Abstract
Cytoplasmic localization of mRNAs is a widespread mechanism for generating cell polarity and can provide the basis for patterning during embryonic development. A prominent example of this is localization of maternal mRNAs in Xenopus oocytes, a process requiring recognition of essential RNA sequences by protein components of the localization machinery. However, it is not yet clear how and when such protein factors associate with localized RNAs to carry out RNA transport. To trace the RNA–protein interactions that mediate RNA localization, we analyzed RNP complexes from the nucleus and cytoplasm. We find that an early step in the localization pathway is recognition of localized RNAs by specific RNA-binding proteins in the nucleus. After transport into the cytoplasm, the RNP complex is remodeled and additional transport factors are recruited. These results suggest that cytoplasmic RNA localization initiates in the nucleus and that binding of specific RNA-binding proteins in the nucleus may act to target RNAs to their appropriate destinations in the cytoplasm.
Keywords: RNA-binding protein; Vg1; VegT; Xenopus; oocyte
Introduction
One means by which regional protein diversification arises within a cell is through the process of cytoplasmic RNA localization (for review see Kloc et al., 2002), and an ever-increasing number of localized mRNAs have been identified in both somatic cells and germ cells (for review see Jansen, 2001; Palacios and St. Johnston, 2001). Localized RNAs function to generate cell polarity in a wide array of cell types, with examples ranging from yeast, where localization of ASH1 mRNA to the tip of budding yeast cells regulates mating type switching (Long et al., 1997; Takizawa et al., 1997), to motile fibroblasts and neuronal growth cones, where localization of β-actin mRNA regulates cell motility and morphology (Kislauskis et al., 1994; Shestakova et al., 2001; Zhang et al., 2001). Asymmetric localization of maternal mRNAs during oogenesis can provide the basis for polarity and patterning during embryogenesis. In Drosophila melanogaster, localized mRNAs underlie both anterior–posterior and dorsal–ventral patterning, and developmental polarity along the animal–vegetal axis in Xenopus laevis can be traced to RNA localization during oogenesis (for review see Palacios and St. Johnston, 2001). Although this process has emerged as a widespread mechanism for generating both somatic and embryonic cellular polarity, the molecular pathway responsible for cytoplasmic RNA localization is not yet defined.
During Xenopus oogenesis, maternal mRNAs localized along the animal–vegetal axis can influence patterning later during embryogenesis (for review see Mowry and Cote, 1999). The vegetal hemisphere is a repository of developmental information; both mesodermal and endodermal determinants reside there (for review see Chan and Etkin, 2001). Indeed, one mRNA localized to the vegetal hemisphere is VegT, a T-box transcription factor (Lustig et al., 1996; Stennard et al., 1996; Zhang and King, 1996; Horb and Thomsen, 1997) required for endoderm and mesoderm specification (Zhang et al., 1998). Also localized to the vegetal hemisphere is Vg1 mRNA, a member of the TGF-β growth factor superfamily (Weeks and Melton, 1987) that has been implicated in mesoderm and endoderm specification (Dale et al., 1993; Thomsen and Melton, 1993; Kessler and Melton, 1995; Joseph and Melton, 1998). Misexpression of either Vg1 or VegT in the animal hemisphere leads to induction of mesoderm in cells that would normally form ectoderm (Dale et al., 1993; Thomsen and Melton, 1993; Zhang and King, 1996), underscoring the importance of regulating the localization of these RNAs.
