Abstract
Accurate chromosome segregation depends on precise regulation of mitosis by the spindle checkpoint. This checkpoint monitors the status of kinetochore–microtubule attachment and delays the metaphase to anaphase transition until all kinetochores have formed stable bipolar connections to the mitotic spindle. Components of the spindle checkpoint include the mitotic arrest defective (MAD) genes MAD1–3, and the budding uninhibited by benzimidazole (BUB) genes BUB1 and BUB3. In animal cells, all known spindle checkpoint proteins are recruited to kinetochores during normal mitoses. In contrast, we show that whereas Saccharomyces cerevisiae Bub1p and Bub3p are bound to kinetochores early in mitosis as part of the normal cell cycle, Mad1p and Mad2p are kinetochore bound only in the presence of spindle damage or kinetochore lesions that interfere with chromosome–microtubule attachment. Moreover, although Mad1p and Mad2p perform essential mitotic functions during every division cycle in mammalian cells, they are required in budding yeast only when mitosis goes awry. We propose that differences in the behavior of spindle checkpoint proteins in animal cells and budding yeast result primarily from evolutionary divergence in spindle assembly pathways.
Keywords: chromosome segregation; kinetochores; mitotic spindle apparatus; mitosis; metaphase
Introduction
The spindle checkpoint ensures the fidelity of chromosome transmission by delaying anaphase until all chromatid pairs have formed proper links to the mitotic spindle. Sister chromatids attach to spindle microtubules (MTs) via kinetochores, which are multiprotein complexes that assemble on centromeric (CEN) DNA. During spindle assembly, a kinetochore must be captured by MTs emanating from one and only one pole of the mitotic spindle, whereas its partner must be captured by MTs emanating from the opposite pole. Sister pairs that have not formed bipolar attachments will not segregate correctly at anaphase. The presence of even a single kinetochore pair that has not achieved bipolar attachment is sufficient to engage the spindle checkpoint and arrest cell-cycle progression (Rieder et al., 1994; Li and Nicklas, 1995).
Spindle checkpoint genes were first identified in budding yeast and include the mitotic arrest defective (MAD) genes MAD1–3 (Li and Murray, 1991) and the budding uninhibited by benzimidazole (BUB) genes BUB1 and BUB3 (Hoyt et al., 1991), all of which are well conserved among eukaryotes. The Bub proteins are thought to be upstream components of the checkpoint pathway, whereas Mad2p and Mad3p (called BubR1 in animal cells) are downstream components that bind to and inhibit the regulatory protein Cdc20p (for review see Yu, 2002). At the metaphase to anaphase transition, Cdc20p activates the anaphase promoting complex, an E3 ubiquitin ligase, thereby promoting ubiquitination and degradation of the securin protein, Pds1p, and subsequent destruction of the cohesin complexes that tether sister chromatids together (for review see Morgan, 1999). Although the spindle checkpoint is not essential in budding yeast under normal growth conditions, it is essential in animal cells (Basu et al., 1999; Kitagawa and Rose, 1999; Dobles et al., 2000; Kalitsis et al., 2000).
Spindle checkpoint proteins have been shown to bind to kinetochores in animal cells and fission yeast (for review see Cleveland et al., 2003), and functional kinetochores are required to generate the checkpoint signal in both animal cells and budding yeast (Rieder et al., 1995; Gardner et al., 2001). However, the exact nature of the kinetochore lesions sensed by the spindle checkpoint remains uncertain. The first possibility is that it is the absence of tension across sister kinetochores that initiates checkpoint signaling (Stern and Murray, 2001), and the second is that it is a lack of MT attachment itself that is responsible (Rieder et al., 1995). Tension-based models are appealing because they link checkpoint silencing to an event that is absolutely dependent on bipolar attachment. However, in higher eukaryotes, tension stabilizes individual kinetochore–MT attachments (Nicklas and Ward, 1994; King and Nicklas, 2000) and disentangling the effects of tension and MT attachment on checkpoint signaling is difficult.
Determining the nature of the events that initiate and silence spindle checkpoint signaling should be less complicated in organisms such as budding yeast in which each kinetochore recruits a single MT. Budding yeast also has the advantage of temperature-sensitive mutants defective in specific steps of kinetochore–MT attachment. Such lesions include mutations in subunits of the Ndc80 complex that cause chromosomes to detach from MTs, mutations in the MT binding component DAM1 and the Aurora B kinase IPL1 that prevent chromosomes from forming bipolar attachments, and mutations in the MT regulator STU2 that allow chromosomes to form bipolar attachments but prevent them from establishing wild-type levels of tension (Biggins et al., 1999; Kim et al., 1999; He et al., 2001; Janke et al., 2001; Wigge and Kilmartin, 2001; Janke et al., 2002; Tanaka et al., 2002).
In budding yeast, only two known kinetochore complexes are required for spindle checkpoint function: CBF3 and Ndc80 (Gardner et al., 2001; McCleland et al., 2003). The CBF3 complex binds directly to CEN DNA and is required for the assembly of all known kinetochore components on CEN DNA (for review see McAinsh et al., 2003). In contrast, the Ndc80 complex is part of a set of “linker” proteins that do not bind directly to DNA or MTs but instead appear to link DNA- and MT-binding components. The Ndc80 complex consists of four essential proteins: Ndc80p, Nuf2p, Spc24p, and Spc25p. Among these, Ndc80p and Nuf2p are well conserved among eukaryotes (Wigge and Kilmartin, 2001) and human Ndc80 (Hec1) can functionally substitute for its yeast counterpart (Zheng et al., 1999). Although loss of function mutations in SPC24 or SPC25 disable the spindle checkpoint (Janke et al., 2001), mutations in NDC80 or NUF2 do not (McCleland et al., 2003). These and other data suggest that the Ndc80 complex may have an important role in relation to spindle checkpoint signaling.
In this paper, we report that four spindle checkpoint proteins—Bub1p, Bub3p, Mad1p, and Mad2p—associate with Saccharomyces cerevisiae kinetochores. Although Bub1p and Bub3p bind to kinetochores during normal mitoses, Mad1p and Mad2p are recruited only in the presence of spindle damage or checkpoint-activating kinetochore lesions. The kinetochore association of Bub1p and Mad2p requires the function of some, but not all, members of the Ndc80 complex. Our findings suggest that budding yeast kinetochores rarely, if ever, detach completely from MTs during normal cell division, and we propose that this aspect of spindle morphogenesis may explain why the checkpoint is not essential for mitosis in budding yeast under normal growth conditions. Our results also suggest that the release of the Bub proteins from kinetochores during normal spindle assembly is likely to be dependent on a transition from immature to mature kinetochore–MT attachment rather than on the establishment of tension across sisters.
Results
Bub1p and Bub3p are recruited to kinetochores during normal cell cycles
To localize spindle checkpoint proteins in S. cerevisiae, endogenous MAD and BUB genes were linked to GFP at their COOH termini via homologous recombination. GFP tagging did not interfere with checkpoint function, as assayed by growth on plates containing the MT-depolymerizing agent benomyl (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200308100/DC1). Spindle pole bodies (SPBs) were visualized by linking the SPB component Spc42p to CFP (Spc42p-CFP; Donaldson et al., 2001; He et al., 2001). Cells expressing GFP-tagged checkpoint proteins and CFP-tagged Spc42p were observed using two-wavelength three-dimensional (3D) deconvolution microscopy (Rines et al., 2002). Cell-cycle state was determined from the length and position of the mitotic spindle.
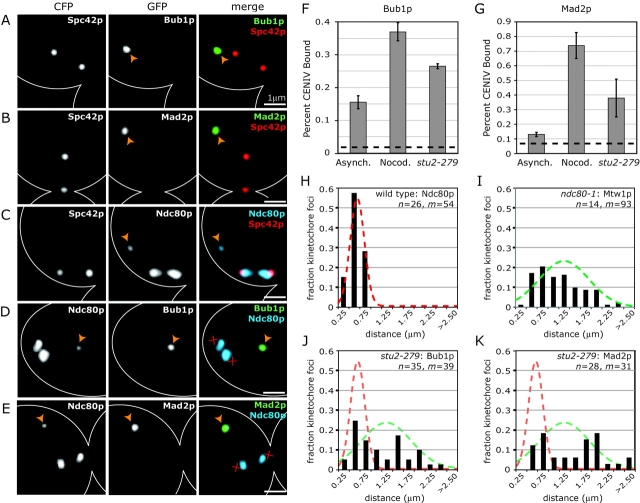
When Bub1p-GFP and Bub3p-GFP were examined in early mitotic cells, a distinct pattern of two GFP lobes lying between the CFP-tagged SPBs was observed (Fig. 1, A and B). This is the classic localization pattern of kinetochore proteins such as Ndc80p and reflects the metaphase clustering of budding yeast kinetochores into two lobes that lie along the spindle axis and between the spindle poles (Fig. 1 C; He et al., 2000). To demonstrate the kinetochore association of Bub1p-GFP and Bub3p-GFP directly, we performed chromatin immunoprecipitation (ChIP) with primers specific for CENIV DNA. In asynchronous cultures, both Bub1p-GFP and Bub3p-GFP exhibited clear CEN binding by ChIP (Fig. 1, D and E). Binding was specific as neither protein cross-linked to DNA at the non-CEN URA3 locus (unpublished data). Moreover, CEN binding required the core kinetochore complex CBF3, as Bub1p-GFP and Bub3p-GFP ChIP signals were negligible in ndc10-1 strains at 37°C (Fig. 1, D and E).
