www.jcb.org/cgi/content/full/jcb.200411012/DC1). (b) Antibody staining patterns of untransfected muscle fibers match GFP fluorescence patterns in muscle fibers transfected with the LL5β-GFP fusion (compare Figs. 4 B and 5 A). (c) There is a similar correspondence for cultured muscle cells. (d) Two of the three antibodies (7E3 and 1H12) detect a single band on immunoblots of C2 myotube lysates (Fig. S1 A). This is the expected size for LL5β. Results using antibody 1H12 (IgG1 subclass) are shown here, but similar results were obtained with 6B9 and 7E3.
Histology
Whole mount in situ hybridization was performed as described by Wilkinson and Nieto (1993) using digoxygenin-labeled cRNA probes. Digoxygenin was detected with alkaline phosphatase-conjugated anti-digoxygenin antibody (Roche), and signal was developed with NBT plus BCIP.
For immunohistochemistry, mouse or rat muscles were fixed in 1% PFA, sunk in 30% sucrose/PBS, frozen, and sectioned at 10 μm in a cryostat. Sections were stained sequentially with anti-LL5β and a mixture of Alexa 488–conjugated secondary antibody and rBTX or Alexa 596–BTX (Molecular Probes).
Electroporation in vivo
Mouse LL5β cDNA was obtained from American Type Culture Collection (IMAGE clone 6405379). Full-length and PH domain–deleted coding sequences (lacking aa 1194–1302) were amplified by RT-PCR and ligated to the BglII–SalI site of pEGFP-C1 vector (CLONTECH Laboratories, Inc.). Inserts were sequenced. Plasmids were introduced into tibialis muscles of P10 mice by electroporation as described by Grady et al. (2003). 4 d after electroporation, muscles were dissected under a fluorescent stereomicroscope, fixed in 4% PFA, counterstained with BTX, and imaged with a confocal laser scanning microscope (model FV500; Olympus) using a 100× oil objective (NA 1.40).
Tissue culture
C2C12 myoblasts (American Type Culture Collection) were plated on Labtek 8-well Permanox chamber slides (Nalge) coated with 0.2% gelatin (Bio-Rad Laboratories) or 10 μg/ml laminin-1 (Invitrogen) as described by Kummer et al. (2004). Where indicated, myoblasts were transfected on the day of plating using FuGENE6 (Roche). One day after plating, cultures were switched to DME plus 2% horse serum to induce fusion. Cultured myotubes were stained live with 1 μg/ml rBTX for 30 min, and then fixed, permeabilized, and stained with anti-LL5β and anti-GFP (Chemicon). Images collected with a CCD camera (model MagnaFire; Optronics) were analyzed with Metamorph software (Universal Imaging Corp.).
To suppress LL5β expression, siRNA sequences were selected using SVM RNAi software (Chang Bioscience), siRNA DESIGN Center (Dharmacon), and the siRNA Selection Server at the Whitehead Institute (Massachusetts Institute of Technology, Cambridge, MA; http://jura.wi.mit.edu/siRNAext/). The mouse genome was searched with selected sequences to ensure their specificity. RNA duplexes of the following target sequences were purchased from Dharmacon: No. 1, AACCUCCUAUUUCUUUCCUCA; No. 2, AAGCCUAAGACAGUCGUCAGA; No. 3, AAGACUUGGAAUUCCAGCAGC; and No. 4, AAGCAAGCCAGUCACAUCGUU. COS-7 cells were cotransfected with an expression vector encoding GFP-LL5β plus one of these RNAs, or control duplexes VII (57% GC Content) or VIII (52% GC Content; Dharmacon), using Lipofectamine 2000 (Invitrogen). RNAs Nos. 1 and 2 suppressed expression of GFP-LL5β dramatically; RNA No. 4 had a less dramatic effect; and RNA No. 3 and control duplexes VII and VIII had no obvious effect on GFP-LL5β expression. RNAs Nos. 1, 2, and 4 had no effect on expression of GFP alone, indicating that their effects were specific; the susceptibility of GFP cDNA to RNAi suppression was demonstrated with RNAi specific for GFP.