Vegetal localization of Vg1 and VegT RNAs is directed by localization elements (LEs) contained within their 3′ untranslated regions (Mowry and Melton, 1992; Bubunenko et al., 2002; Kwon et al., 2002). Within the LEs, repeated sequence elements are critical for proper function (Deshler et al., 1997; Gautreau et al., 1997; Betley et al., 2002; Bubunenko et al., 2002; Kwon et al., 2002; Lewis et al., 2004). Two such elements, VM1 (YYUCU; Gautreau et al., 1997; Cote et al., 1999; Lewis et al., 2004) and E2 (A/U,YCAC; Deshler et al., 1997, 1998), function as binding sites for specific RNA-binding proteins. Clustering of VM1 and E2 sites within LEs appears to be critical for function (Betley et al., 2002; Bubunenko et al., 2002; Kwon et al., 2002; Lewis et al., 2004), perhaps by facilitating interactions between protein components of the localization machinery. Identified protein components include a set of RNA-binding proteins that interact directly with the Vg1 LE (Schwartz et al., 1992; Mowry, 1996; Deshler et al., 1997, 1998; Havin et al., 1998; Cote et al., 1999; Zhao et al., 2001; Kroll et al., 2002). Two of these RNA-binding proteins, hnRNP I (VgRBP60; Cote et al., 1999; Lewis et al., 2004) and Vg1RBP/vera (Deshler et al., 1997, 1998; Havin et al., 1998), bind to VM1 and E2 sites, respectively. Roles in vegetal localization for these proteins were revealed through mutational analyses in which base changes in VM1 or E2 sites both disrupted protein binding in vitro and abolished localization of the RNA in vivo (Deshler et al., 1997, 1998; Cote et al., 1999; Lewis et al., 2004). Both hnRNP I and Vg1RBP/vera colocalize with Vg1 RNA at the vegetal cortex (Cote et al., 1999; Zhang et al., 1999), as do two additional RNA-binding proteins implicated with key roles in vegetal RNA localization, Prrp (Zhao et al., 2001) and XStau (Yoon and Mowry, 2004). Although these proteins bind to and colocalize with Vg1 RNA, when and where they assemble onto Vg1 RNA in vivo is still not known.
Cytoplasmic RNA localization relies on interactions between cis-acting sequences and multiple trans-acting factors, and it has been hypothesized to occur in the context of an RNP complex (Mowry, 1996; Ross et al., 1997; Arn et al., 2003). Indeed, in some instances, large RNP granules have been visualized during RNA transport (Barbarese et al., 1995; Bertrand et al., 1998; Rook et al., 2000; Krichevsky and Kosik, 2001; Wilkie and Davis, 2001). Formation of a localization-specific RNP is arguably an early or initiating event in the localization pathway and, until recently, it has been assumed that assembly of the transport RNP occurs in the cytoplasm. More recent findings have hinted that the process could instead initiate in the nucleus (for review see Farina and Singer, 2002). For example, a growing number of trans-acting localization factors have been identified as hnRNP or otherwise predominantly nuclear proteins. This list includes mammalian hnRNP A2 (Hoek et al., 1998), Drosophila squid (Norvell et al., 1999), Xenopus hnRNP I (Cote et al., 1999), yeast Loc1p (Long et al., 2001), and vertebrate ZBP2/KSRP (Gu et al., 2002). In addition to their roles in cytoplasmic RNA localization, many of these proteins have functions in nuclear events in RNA biogenesis such as splicing and nuclear export (Patton et al., 1991; Matunis et al., 1992; Mayeda et al., 1994; Min et al., 1997; Bilodeau et al., 2001). Intriguingly, a potential link between splicing and mRNA localization has been uncovered through analyses of mRNA localization in Drosophila (for review see Palacios, 2002). In motile fibroblasts, ZBP1 is required for localization of β-actin mRNA to the leading edge (Kislauskis et al., 1994; Ross et al., 1997), and colocalizes with both nuclear and cytoplasmic β-actin transcripts (Oleynikov and Singer, 2003). Although these analyses suggest a link between nuclear factors and cytoplasmic localization, they do not provide insight into the biochemical interactions underlying potential roles for nuclear factors in RNA transport.
Here, we show that hnRNP I and Vg1RBP/vera bind to Vg1 and VegT RNAs in both the nucleus and the cytoplasm, providing biochemical evidence that RNA localization initiates in the nucleus rather than in the cytoplasm. Upon export from the nucleus, the core RNP complex is remodeled and additional factors, including Prrp and XStau, are recruited. These results define distinct nuclear and cytoplasmic steps in the localization pathway, and suggest that the binding of specific RNA-binding proteins in the nucleus can direct an RNA to its final destination in the cytoplasm.