Figure 1.
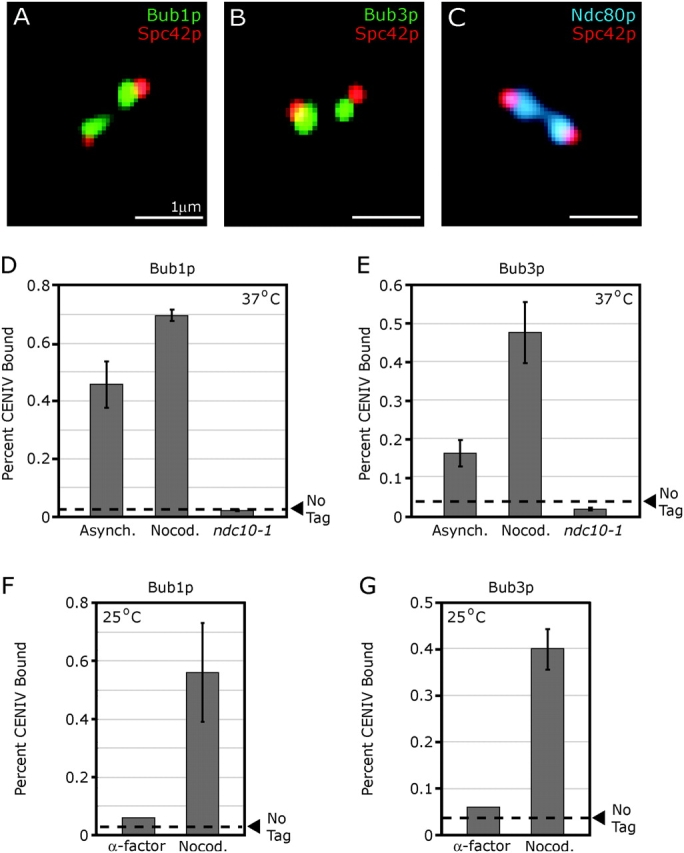
Bub1p-GFP and Bub3p-GFP are associated with kinetochores. (A–C) Typical images of wild-type mitotic cells expressing the SPB marker Spc42p-CFP (red) and the spindle checkpoint proteins Bub1p-GFP or Bub3p-GFP (green) or the kinetochore protein Ndc80p-GFP (blue). Images represent two-dimensional projections of 3D image stacks containing 10–15 0.2-μm sections. (D and E) ChIP of Bub1p-GFP and Bub3p-GFP at CENIV. Cross-linking of Bub proteins to CEN DNA was assayed in asynchronous wild-type cells, nocodazole-treated wild-type cells, and ndc10-1 cells at 37°C. All cells were grown to mid-log phase at 25°C, then shifted to 37°C for 3 h before analysis. The amount of CENIV DNA recovered with immune complexes is shown as a percentage of the amount of CENIV DNA present in each total cell lysate. Dashed lines represent the percentage of CENIV DNA recovered with immune complexes from wild-type cells (a negative control). Error bars show SD for two or more independent immunoprecipitations. Absolute differences in the amount of DNA precipitated among different panels are not considered to be meaningful. (F and G) ChIP of Bub proteins at CENIV is cell-cycle regulated. Wild-type cells expressing Bub1p-GFP or Bub3p-GFP were grown to mid-log phase at 25°C and treated with α-factor (5 μg/ml final) or nocodazole (25 μg/ml final) for 3 h before ChIP analysis.
When cells carrying Bub1p-GFP and Bub3p-GFP were treated with the anti-MT drug nocodazole to activate the spindle checkpoint, the ChIP signals for Bub1p and Bub3p at CENIV rose 1.5- and 3-fold, respectively, relative to untreated asynchronous cells (Fig. 1, D and E). In contrast, in α-factor arrested G1 cells, ChIP signals for Bub1p and Bub3p fell to background levels (Fig. 1, F and G). From these data, we conclude that Bub1p and Bub3p associate with CEN DNA during normal cell divisions, that this association requires functional kinetochores, and that it is cell-cycle regulated, being high in nocodazole-treated mitotic cells and low in G1. Our results with Bub1p in nocodazole-treated cells are consistent with those of Kitagawa et al. (2003) and Kerscher et al. (2003), but unlike Kitagawa, we conclude from imaging and ChIP that little to no Bub1p binds to kinetochores in α-factor–arrested cells.
Kinetochore association by Bub1p occurs early in mitosis
To determine when during the cell cycle Bub proteins are recruited to kinetochores, the localization of Bub1p-GFP was compared with that of the kinetochore protein Ndc80p-GFP. Parallel cell cultures were synchronized using α-factor, released at 25°C, and samples withdrawn and fixed every 15 min. The percentage of cells containing Bub1p-GFP or Ndc80p-GFP foci was determined by analyzing at least 40 individual cells at each time point. Progression through the cell cycle was monitored by examining bud size, spindle length, and spindle position (determined using Spc42p-CFP). In synchronous cultures released from α-factor, very few cells contained kinetochore-localized Bub1-GFP before T = 45 min (Fig. 2, A and C). Kinetochore binding by Bub1p, then rose dramatically, peaking at T = 60 min, and fell again as mitosis progressed (Fig. 2 C). In contrast, kinetochore binding by Ndc80p-GFP was apparent throughout the experiment, giving rise at early time points to a single GFP cluster in close proximity to the newly duplicated SPBs and subsequently resolving into a bi-lobed metaphase configuration (Fig. 2 B).
Figure 2.
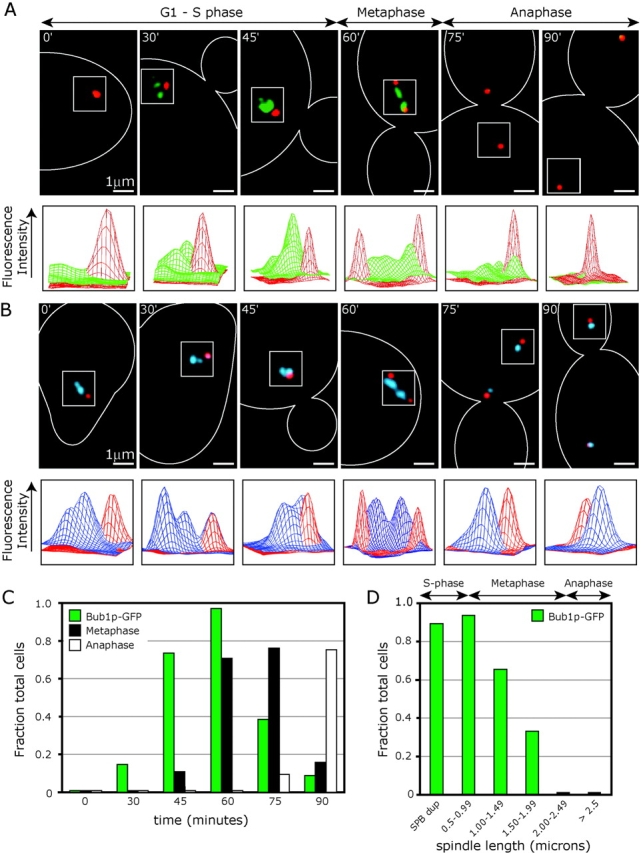
Kinetochore association of Bub proteins is cell-cycle regulated. (A) Wild-type cells expressing Bub1p-GFP (green) and the SPB component Spc42p-CFP (red). Images are representative for each time point after α-factor release at 25°C. The surface plot below each image depicts the distribution of GFP (green) and CFP (red) signal intensities (in arbitrary units) across the boxed regions of each image. For the 30-min time point, we included an image representative of the 15% of cells that contained Bub1p-GFP foci. (B) Images of individual cells expressing the kinetochore protein Ndc80p-GFP (blue) and Spc42-CFP (red). Images and graphs are as described for A. (C) Fraction of total cells containing Bub1p-GFP kinetochore foci, metaphase spindles, and anaphase spindles versus time after α-factor release at 25°C. Metaphase cells were those with spindle lengths between 0.8 and 2.2 μm and anaphase cells those with spindles >2.2 μm. At least 40 individual cells were scored at each time point. (D) Fraction of cells containing Bub1p-GFP kinetochore foci versus spindle length after α-factor release at 25°C (n = 281).