Based on these results, sequences of RNA Nos. 1 and 2 and GFP RNAi were cloned into a plasmid-based siRNA-expression system (pSuper vector; Oligoengine). Plasmids were used rather than oligonucleotides because they were superior for maintaining high expression of the interfering sequences throughout the week-long culture period. Myotubes were transfected at the time of plating using FuGene6 reagent (Roche) according to manufacturer's instructions. A total of 6.6 μl FuGene reagent and 2.2 μg DNA in 100 μl DME was used to transfect every two wells of an 8-well chamber slide. Cells were switched to differentiation medium 1 d after transfection and cultured for an additional 5–6 d. Cells were then stained live with a fluorescent conjugate of BTX plus, where appropriate, anti-GFP.
Online supplemental material
Fig. S1 shows evidence for specificity of anti-LL5β antibodies. Table S1 shows the ratio of synaptic to extrasynaptic signal for each synaptic gene from each microarray experiment. These ratios were then ranked to give the values presented in Table I. Table S2 shows transcripts enriched greater than threefold in synaptic compared with extrasynaptic samples. Rankings and ratios, calculated as in Table I and Table S1, respectively, are shown, along with UniGene number (http://www.ncbi.nlm.nih.gov) and gene name. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200411012/DC1.
Acknowledgments
We thank Catherine Dulac for advice on the sample preparation methods that made our study possible and Jeanette Cunningham for assistance in generation of antibodies. M. Kishi thanks Joe Henry Steinbach for support.
This work was funded by grants to J.R. Sanes from the National Institutes of Health. T.T. Kummer is a student in the Medical Scientist Training Program at Washington University.
M. Kishi's present addresses is Dept. of Anesthesiology, Washington University, St. Louis, MO 63110.
S.J. Eglen's present address is Dept. of Applied Mathematics and Theoretical Physics, University of Cambridge, Cambridge CB3 0WA, UK.
J.R. Sanes' present address is Dept. of Molecular and Cellular Biology, Harvard University, Cambridge, MA 02138.
Abbreviations used in this paper: AChE; acetylcholinesterase; AChR, acetylcholine receptor; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; MuSK, muscle specific kinase; NMJ, neuromuscular junction; PH, pleckstrin homology; PI3K, phosphatidylinositol 3-kinase; PKARIα, protein kinase A RIα subunit; rBTX, rhodamine-α-bungarotoxin; SAM, significance analysis of microarrays.
References
- Altiok, N., J.L. Bessereau, and J.P. Changeux. 1995. ErbB3 and ErbB2/neu mediate the effect of heregulin on acetylcholine receptor gene expression in muscle: differential expression at the endplate. EMBO J. 14:4258–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrantes, F.J. 2002. Lipid matters: nicotinic acetylcholine receptor-lipid interactions. Mol. Membr. Biol. 19:277–284. [DOI] [PubMed] [Google Scholar]
- Belvindrah, R., G. Rougon, and G. Chazal. 2002. Increased neurogenesis in adult mCD24-deficient mice. J. Neurosci. 22:3594–3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch, R.J., and Z.W. Hall. 1983. Cytoskeletal components of the vertebrate neuromuscular junction: vinculin, α-actinin, and filamin. J. Cell Biol. 97:217–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch, R.J., and D.W. Pumplin. 1988. Molecular events in synaptogenesis: nerve-muscle adhesion and postsynaptic differentiation. Am. J. Physiol. 254:C345–C364. [DOI] [PubMed] [Google Scholar]
- Breitling, R., P. Armengaud, A. Amtmann, and P. Herzyk. 2004. Rank products: a simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573:83–92. [DOI] [PubMed] [Google Scholar]
- Calaora, V., G. Chazal, P.J. Nielsen, G. Rougon, and H. Moreau. 1996. mCD24 expression in the developing mouse brain and in zones of secondary neurogenesis in the adult. Neuroscience. 73:581–594. [DOI] [PubMed] [Google Scholar]