Results
hnRNP I protein is distributed in a pattern similar to Vg1 RNA
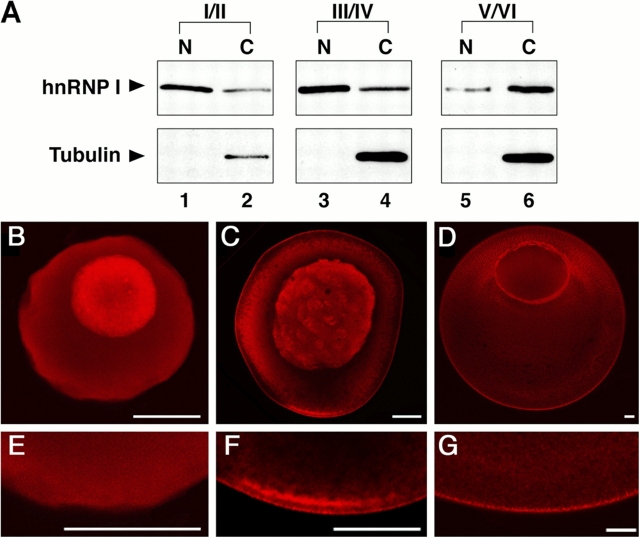
To probe the role of hnRNP I in the vegetal RNA localization pathway, we generated antibodies directed against Xenopus hnRNP I and analyzed the subcellular distribution of the protein at each stage of oogenesis. Western blot analysis of oocyte nuclear and cytoplasmic lysates revealed that hnRNP I is present in both the nucleus and cytoplasm at all stages of oogenesis (Fig. 1 A, top). In stages I/II and III/IV, the protein is more abundant in the nucleus than in the cytoplasm (Fig. 1 A, lanes 1 and 3 vs. lanes 2 and 4), whereas in stage V/VI the majority of the protein is in the cytoplasm (Fig. 1 A, lane 5 vs. lane 6). To confirm that the isolated nuclei were not contaminated by cytoplasm, the lysates were also immunoblotted with anti-tubulin (Fig. 1 A, bottom). Immunofluorescence using anti-hnRNP I also demonstrated the presence of hnRNP I in both the nucleus and cytoplasm at all stages of oogenesis (Fig. 1, B–G). Intriguingly, the cytoplasmic localization pattern of the protein mirrors that determined for Vg1 RNA during oogenesis (Melton, 1987). In early oogenesis (stage I/II), hnRNP I is evenly distributed throughout the oocyte cytoplasm (Fig. 1, B and E), as is Vg1 RNA (Melton, 1987). During mid-oogenesis (stage III/IV) and coincident with the vegetal localization of Vg1 RNA (Melton, 1987), cytoplasmic hnRNP I is localized to the vegetal hemisphere of the oocyte (Fig. 1, C and F). At the end of oogenesis (stage V/VI), hnRNP I is tightly associated with the vegetal cortex (Fig. 1, D and G). The presence of hnRNP I in the nucleus is consistent with nuclear functions that have been characterized in mammalian cells, including roles in alternative splicing (for review see Valcarcel and Gebauer, 1997). However, the decrease in nuclear abundance of hnRNP I in late stages of Xenopus oogenesis, along with its known role in vegetal RNA localization (Cote et al., 1999; Bubunenko et al., 2002; Lewis et al., 2004), raises an important question: does hnRNP I first bind Vg1 and VegT RNAs in the nucleus to direct localization in the cytoplasm?
Figure 1.
Subcellular distribution of hnRNP I during oogenesis. (A) Immunoblot analysis of nuclear (lanes 1, 3, and 5) and cytoplasmic (lanes 2, 4, and 6) lysates from stage I/II (lanes 1 and 2), stage III/IV (lanes 3 and 4), and stage V/VI (lanes 5 and 6) oocytes using anti-hnRNP I (top) and anti-tubulin (bottom). (B–G) Whole-mount immunofluorescence using anti-hnRNP I to detect distribution of hnRNP I in stage I/II (B and E), stage III/IV (C and F), and stage V/VI (D and G) oocytes. E, F, and G are higher magnification images of the vegetal pole of oocytes from B, C, and D, respectively. Confocal images were obtained on a microscope (LSM 410; Carl Zeiss MicroImaging, Inc.). Bars, 50 μm.