Bub1p-GFP foci were first visible around the time of SPB duplication (during S-phase, at T = 30–45 min; Fig. 2, C and D). At this point, the patterns of Bub1p-GFP and Ndc80p-GFP localization were very similar, suggesting that most if not all kinetochores were associated with Bub1p. The peak of Bub1p binding to kinetochores was observed at T = 60 min in cells with spindles that averaged 0.8 μm in length. Cells at this point in the cell cycle contain duplicated SPBs, but kinetochores do not yet exhibit a bi-lobed metaphase configuration (as judged by Ndc80p-GFP). At T = 75 min, 71% of cells contained metaphase-length spindles, but only 38% contained Bub1p-GFP foci (Fig. 2 C), indicating that Bub1p is released from kinetochores as metaphase proceeds. No Bub1p-GFP foci were seen in anaphase cells (Fig. 2 A, 75 and 90 min; Fig. 2 D). Bub1p was also absent from kinetochores arrested in metaphase by cdc23-1 or cdc20-1 mutations (unpublished data). Cells in asynchronous cultures exhibited a pattern of kinetochore association by Bub1p similar to that seen in synchronous cultures, showing that our findings were not an artifact of α-factor release. Moreover, the dynamics of Bub3p binding to kinetochores were indistinguishable from those of Bub1p-GFP (unpublished data). From these results, we conclude that the Bub proteins first associate with kinetochores during S-phase when cells contain monopolar spindles, but dissociate from kinetochores as mature bipolar MT attachments are established early in mitosis.
Kinetochore recruitment of the Mad checkpoint proteins
Next, we examined the kinetochore association of Mad1p, Mad2p, and Mad3p in asynchronous and nocodazole-treated cells. We detected little or no kinetochore-bound Mad1p, Mad2p, or Mad3p in asynchronous cells by imaging or ChIP at any stage of the cell cycle (Fig. 3 A, not depicted; Iouk et al., 2002). However, ChIP signals were high for both Mad1p-GFP and Mad2p-GFP in nocodazole-treated cells (Fig. 3 A). The ChIP signal for Mad3p-GFP was consistently just above background levels in nocodazole-treated cells (Fig. 3 A), but we have been unable to confirm kinetochore association by microscopy (not depicted). From these data we conclude that Mad1p, Mad2p, and Mad3p do not associate significantly with kinetochores in cycling cells but that Mad1p and Mad2p are kinetochore bound in the presence of spindle damage.
Figure 3.
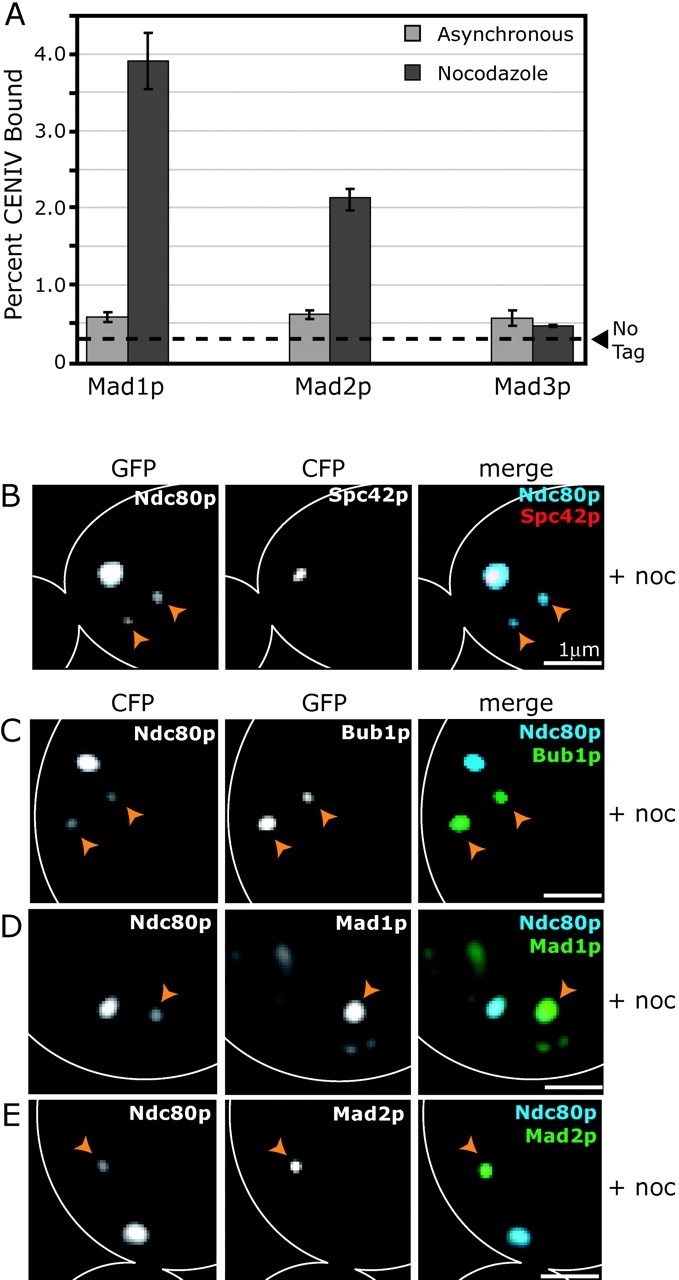
Kinetochore association of spindle checkpoint proteins in nocodazole-treated cells. (A) ChIP of Mad1p-GFP, Mad2p-GFP, and Mad3p-GFP at CENIV in cycling and nocodazole-treated cells. Graphs are as described for Fig. 1 (D–G). (B) Wild-type cell coexpressing the SPB protein Spc42p-CFP and the kinetochore protein Ndc80p-GFP after treatment with 25 μg/ml nocodazole for 1 h at 25°C. Panels show Ndc80p-GFP alone, Spc42p-CFP alone, and Spc42p-CFP (red) merged with Ndc80p-GFP (blue). (C–E) Wild-type cells coexpressing Ndc80p-CFP (blue) and Mad1p-GFP, Mad2p-GFP, or Bub1p-GFP (green) after nocodazole treatment. Panels show CFP channel alone, GFP channel alone, and CFP merged with GFP. Orange arrowheads indicate the locations of unattached kinetochores.
Nocodazole treatment interferes with MT polymerization and causes mitotic spindles to collapse (Jacobs et al., 1988). When we imaged nocodazole-treated cells coexpressing Ndc80p-GFP and Spc42p-CFP, we found that the majority of kinetochores remained in a large cluster close to the collapsed SPBs (Fig. 3 B). However, most cells also contained one or two dim Ndc80p kinetochore foci ≥1 μm away from the SPBs (Fig. 3 B, arrowheads). Data from live-cell chromosome tracking experiments in nocodazole-treated cells suggest that these dim Ndc80p foci represent kinetochores that are detached from spindle MTs (unpublished data). Foci of Mad1p-GFP, Mad2p-GFP, and Bub1p-GFP colocalized specifically with the weaker Ndc80p kinetochore foci that were distant from SPBs (Fig. 3, C–E). Some Mad1p-GFP also remained on the nuclear periphery (Fig. 3 D; Iouk et al., 2002). From these data, we conclude that treating cycling cells with nocodazole causes some, but not all, kinetochores to detach from spindle MTs and that spindle checkpoint proteins are recruited selectively to the detached kinetochores.
A functional checkpoint pathway is required for kinetochore recruitment of Mad1p and Mad2p
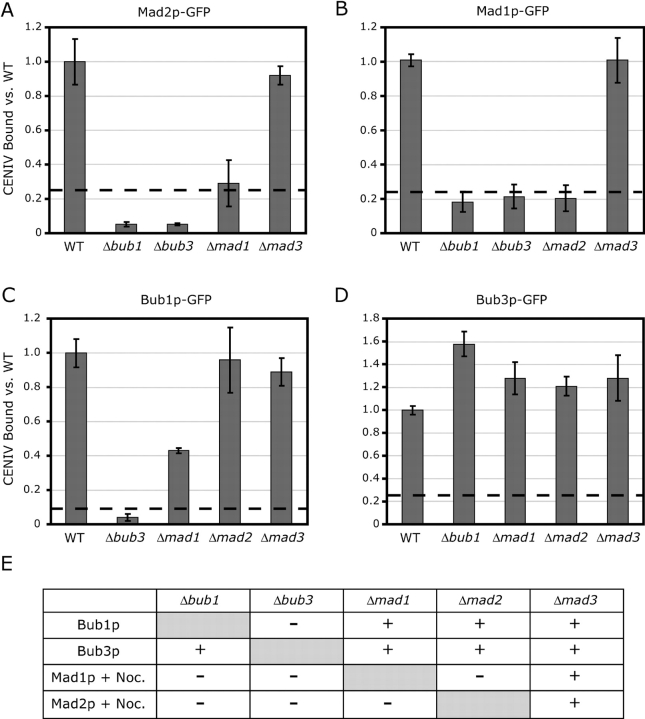
Epistasis analysis has suggested that Bub1p and Bub3p are upstream components of the checkpoint pathway, whereas Mad2p is a downstream effector (Farr and Hoyt, 1998). To determine if interdependencies for CEN binding by checkpoint proteins mirrored their proposed order in the checkpoint signaling pathway, ChIP of Mad1p, Mad2p, Bub1p, and Bub3p was performed in cells deleted for other checkpoint components. CEN association of Mad1p and Mad2p was assayed in cells treated with nocodazole, whereas that of Bub1p and Bub3p was assayed in asynchronous cells. We observed that CEN association by Mad2p-GFP was abolished in Δbub1 and Δbub3 cells, as well as in cells lacking MAD1, but not in cells lacking MAD3 (Fig. 4 A). CEN association by Mad1p-GFP exhibited a similar set of dependencies, requiring BUB1, BUB3, and MAD2, but not MAD3 (Fig. 4 B). In contrast, both Bub1p-GFP and Bub3p-GFP associated with CEN DNA in cells lacking MAD1, MAD2, or MAD3 (Fig. 4, C and D). Finally, although Bub1p-GFP did not bind to kinetochores in cells lacking BUB3, Bub3p-GFP could still be cross-linked to CEN DNA in Δbub1 cells (Fig. 4, C and D). In all but one case (Bub3p-GFP), results from imaging matched those from ChIP (Fig. 4 E). High levels of autofluorescence in Δbub1 cells may have masked Bub3p-GFP kinetochore signals. Overall, our data show that kinetochore binding by checkpoint components is dependent on the presence of proteins upstream in the signaling pathway: kinetochore binding by Mad1p and Mad2p requires BUB1 and BUB3 but not MAD3; Bub1p requires BUB3 but not the MAD genes; and Bub3p is independent of all other checkpoint proteins.