- Carlsson, L., Z. Li, D. Paulin, and L.E. Thornell. 1999. Nestin is expressed during development and in myotendinous and neuromuscular junctions in wild type and desmin knock-out mice. Exp. Cell Res. 251:213–223. [DOI] [PubMed] [Google Scholar]
- Chakkalakal, J.V., and B.J. Jasmin. 2003. Localizing synaptic mRNAs at the neuromuscular junction: it takes more than transcription. Bioessays. 25:25–31. [DOI] [PubMed] [Google Scholar]
- Covault, J., J.P. Merlie, C. Goridis, and J.R. Sanes. 1986. Molecular forms of N-CAM and its RNA in developing and denervated skeletal muscle. J. Cell Biol. 102:731–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coy, J.F., S. Wiemann, I. Bechmann, D. Bachner, R. Nitsch, O. Kretz, H. Christiansen, and A. Poustka. 2002. Pore membrane and/or filament interacting like protein 1 (POMFIL1) is predominantly expressed in the nervous system and encodes different protein isoforms. Gene. 290:73–94. [DOI] [PubMed] [Google Scholar]
- Dmytrenko, G.M., D.W. Pumplin, and R.J. Bloch. 1993. Dystrophin in a membrane skeletal network: localization and comparison to other proteins. J. Neurosci. 13:547–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowler, S., R.A. Currie, D.G. Campbell, M. Deak, G. Kular, C.P. Downes, and D.R. Alessi. 2000. Identification of pleckstrin-homology-domain-containing proteins with novel phosphoinositide-binding specificities. Biochem. J. 351:19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulac, C., and R. Axel. 1995. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 83:195–206. [DOI] [PubMed] [Google Scholar]
- Finn, A.J., G. Feng, and A.M. Pendergast. 2003. Postsynaptic requirement for Abl kinases in assembly of the neuromuscular junction. Nat. Neurosci. 6:717–723. [DOI] [PubMed] [Google Scholar]
- Fromm, L., and S.J. Burden. 1998. Synapse-specific and neuregulin-induced transcription require an ets site that binds GABPalpha/GABPbeta. Genes Dev. 12:3074–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatchalian, C.L., M. Schachner, and J.R. Sanes. 1989. Fibroblasts that proliferate near denervated synaptic sites in skeletal muscle synthesize the adhesive molecules tenascin(J1), N-CAM, fibronectin, and a heparan sulfate proteoglycan. J. Cell Biol. 108:1873–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautam, M., P.G. Noakes, J. Mudd, M. Nichol, G.C. Chu, J.R. Sanes, and J.P. Merlie. 1995. Failure of postsynaptic specialization to develop at neuromuscular junctions of rapsyn-deficient mice. Nature. 377:232–236. [DOI] [PubMed] [Google Scholar]
- Goldman, D., and J. Staple. 1989. Spatial and temporal expression of acetylcholine receptor RNAs in innervated and denervated rat soleus muscle. Neuron. 3:219–228. [DOI] [PubMed] [Google Scholar]
- Goldman, D., H.R. Brenner, and S. Heinemann. 1988. Acetylcholine receptor alpha-, beta-, gamma-, and delta-subunit mRNA levels are regulated by muscle activity. Neuron. 1:329–333. [DOI] [PubMed] [Google Scholar]
- Grady, R.M., M. Akaaboune, A.L. Cohen, M.M. Maimone, J.W. Lichtman, and J.R. Sanes. 2003. Tyrosine-phosphorylated and nonphosphorylated isoforms of α-dystrobrevin: roles in skeletal muscle and its neuromuscular and myotendinous junctions. J. Cell Biol. 160:741–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, Z.W. 1973. Multiple forms of acetylcholinesterase and their distribution in endplate and non-endplate regions of rat diaphragm muscle. J. Neurobiol. 4:343–361. [DOI] [PubMed] [Google Scholar]
- Han, H., P.G. Noakes, and W.D. Phillips. 1999. Overexpression of rapsyn inhibits agrin-induced acetylcholine receptor clustering in muscle cells. J. Neurocytol. 28:763–775. [DOI] [PubMed] [Google Scholar]