hnRNP I and Vg1RBP/vera bind vegetally localized RNAs in the nucleus and cytoplasm
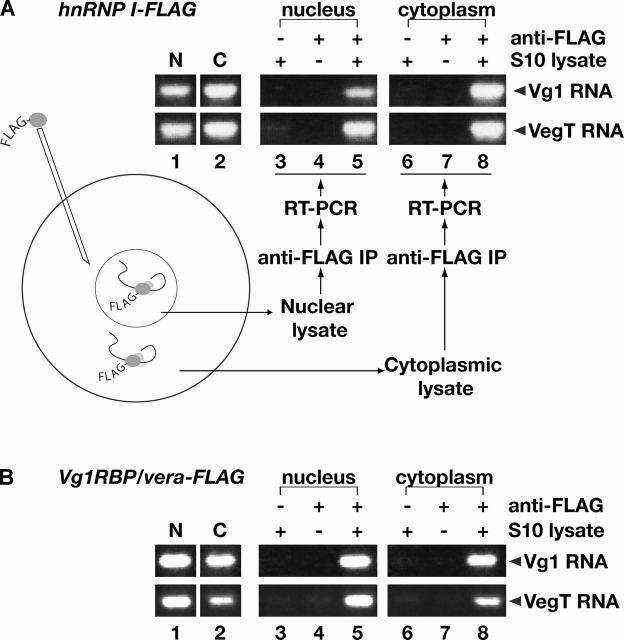
To determine whether hnRNP I associates with vegetal RNAs in the nucleus, we manually isolated nuclear and cytoplasmic fractions and prepared S10 lysates. First, we used RT-PCR to detect endogenous Vg1 and VegT RNA in the nuclear and cytoplasmic S10 lysates and found that both RNAs are present in the nucleus and cytoplasm (Fig. 2, lanes 1 and 2). To test whether hnRNP I binds to these RNAs in the nucleus, a FLAG-tagged version of hnRNP I was expressed in stage III/IV oocytes (Fig. 2 A). Nuclear and cytoplasmic S10 lysates were immunoprecipitated with anti-FLAG antibodies to capture complexes containing tagged hnRNP I, and bound RNAs were detected by RT-PCR (Fig. 2 A, lanes 3–8). Vg1 and VegT RNAs are associated with hnRNP I in both the nucleus (Fig. 2 A, lane 5) and the cytoplasm (lane 8), suggesting that hnRNP I first binds Vg1 and VegT RNAs in the nucleus.
Figure 2.
hnRNP I and Vg1RBP/vera associate with Vg1 and VegT RNAs in the nucleus and cytoplasm. (A) Stage III/IV oocytes were injected with RNA encoding hnRNP I-FLAG, and S10 lysates were prepared from manually isolated nuclei and cytoplasm. Vg1 (top) or VegT (bottom) RNAs in the nucleus (lane 1) or cytoplasm (lane 2) were detected using RT-PCR. Immunoprecipitations were performed using S10 lysates with Sepharose beads (lanes 3 and 6), anti-FLAG beads in the absence of lysate (lanes 4 and 7), and anti-FLAG beads in the presence of nuclear S10 (lane 5) or cytoplasmic S10 (lane 8) lysate. Vg1 and VegT RNAs were detected in each immunoprecipitate by RT-PCR. All samples were run on the same gel, but lane order was changed for presentation in the figure. (B) Stage III/IV oocytes were injected with RNA encoding Vg1RBP/vera-FLAG and S10 lysates were prepared as in A. Vg1 (top) or VegT (bottom) RNAs in the nucleus (lane 1) or cytoplasm (lane 2) were detected by RT-PCR. Immunoprecipitations were performed using S10 lysates with Sepharose beads (lanes 3 and 6), anti-FLAG beads in the absence of lysate (lanes 4 and 7), and anti-FLAG beads in the presence of nuclear (lane 5) or cytoplasmic (lane 8) S10 lysate. Vg1 RNA and VegT RNA were detected in each immunoprecipitate by RT-PCR; all samples were separated on the same gel, but lane order was changed for presentation in the figure.
Like hnRNP I, Vg1RBP/vera specifically binds to both the Vg1 and VegT LEs in an interaction that is critical for proper vegetal RNA localization (Schwartz et al., 1992; Deshler et al., 1997, 1998; Havin et al., 1998; Bubunenko et al., 2002; Kwon et al., 2002). To test whether Vg1RBP/vera, like hnRNP I, could first associate with vegetal RNAs in the nucleus, we expressed FLAG-tagged Vg1RBP/vera in stage III/IV oocytes. Complexes containing Vg1RBP/vera-FLAG were immunoprecipitated from nuclear and cytoplasmic lysates with anti-FLAG antibodies, and were assayed for bound RNA by RT-PCR. As shown in Fig. 2 B, Vg1RBP/vera interacts with Vg1 and VegT RNAs in both the nucleus and the cytoplasm (lanes 5 and 8). These data suggest that Vg1RBP/vera and hnRNP I bind vegetally localized RNAs in the nucleus and may form a core localization RNP.