Figure 4.
Interdependencies of checkpoint proteins for kinetochore binding. (A) ChIP of Mad2p-GFP at CENIV in wild-type and checkpoint-delete cells in the presence of 25 μg/ml nocodazole. (B) ChIP of Mad1p-GFP at CENIV in wild-type and checkpoint-delete cells in the presence of nocodazole. (C and D) ChIP of Bub1p-GFP or Bub3p-GFP at CENIV in wild-type and checkpoint delete cells. ChIP signals from deletion strains were normalized to the ChIP signal obtained from the wild-type strain. Dashed lines show the amount of CEN DNA precipitated using untagged wild-type cells (negative control). (E) Summary of the interdependencies of checkpoint protein kinetochore binding as assayed by imaging. Mad1p-GFP and Mad2p-GFP were examined in the presence of nocodazole, whereas Bub1p-GFP and Bub3p-GFP were examined in asynchronous cells.
Bub1p and Mad2p bind to kinetochores in ndc80-1 cells but not in spc25-7 cells
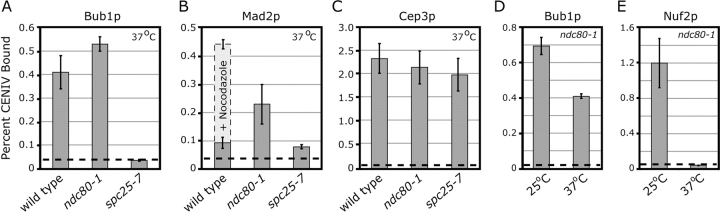
The structural proteins that recruit checkpoint components to kinetochores are unknown. The best candidates are those kinetochore components whose loss disables spindle checkpoint signaling. One such protein is Spc25p, a component of the Ndc80 complex. Kinetochores detach from spindle MTs in spc25-7 cells but the spindle checkpoint is not activated (He et al., 2001; Janke et al., 2001; Wigge and Kilmartin, 2001; McCleland et al., 2003). Consistent with this, neither Bub1p-GFP nor Mad2p-GFP is associated with CEN DNA in spc25-7 cells at 37°C (Fig. 5, A and B). In contrast, kinetochores also detach from spindle MTs in ndc80-1 cells at 37°C, but the checkpoint is engaged (Janke et al., 2001; McCleland et al., 2003) and we found that Bub1p-GFP and Mad2p-GFP are associated with CEN DNA in this mutant (Fig. 5, A and B). In a control experiment, we observed that CEN binding by the Cep3p component of CBF3 was equally high in wild-type, spc25-7, and ndc80-1 cells (Fig. 5 C).
Figure 5.
ChIP of Bub1p and Mad2p in Ndc80 complex mutants. (A–C) ChIP of (A) Bub1p-GFP, (B) Mad2-GFP (+/− nocodazole) and (C) Cep3p at CENIV in wild-type, ndc80-1, and spc25-7 cells at 37°C. ChIP of (D) Bub1p-GFP and (E) Nuf2p-myc in an ndc80-1 background at 25°C and 37°C. Graphs are as described in Fig. 1 D.
To confirm that the ndc80-1 mutant was effectively disrupting kinetochore structure under our experimental conditions, we performed ChIP experiments using ndc80-1 cells coexpressing Bub1p-GFP and myc-tagged Nuf2p, a protein known to require functional Ndc80p for CEN association (He et al., 2001). Although Bub1p-GFP and Nuf2p-myc could be cross-linked to CEN DNA in ndc80-1 cells at permissive temperature, only Bub1p-GFP remained CEN bound at 37°C (Fig. 5, D and E). From these results, we conclude that the association of Bub1p and Mad2p with unattached kinetochores in budding yeast is dependent on kinetochore components that assemble properly in ndc80-1 cells but not in spc25-7 cells. Differences between kinetochores in ndc80-1 and spc25-7 cells are likely to be quite subtle, and it is possible that Spc25p or other subunits of the Ndc80 complex may directly bind to Mad and Bub proteins.
Mad2p is recruited to kinetochores in dam1-1 but not ipl1-321 cells
The existence of kinetochore mutants with distinct effects on chromosome dynamics affords an opportunity to investigate which types of lesions recruit checkpoint proteins to kinetochores. In dam1-1 and ipl1-321 cells, kinetochores cannot form stable bipolar attachments to spindle MTs, sister chromatid pairs each remain associated with a single SPB, and chromosome congression fails (Biggins et al., 1999; Kim et al., 1999; He et al., 2001; Janke et al., 2002; Tanaka et al., 2002). Interestingly, although dam1-1 mutants engage the spindle checkpoint, ipl1-321 mutants do not (Biggins and Murray, 2001; Cheeseman et al., 2001; He et al., 2001; Jones et al., 2001; Janke et al., 2002). To determine whether checkpoint proteins are recruited to kinetochores in dam1-1 and ipl1-321 mutants, we examined the localization of Ndc80p-GFP, Bub1p-GFP, and Mad2p-GFP in mutant cells coexpressing the SPB marker, Spc42p-CFP. Although it has been reported previously that kinetochores preferentially associate with the old SPB when subunits of the Dam1 complex are inactivated (Janke et al., 2002), we find the asymmetric distribution of kinetochores in dam1-1 cells to be somewhat variable. In many cells, similar numbers of chromosomes were bound to each SPB (Fig. 6 A). In contrast, the asymmetric distribution of kinetochores in ipl1-321 cells was dramatic and consistent (Fig. 6 D). By imaging, we found that Bub1p-GFP was present on kinetochores at nonpermissive temperature in both dam1-1 and ipl1-321 cells (Fig. 6, B, E, G, and H). Although Mad2p-GFP appeared to be kinetochore bound in the majority of dam1-1 cells after 1 h at 37°C (Fig. 6, C and G), Mad2p-GFP was rarely detected on kinetochores in ipl1-321 cells at nonpermissive temperature (Fig. 6, F and H). ChIP analysis confirmed these findings (unpublished data).
Figure 6.
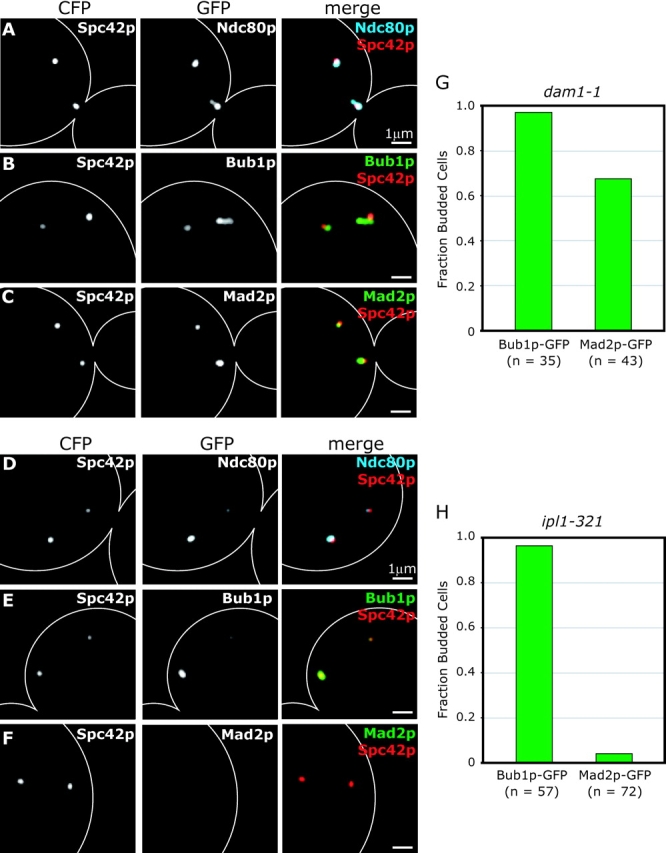
Bub1p and Mad2p localization in dam1-1 and ipl1-321 cells. (A–C) dam1-1 cells expressing the SPB protein Spc42p-CFP and Ndc80p-GFP, Bub1p-GFP or Mad2p-GFP at nonpermissive temperature. Cells were grown at 25°C to mid-log phase and shifted to 37°C for 1 h before fixation. Panels show Spc42p-CFP alone; Ndc80p-GFP, Bub1p-GFP, or Mad2p-GFP; and Spc42p-CFP (red) merged with Ndc80p-CFP (blue), Bub1p-GFP (green), or Mad2p-GFP (green). (D–F) ipl1-321 cells expressing Spc42p-CFP and Ndc80p-GFP, Bub1p-GFP, or Mad2p-GFP at nonpermissive temperature. Cells were arrested in α-factor for 2 h, shifted to 37°C for 10 min and released at 37°C for 2 h before fixation. (G and H) Fraction of budded cells containing Bub1p-GFP and Mad2p-GFP for dam1-1 cells after 1 h at 37°C and ipl1-321 cells after 2 h at 37°C after α-factor release. n = number of budded cells counted.