- Hekimi, S., and D. Kershaw. 1993. Axonal guidance defects in a Caenorhabditis elegans mutant reveal cell-extrinsic determinants of neuronal morphology. J. Neurosci. 13:4254–4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippenmeyer, S., N.A. Shneider, C. Birchmeier, S.J. Burden, T.M. Jessell, and S. Arber. 2002. A role for neuregulin1 signaling in muscle spindle differentiation. Neuron. 36:1035–1049. [DOI] [PubMed] [Google Scholar]
- Imaizumi-Scherrer, T., D.M. Faust, J.C. Benichou, R. Hellio, and M.C. Weiss. 1996. Accumulation in fetal muscle and localization to the neuromuscular junction of cAMP-dependent protein kinase A regulatory and catalytic subunits RIα and Cα. J. Cell Biol. 134:1241–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasmin, B.J., R.K. Lee, and R.L. Rotundo. 1993. Compartmentalization of acetylcholinesterase mRNA and enzyme at the vertebrate neuromuscular junction. Neuron. 11:467–477. [DOI] [PubMed] [Google Scholar]
- Jessen, K.R., and R. Mirsky. 2002. Signals that determine Schwann cell identity. J. Anat. 200:367–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, M.A., and M.J. Werle. 2000. Nitric oxide is a downstream mediator of agrin-induced acetylcholine receptor aggregation. Mol. Cell. Neurosci. 16:649–660. [DOI] [PubMed] [Google Scholar]
- Klarsfeld, A., J.L. Bessereau, A.M. Salmon, A. Triller, C. Babinet, and J.P. Changeux. 1991. An acetylcholine receptor alpha-subunit promoter conferring preferential synaptic expression in muscle of transgenic mice. EMBO J. 10:625–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleene, R., H. Yang, M. Kutsche, and M. Schachner. 2001. The neural recognition molecule L1 is a sialic acid-binding lectin for CD24, which induces promotion and inhibition of neurite outgrowth. J. Biol. Chem. 276:21656–21663. [DOI] [PubMed] [Google Scholar]
- Koike, S., L. Schaeffer, and J.P. Changeux. 1995. Identification of a DNA element determining synaptic expression of the mouse acetylcholine receptor delta-subunit gene. Proc. Natl. Acad. Sci. USA. 92:10624–10628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kummer, T.T., T. Misgeld, J.W. Lichtman, and J.R. Sanes. 2004. Nerve-independent formation of a topologically complex postsynaptic apparatus. J. Cell Biol. 164:1077–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon, M.A., and K.M. Ferguson. 2000. Signal-dependent membrane targeting by pleckstrin homology (PH) domains. Biochem. J. 350:1–18. [PMC free article] [PubMed] [Google Scholar]
- Lipshutz, R.J., S.P. Fodor, T.R. Gingeras, and D.J. Lockhart. 1999. High density synthetic oligonucleotide arrays. Nat. Genet. 21:20–24. [DOI] [PubMed] [Google Scholar]
- Lockhart, D.J., H. Dong, M.C. Byrne, M.T. Follettie, M.V. Gallo, M.S. Chee, M. Mittmann, C. Wang, M. Kobayashi, H. Horton, and E.L. Brown. 1996. Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat. Biotechnol. 14:1675–1680. [DOI] [PubMed] [Google Scholar]
- Luo, Z.G., Q. Wang, J.Z. Zhou, J. Wang, Z. Luo, M. Liu, X. He, A. Wynshaw-Boris, W.C. Xiong, B. Lu, and L. Mei. 2002. Regulation of AChR clustering by Dishevelled interacting with MuSK and PAK1. Neuron. 35:489–505. [DOI] [PubMed] [Google Scholar]
- Luo, Z.G., H.S. Je, Q. Wang, F. Yang, G.C. Dobbins, Z.H. Yang, W.C. Xiong, B. Lu, and L. Mei. 2003. Implication of geranylgeranyltransferase I in synapse formation. Neuron. 40:703–717. [DOI] [PubMed] [Google Scholar]
- Maes, T., A. Barcelo, and C. Buesa. 2002. Neuron navigator: a human gene family with homology to unc-53, a cell guidance gene from Caenorhabditis elegans. Genomics. 80:21–30. [DOI] [PubMed] [Google Scholar]
- Martin, K.C., and K.S. Kosik. 2002. Synaptic tagging—who's it? Nat. Rev. Neurosci. 3:813–820. [DOI] [PubMed] [Google Scholar]
- Merlie, J.P., and J.R. Sanes. 1985. Concentration of acetylcholine receptor mRNA in synaptic regions of adult muscle fibres. Nature. 317:66–68. [DOI] [PubMed] [Google Scholar]