hnRNP I and Vg1RBP/vera interact both in vitro and in vivo
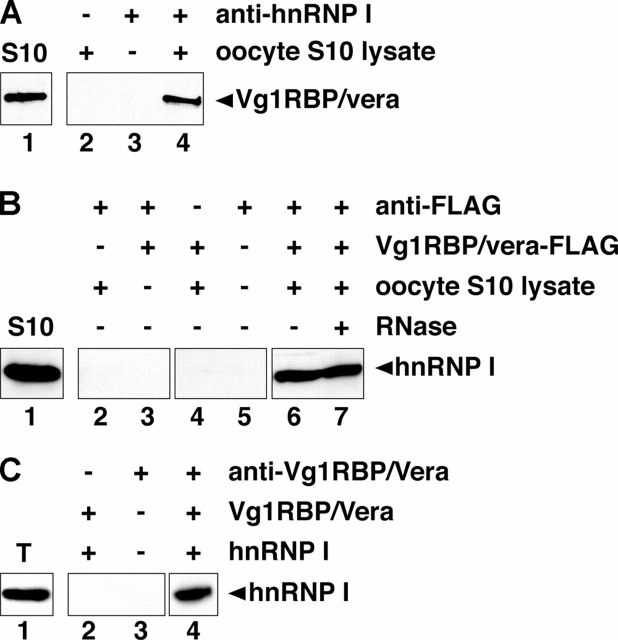
Interaction between Vg1RBP/vera and hnRNP I may be important for vegetal RNA transport; both proteins are required for localization of Vg1 and VegT RNAs (Deshler et al., 1997, 1998; Havin et al., 1998; Cote et al., 1999; Bubunenko et al., 2002; Kwon et al., 2002), they interact with these RNAs in the nucleus (Fig. 2), and their binding sites are clustered in close proximity (Bubunenko et al., 2002; Lewis et al., 2004). To test whether Vg1RBP/vera and hnRNP I can interact with one another, we immunoprecipitated hnRNP I from S10 lysates and tested for the presence of Vg1RBP/vera by immunoblot (Fig. 3 A). Indeed, Vg1RBP/vera is associated with hnRNP I (Fig. 3 A, lane 4). To further test this interaction, FLAG-tagged Vg1RBP/vera was translated in vitro and incubated with oocyte S10 lysate. After immunoprecipitation with anti-FLAG antibodies, immunoblot analysis with anti-hnRNP I showed that Vg1RBP/vera complexes contain hnRNP I (Fig. 3 B, lane 6). To determine whether the Vg1RBP/vera–hnRNP I interaction relied on binding to the same target RNA, binding reactions were treated with RNase A before immunoprecipitation. The interaction was insensitive to RNase in vitro (Fig. 3 B, lane 7), suggesting the possibility of a direct interaction between the two proteins. To test this, in vitro–translated Vg1RBP/vera and hnRNP I were mixed and were immunoprecipitated with anti-Vg1RBP/vera, and bound hnRNP I was detected by immunoblotting with anti-hnRNP I. Indeed, the two in vitro–translated proteins can interact in the absence of any other Xenopus proteins or mRNA (Fig. 3 C, lane 4). These results indicate that hnRNP I and Vg1RBP/vera are capable of interacting with one another in vitro, perhaps directly.
Figure 3.