Why do dam1-1 kinetochores recruit Mad2p, whereas ipl1-321 kinetochores do not? One possibility is that Ipl1p is an upstream component of the checkpoint pathway required for the activity of Mad2p (Biggins and Murray, 2001). This is not strictly true, however, as Mad2p binding to CEN DNA could be detected by imaging and ChIP in ipl1-321 cells treated with nocodazole (unpublished data). A second possibility is that kinetochore–MT links in ipl1-321 cells prevent Mad2p binding. It has been proposed that Ipl1p plays an essential role in releasing syntelic attachments that form early in the cell cycle when both kinetochores in a pair of sister chromatids bind to MTs emanating from the same SPB (Tanaka et al., 2002). We speculate that yeast Mad2p is not recruited to kinetochores in ipl1-321 cells because they have syntelic MT attachments. In contrast, monotelic attachments (in which one kinetochore is attached, whereas its partner is unattached) likely predominate in dam1-1 cells, and Mad2p is therefore recruited to the unattached kinetochore. By this reasoning, the inability of ipl1-321 cells to engage the spindle checkpoint does not reflect a role for IPL1 in checkpoint signaling, but rather the failure of ipl1-321 cells to generate a kinetochore structure that the checkpoint can recognize.
Loss of tension is not sufficient to recruit Bub1p or Mad2p to kinetochores in stu2-279 cells
A major question in the study of mitosis is whether it is the absence of tension or the loss of MT attachment that is ultimately responsible for activating checkpoint signaling. Our data show that kinetochores that remain attached to collapsed spindles in nocodazole-treated cells do not recruit Mad and Bub proteins (Fig. 3, C–E). As it is mechanically impossible for collapsed spindles to impose tension on chromatids, these results suggest that loss of tension does not recruit high levels of Mad or Bub proteins to kinetochores. To determine if checkpoint proteins are kinetochore bound in cells in which tension has been eliminated by other means, we examined cells carrying mutations in the MT-associated protein Stu2p (He et al., 2001). stu2-279 cells arrest in a checkpoint-dependent fashion with kinetochores that have bipolar attachments but are not under detectable tension (He et al., 2001; Severin et al., 2001a). When stu2-279 cells coexpressing the SPB marker Spc42p-CFP and Ndc80p-GFP, Mad2p-GFP, or Bub1p-GFP were examined by imaging and ChIP at nonpermissive temperatures, one or two bright GFP foci were visible (Fig. 7, A and B) and both Mad2p and Bub1p were CEN associated by ChIP (Fig. 7, F and G). However, almost all Mad2p-GFP and Bub1p-GFP foci lay ≥1 μm from the spindle axis (Fig. 7, A and B), whereas the majority of kinetochores, as monitored by Ndc80p-GFP, lay between the SPBs (Fig. 7 C). In most cells, one or two dim Ndc80p-GFP foci were also visible ≥1 μm from the spindle axis (Fig. 7 C). The analysis of stu2-279 cells coexpressing Ndc80p-CFP and either Bub1p-GFP or Mad2p-GFP made it clear that the dim Ndc80p-CFP foci distant from the spindle axis were coincident with the bright Bub1p-GFP and Mad2p-GFP foci (Fig. 7, D and E). Thus, it appears that Bub1p and Mad2p are specifically recruited only to a subset of kinetochores in stu2-279 cells. Similar results were obtained with a stu2-277 mutant (unpublished data).
Figure 7.
Bub1p and Mad2p localization in stu2-279 cells. (A–C) stu2-279 cells coexpressing the SPB protein Spc42p-CFP and Bub1p-GFP, Mad2p-GFP, or Ndc80p-GFP. Panels show Spc42p-CFP alone; Bub1p-GFP, Mad2p-GFP, or Ndc80p-GFP alone; and Spc42p-CFP (red) merged with Bub1p-GFP (green), Mad2p-GFP (green), or Ndc80p-GFP (blue). Cells were grown at 25°C to mid-log phase and shifted to 37°C for 2 h before fixation. Orange arrowheads denote unattached kinetochores. (D and E) stu2-279 cells coexpressing the kinetochore protein Ndc80p-CFP and Bub1p-GFP or Mad2p-GFP. Panels show Ndc80p-CFP alone; Bub1p-GFP or Mad2p-GFP alone; and Ndc80p-CFP (blue) merged with Bub1p-GFP (green) or Mad2p-GFP (green). Images are as described in Fig. 1 A. Red X's denote the inferred positions of SPBs. (F and G) ChIP of Bub1p-GFP and Mad2p-GFP at CENIV in asynchronous wild-type cells, nocodazole-treated wild-type cells, and stu2-279 cells, all at 37°C. Graphs are as described in Fig. 1 (D–G). (H–K) Spatial distribution of kinetochore protein foci for (H) Ndc80p-GFP in wild-type cells with attached kinetochores, (I) Mtw1p-GFP in ndc80-1 cells with unattached kinetochores (at 37°C), and (J and K) Bub1p-GFP or Mad2p-GFP in stu2-279 cells (also at 37°C). Distances were measured from each GFP focus to the center of the spindle. Spindle orientation and length was determined using Spc42p-CFP. Only cells with spindles between 0.75 and 1.50 μm were included. Lines represent normal distributions for attached (red, μ = 0.40 μm; σ = 0.15) or unattached kinetochores (green, μ = 1.01 μm; σ = 0.52). The number of cells (n) and number of kinetochore foci (m) analyzed are listed on each graph.
What distinguishes kinetochores that recruit Bub1p and Mad2p in stu2 cells from those that do not? One possibility is that kinetochores that lie off of the spindle axis, and that bind to Bub1p and Mad2p, are not correctly attached to MTs. Although we had not anticipated that stu2 cells would contain unattached kinetochores, MTs are known to be fewer in number and less dynamic in stu2 mutants (Kosco et al., 2001) and it is likely that the spindle's ability to capture kinetochores and maintain kinetochore–MT attachments is compromised in these cells. Moreover, although we only detected attached chromosomes in our initial studies of stu2 cells (He et al., 2001), recent live-cell data indicate that a subset of kinetochores do detach from spindle MTs in stu2 mutants (unpublished data).
To better characterize the state of chromosome–MT attachment in stu2 cells, we profiled the spatial distributions of Bub1p and Mad2p foci within the nuclei of these cells and compared them to the spatial distribution of kinetochores known to be attached (as determined from the positions of Ndc80p-GFP foci in metaphase wild-type cells) and those known to be unattached (as determined from the positions of Mtw1p-GFP foci in ndc80-1 cells). In each case, spatial kinetochore distributions were profiled by measuring the distances from each GFP focus to the center of the spindle. Although attached kinetochores exhibited a narrow distribution with a mean of 0.4 μm (Fig. 7 H), unattached kinetochores showed a broad distribution with a mean of 1.0 μm and a maximum of 2.3 μm (Fig. 7 I). Importantly, the distribution of Bub1p-GFP and Mad2p-GFP foci in stu2-279 cells was very similar to that of unattached kinetochores, strongly suggesting that checkpoint proteins are recruited to kinetochores that have become detached from the spindle in stu2-279 cells (Fig. 7, J and K). We conclude that, in stu2 mutants, the majority of kinetochores are attached to MTs and lack detectable Bub1p and Mad2p, despite a lack of tension. However, a subset of kinetochores, perhaps one or two per cell, are not attached to MTs, and these kinetochores selectively recruit high levels of Bub1p and Mad2p.
Bub1p binds kinetochores in the absence of sister cohesion but Mad2p does not
Another method by which tension across kinetochores can be eliminated is by inactivating sister cohesion. A temperature-sensitive mcd1-1 cohesin mutant disables sister pairing and allows chromatids to segregate independently of one another (Guacci et al., 1997). Although mcd1-1 cells experience a slight checkpoint-dependent cell-cycle delay, they appear to undergo a morphologically normal anaphase (Biggins and Murray, 2001; Severin et al., 2001b). We were unable to detect Mad2p on kinetochores in mcd1-1 cells by ChIP or imaging (unpublished data), even though the cell-cycle delay in mcd1-1 cells is known to be MAD2 dependent. We cannot tell if this reflects an off-kinetochore function for Mad2p in response to lack of tension (Martin-Lluesma et al., 2002), or if Mad2p is present transiently at kinetochores below our limit of detection. However, it is clear that the lack of tension on kinetochores in mcd1-1 cells is not sufficient to recruit the high levels of Mad2p seen on unattached kinetochores.