- Merlie, J.P., K.E. Isenberg, S.D. Russell, and J.R. Sanes. 1984. Denervation supersensitivity in skeletal muscle: analysis with a cloned cDNA probe. J. Cell Biol. 99:332–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill, R.A., L.A. Plum, M.E. Kaiser, and M. Clagett-Dame. 2002. A mammalian homolog of unc-53 is regulated by all-trans retinoic acid in neuroblastoma cells and embryos. Proc. Natl. Acad. Sci. USA. 99:3422–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miner, J.H., and J.R. Sanes. 1994. Collagen IV α3, α4, and α5 chains in rodent basal laminae: sequence, distribution, association with laminins, and developmental switches. J. Cell Biol. 127:879–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, T.B., J.J. Weis, and C.T. Wittwer. 1998. Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. Biotechniques. 24:954–958. [PubMed] [Google Scholar]
- Moscoso, L.M., G.C. Chu, M. Gautam, P.G. Noakes, J.P. Merlie, and J.R. Sanes. 1995. a. Synapse-associated expression of an acetylcholine receptor-inducing protein, ARIA/heregulin, and its putative receptors, ErbB2 and ErbB3, in developing mammalian muscle. Dev. Biol. 172:158–169. [DOI] [PubMed] [Google Scholar]
- Moscoso, L.M., J.P. Merlie, and J.R. Sanes. 1995. b. N-CAM, 43K-rapsyn, and S-laminin mRNAs are concentrated at synaptic sites in muscle fibers. Mol. Cell. Neurosci. 6:80–89. [DOI] [PubMed] [Google Scholar]
- Niehrs, C., and N. Pollet. 1999. Synexpression groups in eukaryotes. Nature. 402:483–487. [DOI] [PubMed] [Google Scholar]
- Paranavitane, V., W.J. Coadwell, A. Eguinoa, P.T. Hawkins, and L. Stephens. 2003. LL5beta is a phosphatidylinositol (3,4,5)-trisphosphate sensor that can bind the cytoskeletal adaptor, gamma-filamin. J. Biol. Chem. 278:1328–1335. [DOI] [PubMed] [Google Scholar]
- Patton, B.L., J.H. Miner, A.Y. Chiu, and J.R. Sanes. 1997. Distribution and function of laminins in the neuromuscular system of developing, adult, and mutant mice. J. Cell Biol. 139:1507–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters, P.J., A. Baker, I. Goris, G. Daneels, P. Verhasselt, W.H. Luyten, J.J. Geysen, S.U. Kass, and D.W. Moechars. 2004. Sensory deficits in mice hypomorphic for a mammalian homologue of unc-53. Brain Res. Dev. Brain Res. 150:89–101. [DOI] [PubMed] [Google Scholar]
- Peters, M.F., N.R. Kramarcy, R. Sealock, and S.C. Froehner. 1994. beta 2-Syntrophin: localization at the neuromuscular junction in skeletal muscle. Neuroreport. 5:1577–1580. [PubMed] [Google Scholar]
- Pumplin, D.W., and R.J. Bloch. 1983. Lipid domains of acetylcholine receptor clusters detected with saponin and filipin. J. Cell Biol. 97:1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes, J.R., and J.W. Lichtman. 1999. Development of the vertebrate neuromuscular junction. Annu. Rev. Neurosci. 22:389–442. [DOI] [PubMed] [Google Scholar]
- Sanes, J.R., and J.W. Lichtman. 2001. Induction, assembly, maturation and maintenance of a postsynaptic apparatus. Nat. Rev. Neurosci. 2:791–805. [DOI] [PubMed] [Google Scholar]
- Sanes, J.R., Y.R. Johnson, P.T. Kotzbauer, J. Mudd, T. Hanley, J.C. Martinou, and J.P. Merlie. 1991. Selective expression of an acetylcholine receptor-lacZ transgene in synaptic nuclei of adult muscle fibers. Development. 113:1181–1191. [DOI] [PubMed] [Google Scholar]
- Schaeffer, L., N. Duclert, M. Huchet-Dymanus, and J.P. Changeux. 1998. Implication of a multisubunit Ets-related transcription factor in synaptic expression of the nicotinic acetylcholine receptor. EMBO J. 17:3078–3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer, L., de Kerchove d'Exaerde, A., and J.P. Changeux. 2001. Targeting transcription to the neuromuscular synapse. Neuron. 31:15–22. [DOI] [PubMed] [Google Scholar]
- Scher, M.G., and R.J. Bloch. 1991. The lipid bilayer of acetylcholine receptor clusters of cultured rat myotubes is organized into morphologically distinct domains. Exp. Cell Res. 195:79–91. [DOI] [PubMed] [Google Scholar]
- Shadiack, A.M., and R.M. Nitkin. 1991. Agrin induces alpha-actinin, filamin, and vinculin to co-localize with AChR clusters on cultured chick myotubes. J. Neurobiol. 22:617–628. [DOI] [PubMed] [Google Scholar]