Vg1RBP/vera and hnRNP I interact in vitro and in vivo. (A) Anti-hnRNP I was used to immunoprecipitate endogenous hnRNP I from oocyte S10 lysate. Bound Vg1RBP/vera was detected by immunoblotting (lane 4). Shown in lane 1 is the total Vg1RBP/vera in the lysate, and immunoprecipitation with protein G beads with or without lysate is shown in lanes 2 and 3, respectively. (B) In vitro–translated Vg1RBP/vera-FLAG was incubated with oocyte S10 lysate. Immunoprecipitations were performed using anti-FLAG beads with S10 lysate (lane 2), anti-FLAG beads with in vitro–translated Vg1RBP/vera-FLAG (lane 3), Sepharose beads with S10 lysate plus in vitro–translated Vg1RBP/vera-FLAG (lane 4), anti-FLAG beads alone (lane 5), and anti-FLAG beads with S10 lysate plus in vitro–translated Vg1RBP/vera-FLAG in the absence (lane 6) or presence (lane 7) of RNase A. Total (lane 1) and bound (lanes 2–7) proteins were separated by 10% SDS-PAGE and immunoblotted with anti-hnRNP I. (C) Recombinant Vg1RBP/vera and hnRNP I were translated in vitro and mixed; shown in lane 1 is the input amount of in vitro–translated hnRNP I. Immunoprecipitation reactions were performed using protein G beads in the presence of recombinant Vg1RBP/vera and hnRNP I (lane 2) and anti-Vg1RBP/vera bound to protein G beads either alone (lane 3) or with recombinant Vg1RBP/vera and hnRNP I (lane 4). Total (lane 1) and bound (lanes 2–4) proteins were separated by 10% SDS-PAGE, and were immunoblotted with anti-hnRNP I. For each panel (A–C), samples were run on the same gel, but lane order was changed for presentation in the figure; adjacent lanes are boxed.
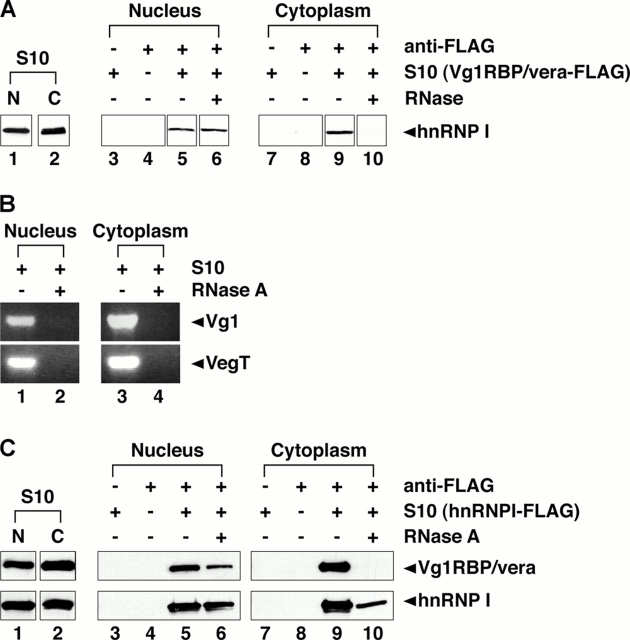
Next, we tested whether Vg1RBP/vera and hnRNP I interact in the nucleus or in the cytoplasm in vivo. FLAG-tagged Vg1RBP/vera was expressed in stage III/IV oocytes, and nuclear and cytoplasmic fractions were isolated. After immunoprecipitation with anti-FLAG antibodies, the presence of hnRNP I was assessed by immunoblot. As shown in Fig. 4 A, hnRNP I binds Vg1RBP/vera in both the nucleus and cytoplasm (lanes 5 and 9); however, the nature of the interaction is distinct between the compartments. When complexes were subjected to treatment with RNase A before immunoprecipitation, we observed that nuclear complexes are not sensitive to RNase A (Fig. 4 A, lane 6), whereas the cytoplasmic complexes are sensitive (Fig. 4 A, lane 10). Efficacy of the RNase treatment was assessed by RT-PCR for known targets of hnRNP I and Vg1RBP/vera, Vg1 and VegT (Fig. 4 B); both RNAs are undetectable in oocyte S10 lysates treated with RNase A (Fig. 4 B, lanes 2 and 4). Interaction between hnRNP I and Vg1RBP/vera was further confirmed in both the nucleus and cytoplasm in oocytes expressing hnRNP I-FLAG (Fig. 4 C, top). Vg1RBP/vera binds hnRNP I in both the nucleus (Fig. 4 C, lane 5) and the cytoplasm (Fig. 4 C, lane 9), and the interaction in the cytoplasm is sensitive to RNase A (Fig. 4 C, lane 10), whereas the interaction in the nucleus is not (Fig. 4 C, lane 6). As expected, immunoprecipitation of hnRNP I-FLAG by anti-FLAG is unaffected by treatment with RNase A (Fig. 4 C, bottom). These data suggest that Vg1RBP/vera and hnRNP I may interact directly in the nucleus; indeed, the in vitro interaction between the two proteins supports this possibility (Fig. 3 C). The Vg1RBP/vera–hnRNP I association is altered upon export to the cytoplasm, as the cytoplasmic interaction is dependent on RNA binding. These results suggest a remodeling of the RNP complex, raising the question of whether other factors are recruited to the Vg1 RNP in the cytoplasm.