A comparison of wild-type and mcd1-1 cells coexpressing Bub1p-GFP and Ndc80p-CFP revealed that Bub1p binding to kinetochores was very similar from 0 to 60 min after α-factor release (Fig. 8, A, B, and G). However, the dissociation of Bub1p from kinetochores was delayed ∼15 min relative to wild-type cells (Fig. 8 G). Interestingly, mcd1-1 cells with longer spindles almost always contained a heterogeneous population of Bub1p-positive and -negative kinetochores (Fig. 8 B, compare Ndc80p with Bub1p), suggesting that Bub1p binding is likely to depend on the attachment status of individual kinetochores. From these data, we conclude that Bub1p is recruited properly to kinetochores in mcd1-1 mutants early in mitosis and is then lost as mitosis progresses. Thus, bipolar attachment and tension are not absolutely required to release Bub1p from kinetochores. At this point, it is not clear if delayed release of Bub1p from kinetochores in mcd1-1 cells is a consequence of lack of tension per se, or rather of problems in establishing mature chromosome–MT attachments due to a lack of sister pairing.
Figure 8.
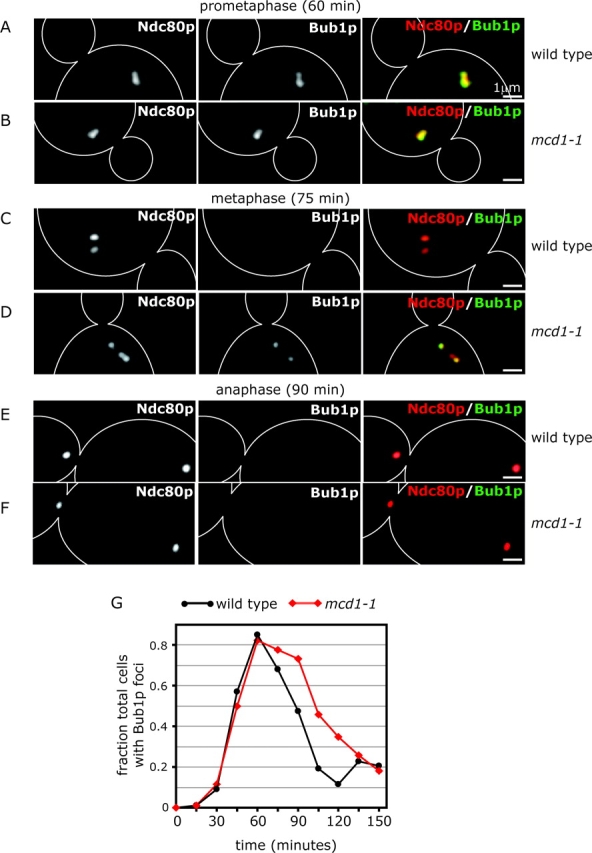
Bub1p localization in mcd1-1 cells. (A–F) Typical images of mcd1-1 and wild-type cells coexpressing Bub1p-GFP (green) and Ndc80p-CFP (red) at 60, 75, and 90 min after α-factor release at 37°C. (G) Fraction of mcd1-1 and wild-type cells containing Bub1p kinetochore foci at 15-min time points after α-factor release at 37°C. At least 60 individual cells were analyzed at each time point.
Discussion
In this paper, we show that spindle checkpoint proteins in S. cerevisiae are recruited to centromeres in a kinetochore-dependent manner, just as they are in animal cells. Despite the high degree of conservation in Mad and Bub proteins through evolution, however, our data also show that interactions between kinetochores and spindle checkpoint proteins in yeast and animal cells differ in several significant ways. Budding yeast Bub1p and Bub3p are like their mammalian counterparts in that they bind to kinetochores during normal cell division. This binding is cell-cycle regulated, being highest early in mitosis around the time of SPB duplication and falling as mitosis proceeds. In contrast, although mammalian Mad1 and Mad2 are bound to kinetochores during prometaphase in normally dividing cells, yeast Mad1p and Mad2p are kinetochore bound only in cells in which chromosome–MT attachment is inhibited. We propose that organism-specific differences in the behavior of spindle checkpoint proteins are likely to reflect evolutionary divergence in the mechanics of spindle assembly rather than extensive differences in the pathways of checkpoint signaling.
Several key features distinguish spindle assembly in animal cells and budding yeast. Animal cells undergo an open mitosis and prometaphase chromosomes are initially free of spindle MTs after nuclear envelope breakdown. High levels of Mad and Bub proteins are present on these unattached kinetochores, but Mad1 and Mad2, in particular, dissociate as chromosome–MT attachments form (Waters et al., 1998). In contrast, budding yeast cells undergo a closed mitosis in which kinetochores remain closely associated with SPBs throughout the cell cycle (Jin et al., 2000; unpublished data). Although we find Mad1p and Mad2p on unattached S. cerevisiae kinetochores in cells with spindle damage or kinetochore lesions, yeast kinetochores do not recruit high levels of these proteins during normal mitosis, which is consistent with the idea that yeast chromosomes are continuously linked to MTs. The maintenance of kinetochore–MT attachments throughout the yeast cell cycle may make spindle assembly more efficient, a property that could explain why yeast MAD2 is not required for normal cell growth (Li and Murray, 1991), whereas murine Mad2 is essential (Dobles et al., 2000). Interestingly, yeast Mad2p appears to be important for chromosome biorientation during the first meiotic division (Shonn et al., 2000, 2003), which implies that kinetochore binding by Mad2p might be a normal feature of meiosis. Therefore, it will be interesting to determine if Mad2p-positive chromosomes are generated during meiotic bouquet formation (Trelles-Sticken et al., 1999).
The Ndc80 complex and spindle checkpoint signaling
An important issue in the study of spindle checkpoint signaling is determining how spindle checkpoint proteins bind to kinetochores. The best candidates for proteins that link Mad and Bub proteins to kinetochores are those whose inactivation disrupts checkpoint signaling without completely disrupting kinetochore assembly. Although mutations in almost all known kinetochore components engage the checkpoint (Gardner et al., 2001), loss of function mutations in subunits of the CBF3 complex (which consists of Ndc10p, Cep3p, Ctf13p, and Skp1p) and some subunits of the Ndc80 complex (which consists of Spc24p, Spc25p, Ndc80p, and Nuf2p) have the special property of abolishing the checkpoint (Goh and Kilmartin, 1993; Gardner et al., 2001; Janke et al., 2001; McCleland et al., 2003). However, protein–protein and protein–DNA associations among kinetochore proteins are hierarchical; whereas loss of CBF3 function prevents all known kinetochore proteins from associating with CEN DNA (Goh and Kilmartin, 1993; He et al., 2001), loss of Ndc80 function disrupts the assembly of only a small subset of kinetochore components (He et al., 2001; Janke et al., 2001; De Wulf et al., 2003). It has been suggested that the CBF3 subunit, Skp1p, mediates the binding of Bub1p to kinetochores (Kitagawa et al., 2003), but our data show that the spc25-7 mutation prevents Bub1p and Mad2p from binding to kinetochores at nonpermissive temperature without altering the level of CEN-bound CBF3 (Fig. 5 C, as measured using the CBF3 component, Cep3p). This evidence strongly suggests that CBF3, and hence Skp1p, cannot be sufficient for the recruitment of Bub1p to kinetochores.
Mutant analysis suggests the link between checkpoint signaling and mutations in subunits of the Ndc80 complex is fairly complex: spc24-2 and spc25-7 mutants abrogate the checkpoint, whereas ndc80-1 and nuf2-457 mutants engage the checkpoint (He et al., 2001; Janke et al., 2001; Wigge and Kilmartin, 2001; McCleland et al., 2003). We have found that these functional differences are reflected in the extent to which Mad and Bub proteins are recruited to kinetochores. Gene and allele-specific differences among spc24, spc25, ndc80, and nuf2 mutations may be a simple consequence of differences in allelic strength: in the case of CBF3, Burke and colleagues have elegantly demonstrated that hypomorphic alleles engage the checkpoint, whereas complete loss of function mutations inactivate it (Doheny et al., 1993; Strunnikov et al., 1995; Connelly and Hieter, 1996; Tavormina and Burke, 1998; Gardner et al., 2001); and the results of McCleland et al. (2003) suggest that ndc80-1 may indeed be a hypomorphic allele. Alternatively, it is also possible that some subunits of the Ndc80 complex are required for the recruitment of Mad and Bub proteins to kinetochores, whereas other subunits are not. Either way, the requirement for a functional Ndc80 complex in checkpoint signaling and the evolutionary conservation of the Ndc80 complex (human Ndc80/HEC1 can functionally substitute for yeast NDC80; Zheng et al., 1999) are suggestive of important functional connections between the Ndc80 complex and the spindle checkpoint.