- Sharrocks, A.D. 2001. The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2:827–837. [DOI] [PubMed] [Google Scholar]
- Sheng, M., and M.J. Kim. 2002. Postsynaptic signaling and plasticity mechanisms. Science. 298:776–780. [DOI] [PubMed] [Google Scholar]
- Shewan, D., V. Calaora, P. Nielsen, J. Cohen, G. Rougon, and H. Moreau. 1996. mCD24, a glycoprotein transiently expressed by neurons, is an inhibitor of neurite outgrowth. J. Neurosci. 16:2624–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon, A.M., P. Hoppe, and S.J. Burden. 1992. Spatial restriction of AChR gene expression to subsynaptic nuclei. Development. 114:545–553. [DOI] [PubMed] [Google Scholar]
- Steward, O. 1983. Polyribosomes at the base of dendritic spines of central nervous system neurons—their possible role in synapse construction and modification. Cold Spring Harb. Symp. Quant. Biol. 48:745–759. [DOI] [PubMed] [Google Scholar]
- Steward, O., and E.M. Schuman. 2001. Protein synthesis at synaptic sites on dendrites. Annu. Rev. Neurosci. 24:299–325. [DOI] [PubMed] [Google Scholar]
- Stringham, E., N. Pujol, J. Vandekerckhove, and T. Bogaert. 2002. unc-53 controls longitudinal migration in C. elegans. Development. 129:3367–3379. [DOI] [PubMed] [Google Scholar]
- Tietjen, I., J.M. Rihel, Y. Cao, G. Koentges, L. Zakhary, and C. Dulac. 2003. Single-cell transcriptional analysis of neuronal progenitors. Neuron. 38:161–175. [DOI] [PubMed] [Google Scholar]
- Tusher, V.G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA. 98:5116–5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaittinen, S., R. Lukka, C. Sahlgren, J. Rantanen, T. Hurme, U. Lendahl, J.E. Eriksson, and H. Kalimo. 1999. Specific and innervation-regulated expression of the intermediate filament protein nestin at neuromuscular and myotendinous junctions in skeletal muscle. Am. J. Pathol. 154:591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela, D.M., T.N. Stitt, P.S. DiStefano, E. Rojas, K. Mattsson, D.L. Compton, L. Nunez, J.S. Park, J.L. Stark, D.R. Gies, et al. 1995. Receptor tyrosine kinase specific for the skeletal muscle lineage: expression in embryonic muscle, at the neuromuscular junction, and after injury. Neuron. 15:573–584. [DOI] [PubMed] [Google Scholar]
- Velleca, M.A., M.C. Wallace, and J.P. Merlie. 1994. A novel synapse-associated noncoding RNA. Mol. Cell. Biol. 14:7095–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston, C., B. Yee, E. Hod, and J. Prives. 2000. Agrin-induced acetylcholine receptor clustering is mediated by the small guanosine triphosphatases Rac and Cdc42. J. Cell Biol. 150:205–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson, D.G., and M.A. Nieto. 1993. Detection of messenger RNA by in situ hybridization to tissue sections and whole mounts. Methods Enzymol. 225:361–373. [DOI] [PubMed] [Google Scholar]
- Wilson, M.D., C. Riemer, D.W. Martindale, P. Schnupf, A.P. Boright, T.L. Cheung, D.M. Hardy, S. Schwartz, S.W. Scherer, L.C. Tsui, et al. 2001. Comparative analysis of the gene-dense ACHE/TFR2 region on human chromosome 7q22 with the orthologous region on mouse chromosome 5. Nucleic Acids Res. 29:1352–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittwer, C.T., M.G. Herrmann, A.A. Moss, and R.P. Rasmussen. 1997. Continuous fluorescence monitoring of rapid cycle DNA amplification. Biotechniques. 22:130–131, 134–138. [DOI] [PubMed]
- Yoshihara, C.M., and Z.W. Hall. 1993. Increased expression of the 43-kD protein disrupts acetylcholine receptor clustering in myotubes. J. Cell Biol. 122:169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, X., C. Lai, S. Thomas, and S.J. Burden. 1995. Neuregulin receptors, erbB3 and erbB4, are localized at neuromuscular synapses. EMBO J. 14:5842–5848. [DOI] [PMC free article] [PubMed] [Google Scholar]