Figure 4.
Vg1RBP/vera and hnRNP I interact in both the nucleus and cytoplasm. (A) Vg1RBP/vera-FLAG was expressed in stage III/IV oocytes, and nuclear (lanes 1 and 3–6) or cytoplasmic (lanes 2 and 7–10) lysates were prepared. Immunoprecipitations were performed using Sepharose beads plus Vg1RBP/vera S10 lysates (lanes 3 and 7), anti-FLAG beads alone (lanes 4 and 8), anti-FLAG beads with Vg1RBP/vera-FLAG, and nuclear (lanes 5 and 6) or cytoplasmic (lanes 9 and 10) S10 lysate in the presence (lanes 6 and 10) or absence (lanes 5 and 9) of RNase A. Total (lanes 1 and 2) and bound (lanes 3–10) proteins were separated by 10% SDS-PAGE and immunoblotted with anti-hnRNP I. All samples were run on the same gel, but lane order was changed for presentation in the figure, with adjacent lanes boxed together. (B) Nuclear (lanes 1 and 2) and cytoplasmic (lanes 3 and 4) S10 lysates were prepared and were subjected to digestion with RNase A (lanes 2 and 4) or mock digestion (lanes 1 and 3). Vg1 (top) and VegT (bottom) RNAs present in the lysates were detected by RT-PCR. All samples were run on the same gel; adjacent lanes are grouped together. (C) Nuclear (lanes 1 and 3–6) and cytoplasmic (lanes 2 and 7–10) lysates were prepared from oocytes expressing hnRNP I-FLAG. Immunoprecipitations were performed using Sepharose beads plus lysate (lanes 3 and 7), anti-FLAG beads alone (lanes 4 and 8), and anti-FLAG beads and nuclear (lanes 5 and 6) or cytoplasmic (lanes 9 and 10) S10 lysate either with (lanes 6 and 10) or without (lanes 5 and 9) RNase A treatment. Total (lanes 1 and 2) and bound (lanes 3–10) proteins were separated by SDS-PAGE. Bound Vg1RBP/vera was detected by immunoblot with anti-Vg1RBP/vera (top panels), and immunoprecipitated hnRNP I was detected by immunoblot with anti-hnRNP I (bottom panels). All samples were run on the same gel, but lane order was changed for presentation in the figure, with adjacent lanes boxed together.
Prrp associates with vegetal RNAs, hnRNP I, and Vg1RBP/vera in the cytoplasm
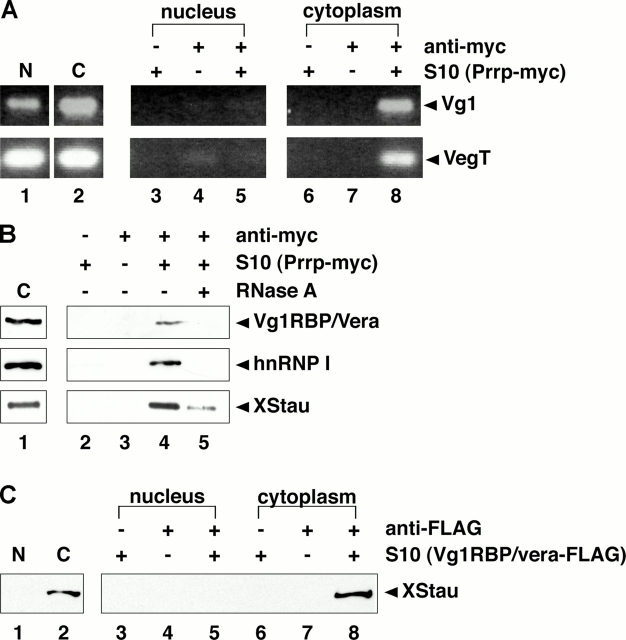
Prrp, which binds to the Vg1 LE and colocalizes with the RNA in the cytoplasm (Zhao et al., 2001), has been suggested to associate with factors involved in actin polymerization. Interestingly, anchoring of Vg1 RNA to the vegetal cortex is dependent upon an intact microfilament network (Yisraeli et al., 1990), raising the possibility that Prrp could be a component of the vegetal RNA localization RNP during the later steps in the pathway. In such a model, Prrp might be expected to associate with the vegetal RNP in the cytoplasm rather than the nucleus. To test this, we first determined when and where Prrp binds to vegetal RNAs. Myc-tagged Prrp was expressed in stage III/IV oocytes, and nuclear and cytoplasmic fractions were isolated. RT-PCR was used to confirm the presence of Vg1 and VegT RNAs in the lysates (Fig. 5 A, lanes 1 and 2). Nuclear or cytoplasmic lysates were immunoprecipitated with anti-myc followed by RT-PCR to detect vegetal RNAs present in Prrp-containing complexes. As shown in Fig. 5 A, Prrp binds to Vg1 and VegT RNAs in the cytoplasm (lane 8), but not in the nucleus (lane 5).