Attachment, tension, and the spindle checkpoint in budding yeast
Two main hypotheses exist regarding what features of kinetochore–MT attachment are monitored by the spindle checkpoint. The tension hypothesis posits that the checkpoint monitors tension across paired sister kinetochores (Stern and Murray, 2001), whereas the attachment hypothesis suggests that the checkpoint monitors the occupancy of kinetochore–MT attachment sites (Rieder et al., 1995). In budding yeast, Mad1p, Mad2p, Bub1p, and Bub3p are recruited to unattached kinetochores in ndc80-1 cells and to kinetochores with monopolar attachments in dam1-1 cells. However, in no context have we observed high levels of checkpoint proteins bound to kinetochores that have achieved bipolar attachment but lack tension. Although cells carrying a mutation in the kinetochore-associated MAP, Stu2p, contain attached tension-free kinetochores as well as unattached kinetochores, high levels of Bub1p and Mad2p are recruited only to the latter. Similarly, although a few kinetochores detach from spindle MTs in cells treated with the anti-MT drug nocodazole, the majority of kinetochores remain attached to very short MTs and in close proximity to the collapsed SPBs. Although the collapsed spindles in nocodazole-treated cells cannot generate tension across sister kinetochores, Bub1p and Mad2p are found only on unattached kinetochores. Finally, Mad2p is not detectable on kinetochores in mcd1-1 cells that lack sister cohesion and bipolar tension. Thus, the absence of tension on paired sister chromosomes is not sufficient to recruit high levels of Mad or Bub proteins to kinetochores. Overall, our data are most consistent with the attachment hypothesis, but it remains possible that lack of tension may cause the transient binding of Bub and Mad proteins to kinetochores at levels that are below our limit of detection.
Role of the Bub proteins during normal spindle assembly
High levels of Bub1p and Bub3p, but not Mad1p or Mad2p, are recruited to kinetochores during normal mitosis, suggesting that Bub1p and Bub3p play a role in spindle assembly that the Mad proteins do not share. Several additional pieces of evidence support this hypothesis. First, budding yeast cells deleted for BUB1 or BUB3 experience much more severe chromosome loss than do cells deleted for MAD1, MAD2, or MAD3 (Warren et al., 2002). Second, extra copies of BUB1 or BUB3 suppress the chromosome–MT attachment defects generated by tub1-729 mutant, independent of MAD2-dependent signaling (Abruzzi et al., 2002). Third, although the conserved kinase domain of Bub1p is not required for nocodazole arrest in yeast (Sharp-Baker and Chen, 2001; Warren et al., 2002) or the recruitment of downstream checkpoint proteins to kinetochores in Xenopus (Sharp-Baker and Chen, 2001; Warren et al., 2002), it is required for suppression of attachment defects in tub1-729 cells (Abruzzi et al., 2002) and for accurate chromosome transmission in wild-type cells (Warren et al., 2002).
We find selective binding of Bub proteins, but not Mad proteins, to kinetochores in three contexts: wild-type cells early in mitosis, ipl1-321 cells, and mcd1-1 cells. Early during spindle assembly, kinetochores are thought to form transient syntelic attachments in which both sister kinetochores are linked to the old SPB. Syntelic attachments resolve to bipolar attachments early in spindle assembly in wild-type cells, but persist in ipl1-321 cells (Tanaka et al., 2002). Although Bub1p is recruited to kinetochores with syntelic attachments in ipl1-321 cells, it is also recruited to kinetochores in mcd1-1 cells, which are necessarily unpaired and therefore unable to form syntelic attachments. What feature is common to ipl1-321 and mcd1-1 chromosome–MT attachments as well as to wild-type attachments early in the cell cycle? It is known that kinetochores in animal cells initially bind to the sides of MTs during spindle assembly (Merdes and De Mey, 1990), and MT binding assays have demonstrated that reconstituted budding yeast kinetochores form “lateral” attachments to the sides of MTs in vitro (Sorger et al., 1994). Therefore, we propose that Bub proteins are recruited in yeast to kinetochores that have attached to the sides rather than the ends of MTs, as well as to kinetochores that lack MT attachment altogether.
Summary
In summary, our analysis of spindle checkpoint proteins in budding yeast reinforces the idea that Bub1p and Bub3p have a role during spindle assembly that Mad1p and Mad2p do not share. Although the Bub proteins appear to respond to changes in chromosome–MT attachment that occur during the course of normal spindle assembly, Mad proteins respond primarily to chromosome–MT detachment, a condition that does not exist in normally growing yeast cells. Our data help to explain why the spindle checkpoint is nonessential in budding yeast as well as why deletions of BUB1 or BUB3 have more dramatic effects on cell growth and chromosome loss than do deletions of MAD1–3. More broadly, our findings support the hypothesis that it is changes in the state of chromosome–MT attachment rather than in tension across sister kinetochores that is responsible for recruiting checkpoint proteins to kinetochores and, presumably, for initiating checkpoint signaling.
Materials and methods
Yeast strains and manipulations
Strains were derived from W303 or S288C parental stocks. Proteins were tagged with GFP or CFP by linking a 300–800-bp COOH-terminal PCR gene fragment to the coding sequence for EGFP or ECFP in pRS306 or pRS304. Endogenous genes were replaced using one-step gene replacement and correct integrants were verified by PCR.
Microscopy analysis
Images of fixed cells carrying CFP and GFP fusion proteins were collected at RT using a fluorescence microscope (Deltavision with Nikon TE200 base), Plan Apo 100X/1.40 oil objective, and a camera (model CoolSnap HQ; Photometrics) with Chroma 86002 JP4 (CFP) and 41018 (GFP) filters. 3D image acquisition, deconvolution, and maximum intensity two-dimensional projections were done using softWoRx software. Fixed cells were treated with 2% formaldehyde for 5–10 min followed by 0.1 M phosphate buffer, pH 6.6, for at least 5 min and imaged at RT.
ChIP
ChIP was performed as described previously (Megee et al.,1999) except that cells were cross-linked with formaldehyde for 2 h at RT, lysed using glass beads in a Bio101 FastPrep FP120, sonicated until DNA was an average of 200–500 bp in length, and centrifuged to remove cellular debris. Immunoprecipitations were performed using anti-GFP (CLONTECH Laboratories, Inc.), anti-myc (Santa Cruz Biotechnology, Inc.), and anti-Cep3p (Sorger laboratory) antibodies. PCR amplifications of 200-bp fragments of URA3 and CENIV were performed on serial dilutions (to determine linearity) of two or more independent immunoprecipitations; error bars show SDs (Figs. 1, 3–5, and 7).
Online supplemental material
Benomyl sensitivity assays of strains expressing GFP-tagged proteins are shown in Fig. S1. A summary of kinetochore localization by spindle checkpoint proteins can be found in Table S1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200308100/DC1.
Acknowledgments
We thank A. Amon, G. Barnes, S. Biggins, J. Kilmartin, A. Murray, E. Schiebel, P. Silver, A. McAinsh, and P. De Wulf.
This work was supported by National Institutes of Health grants GM51464 and CA84179.
E.S. Gillett and C.W. Espelin contributed equally to this work.
The online version of this article contains supplemental material.
Abbreviations used in this paper: 3D, three-dimensional; BUB, budding uninhibited by benzimidazole; CEN, centromeric; ChIP, chromatin immunoprecipitation; MAD, mitotic arrest defective; MT, microtubule; SPB, spindle pole body.
References
- Abruzzi, K.C., M. Magendantz, and F. Solomon. 2002. An alpha-tubulin mutant demonstrates distinguishable functions among the spindle assembly checkpoint genes in Saccharomyces cerevisiae. Genetics. 161:983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu, J., H. Bousbaa, E. Logarinho, Z. Li, B.C. Williams, C. Lopes, C.E. Sunkel, and M.L. Goldberg. 1999. Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. J. Cell Biol. 146:13–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins, S., and A.W. Murray. 2001. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 15:3118–3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins, S., F.F. Severin, N. Bhalla, I. Sassoon, A.A. Hyman, and A.W. Murray. 1999. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 13:532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., M. Enquist-Newman, T. Muller-Reichert, D.G. Drubin, and G. Barnes. 2001. Mitotic spindle integrity and kinetochore function linked by the Duo1p–Dam1p complex. J. Cell Biol. 152:197–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland, D.W., Y. Mao, and K.F. Sullivan. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell. 112:407–421. [DOI] [PubMed] [Google Scholar]
- Connelly, C., and P. Hieter. 1996. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell. 86:275–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wulf, P., A.D. McAinsh, and P.K. Sorger. 2003. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 17:2902–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobles, M., V. Liberal, M.L. Scott, R. Benezra, and P.K. Sorger. 2000. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell. 101:635–645. [DOI] [PubMed] [Google Scholar]
- Doheny, K.F., P.K. Sorger, A.A. Hyman, S. Tugendreich, F. Spencer, and P. Hieter. 1993. Identification of essential components of the S. cerevisiae kinetochore. Cell. 73:761–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson, M.M., A.A. Tavares, H. Ohkura, P. Deak, and D.M. Glover. 2001. Metaphase arrest with centromere separation in polo mutants of Drosophila. J. Cell Biol. 153:663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr, K.A., and M.A. Hoyt. 1998. Bub1p kinase activates the Saccharomyces cerevisiae spindle assembly checkpoint. Mol. Cell. Biol. 18:2738–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner, R.D., A. Poddar, C. Yellman, P.A. Tavormina, M.C. Monteagudo, and D.J. Burke. 2001. The spindle checkpoint of the yeast Saccharomyces cerevisiae requires kinetochore function and maps to the CBF3 domain. Genetics. 157:1493–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh, P.Y., and J.V. Kilmartin. 1993. NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 121:503–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci, V., D. Koshland, and A. Strunnikov. 1997. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 91:47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X., S. Asthana, and P.K. Sorger. 2000. Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell. 101:763–775. [DOI] [PubMed] [Google Scholar]
- He, X., D.R. Rines, C.W. Espelin, and P.K. Sorger. 2001. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 106:195–206. [DOI] [PubMed] [Google Scholar]
- Hoyt, M.A., L. Totis, and B.T. Roberts. 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 66:507–517. [DOI] [PubMed] [Google Scholar]
- Iouk, T., O. Kerscher, R.J. Scott, M.A. Basrai, and R.W. Wozniak. 2002. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint. J. Cell Biol. 159:807–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs, C.W., A.E. Adams, P.J. Szaniszlo, and J.R. Pringle. 1988. Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 107:1409–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, C., J. Ortiz, J. Lechner, A. Shevchenko, M.M. Magiera, C. Schramm, and E. Schiebel. 2001. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 20:777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, C., J. Ortiz, T.U. Tanaka, J. Lechner, and E. Schiebel. 2002. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 21:181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Q.W., J. Fuchs, and J. Loidl. 2000. Centromere clustering is a major determinant of yeast interphase nuclear organization. J. Cell Sci. 113:1903–1912. [DOI] [PubMed] [Google Scholar]
- Jones, M.H., X. He, T.H. Giddings, and M. Winey. 2001. Yeast Dam1p has a role at the kinetochore in assembly of the mitotic spindle. Proc. Natl. Acad. Sci. USA. 98:13675–13680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalitsis, P., E. Earle, K.J. Fowler, and K.H. Choo. 2000. Bub3 gene disruption in mice reveals essential mitotic spindle checkpoint function during early embryogenesis. Genes Dev. 14:2277–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerscher, O., L.B. Crotti, and M.A. Basrai. 2003. Recognizing chromosomes in trouble: association of the spindle checkpoint protein Bub3p with altered kinetochores and a unique defective centromere. Mol. Cell. Biol. 23:6406–6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.H., J.S. Kang, and C.S. Chan. 1999. Sli15 associates with the ipl1 protein kinase to promote proper chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 145:1381–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King, J.M., and R.B. Nicklas. 2000. Tension on chromosomes increases the number of kinetochore microtubules but only within limits. J. Cell Sci. 113:3815–3823. [DOI] [PubMed] [Google Scholar]
- Kitagawa, K., R. Abdulle, P.K. Bansal, G. Cagney, S. Fields, and P. Hieter. 2003. Requirement of skp1-bub1 interaction for kinetochore-mediated activation of the spindle checkpoint. Mol. Cell. 11:1201–1213. [DOI] [PubMed] [Google Scholar]
- Kitagawa, R., and A.M. Rose. 1999. Components of the spindle-assembly checkpoint are essential in Caenorhabditis elegans. Nat. Cell Biol. 1:514–521. [DOI] [PubMed] [Google Scholar]
- Kosco, K.A., C.G. Pearson, P.S. Maddox, P.J. Wang, I.R. Adams, E.D. Salmon, K. Bloom, and T.C. Huffaker. 2001. Control of microtubule dynamics by Stu2p is essential for spindle orientation and metaphase chromosome alignment in yeast. Mol. Biol. Cell. 12:2870–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, R., and A.W. Murray. 1991. Feedback control of mitosis in budding yeast. Cell. 66:519–531. (published erratum in Cell. 1994. 79:following 388). [DOI] [PubMed] [Google Scholar]
- Li, X., and R.B. Nicklas. 1995. Mitotic forces control a cell-cycle checkpoint. Nature. 373:630–632. [DOI] [PubMed] [Google Scholar]
- Martin-Lluesma, S., V.M. Stucke, and E.A. Nigg. 2002. Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science. 297:2267–2270. [DOI] [PubMed] [Google Scholar]
- McAinsh, A.D., J.D. Tytell, and P.K. Sorger. 2003. Structure, function, and regulation of budding yeast kinetochores. Annu. Rev. Cell Dev. Biol. 19:519–539. [DOI] [PubMed] [Google Scholar]
- McCleland, M.L., R.D. Gardner, M.J. Kallio, J.R. Daum, G.J. Gorbsky, D.J. Burke, and P.T. Stukenberg. 2003. The highly conserved Ndc80 complex is required for kinetochore assembly, chromosome congression, and spindle checkpoint activity. Genes Dev. 17:101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megee, P.C., C. Mistrot, V. Guacci, and D. Koshland. 1999. The centromeric sister chromatid cohesion site directs Mcd1p binding to adjacent sequences. Mol. Cell. 4:445–450. [DOI] [PubMed] [Google Scholar]
- Merdes, A., and J. De Mey. 1990. The mechanism of kinetochore-spindle attachment and polewards movement analyzed in PtK2 cells at the prophase-prometaphase transition. Eur. J. Cell Biol. 53:313–325. [PubMed] [Google Scholar]
- Morgan, D.O. 1999. Regulation of the APC and the exit from mitosis. Nat. Cell Biol. 1:E47–E53. [DOI] [PubMed] [Google Scholar]
- Nicklas, R.B., and S.C. Ward. 1994. Elements of error correction in mitosis: microtubule capture, release, and tension. J. Cell Biol. 126:1241–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, C.L., A. Schultz, R. Cole, and G. Sluder. 1994. Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J. Cell Biol. 127:1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder, C.L., R.W. Cole, A. Khodjakov, and G. Sluder. 1995. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J. Cell Biol. 130:941–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rines, D.R., X. He, and P.K. Sorger. 2002. Quantitative microscopy of green fluorescent protein-labeled yeast. Methods Enzymol. 351:16–34. [DOI] [PubMed] [Google Scholar]
- Severin, F., B. Habermann, T. Huffaker, and T. Hyman. 2001. a. Stu2 promotes mitotic spindle elongation in anaphase. J. Cell Biol. 153:435–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin, F., A.A. Hyman, and S. Piatti. 2001. b. Correct spindle elongation at the metaphase/anaphase transition is an APC-dependent event in budding yeast. J. Cell Biol. 155:711–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp-Baker, H., and R.H. Chen. 2001. Spindle checkpoint protein Bub1 is required for kinetochore localization of Mad1, Mad2, Bub3, and CENP-E, independently of its kinase activity. J. Cell Biol. 153:1239–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shonn, M.A., R. McCarroll, and A.W. Murray. 2000. Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science. 289:300–303. [DOI] [PubMed] [Google Scholar]
- Shonn, M.A., A.L. Murray, and A.W. Murray. 2003. Spindle checkpoint component Mad2 contributes to biorientation of homologous chromosomes. Curr. Biol. 13:1979–1984. [DOI] [PubMed] [Google Scholar]
- Sorger, P.K., F.F. Severin, and A.A. Hyman. 1994. Factors required for the binding of reassembled yeast kinetochores to microtubules in vitro. J. Cell Biol. 127:995–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, B.M., and A.W. Murray. 2001. Lack of tension at kinetochores activates the spindle checkpoint in budding yeast. Curr. Biol. 11:1462–1467. [DOI] [PubMed] [Google Scholar]
- Strunnikov, A.V., J. Kingsbury, and D. Koshland. 1995. CEP3 encodes a centromere protein of Saccharomyces cerevisiae. J. Cell Biol. 128:749–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, T.U., N. Rachidi, C. Janke, G. Pereira, M. Galova, E. Schiebel, M.J. Stark, and K. Nasmyth. 2002. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 108:317–329. [DOI] [PubMed] [Google Scholar]
- Tavormina, P.A., and D.J. Burke. 1998. Cell cycle arrest in cdc20 mutants of Saccharomyces cerevisiae is independent of Ndc10p and kinetochore function but requires a subset of spindle checkpoint genes. Genetics. 148:1701–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trelles-Sticken, E., J. Loidl, and H. Scherthan. 1999. Bouquet formation in budding yeast: initiation of recombination is not required for meiotic telomere clustering. J. Cell Sci. 112:651–658. [DOI] [PubMed] [Google Scholar]
- Warren, C.D., D.M. Brady, R.C. Johnston, J.S. Hanna, K.G. Hardwick, and F.A. Spencer. 2002. Distinct chromosome segregation roles for spindle checkpoint proteins. Mol. Biol. Cell. 13:3029–3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters, J.C., R.H. Chen, A.W. Murray, and E.D. Salmon. 1998. Localization of Mad2 to kinetochores depends on microtubule attachment, not tension. J. Cell Biol. 141:1181–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigge, P.A., and J.V. Kilmartin. 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152:349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H. 2002. Regulation of APC-Cdc20 by the spindle checkpoint. Curr. Opin. Cell Biol. 14:706–714. [DOI] [PubMed] [Google Scholar]
- Zheng, L., Y. Chen, and W.H. Lee. 1999. Hec1p, an evolutionarily conserved coiled-coil protein, modulates chromosome segregation through interaction with SMC proteins. Mol. Cell. Biol. 19:5417–5428. [DOI] [PMC free article] [PubMed] [Google Scholar]