Figure 5.
Prrp and XStau interact with the localization RNP in the cytoplasm. (A) Stage III/IV oocytes were injected with RNA encoding Prrp-myc, and S10 lysates were prepared from isolated nuclear (lane 1 and lanes 3–5) and cytoplasmic (lane 2 and lanes 6–8) fractions. Immunoprecipitations were performed using control antibody plus S10 lysates (lanes 3 and 6), anti-myc in the absence of lysate (lanes 4 and 7), and anti-myc in the presence of nuclear (lane 5) or cytoplasmic (lane 8) lysates. Total (lanes 1 and 2) and immunoprecipitated (lanes 3–8) Vg1 (top) and VegT (bottom) RNAs were detected by RT-PCR. All samples were run on the same gel, but lane order was changed for presentation in the figure. (B) After expression of Prrp-myc in stage III/IV oocytes, immunoprecipitations were performed using control antibody with Prrp-myc cytoplasmic S10 lysate (lane 2), anti-myc alone (lane 3), and anti-myc plus lysate without (lane 4) or with (lane 5) RNase A treatment. Total (lane 1) and bound proteins (lanes 2–5) were separated by 10% SDS-PAGE and were immunoblotted for Vg1RBP/vera (top), hnRNP I (middle), or XStau (bottom). For each panel all samples were run on the same gel, and adjacent lanes are boxed together. (C) Vg1RBP/vera-FLAG was expressed in stage III/IV oocytes and S10 lysates were prepared from isolated nuclear (lanes 1 and 3–5) and cytoplasmic (lanes 2 and 6–8) fractions. Immunoprecipitations were performed using Sepharose beads plus Vg1RBP/vera S10 lysates (lanes 3 and 6), anti-FLAG beads alone (lanes 4 and 7), and anti-FLAG beads in the presence of nuclear (lane 5) or cytoplasmic (lane 8) lysates. Total (lanes 1 and 2) and bound (lanes 3–8) proteins were separated by 10% SDS-PAGE and immunoblotted for XStau. All samples were run on the same gel, with adjacent lanes boxed together for presentation in the figure.
To address whether Prrp and Vg1RBP/vera or hnRNP I are present in the same vegetal RNP complex, cytoplasmic Prrp-myc–containing complexes were immunoprecipitated with anti-myc and assayed by immunoblot to detect the association of either Vg1RBP/vera or hnRNP I. Both Vg1RBP/vera and hnRNP I were present in cytoplasmic complexes containing Prrp (Fig. 5 B, lane 4). However, the Prrp–Vg1RBP/vera and Prrp–hnRNP I interactions are indirect and are likely to depend upon binding to the same RNA target, as they are abolished by treatment with RNase A (Fig. 5 B, lane 5). Association of either protein with Prrp was not detected in nuclear lysates (unpublished data). Thus, it is likely that Prrp associates with the Vg1 and VegT RNPs after export from the nucleus of the core RNP that contains hnRNP I and Vg1RBP/vera.
XStau associates with the vegetal RNA localization RNP in the cytoplasm
Recent results have implicated an additional RNA-binding protein, XStau, with a role in Vg1 RNA transport (Yoon and Mowry, 2004). To ask where XStau associates with the vegetal localization RNP, we probed interactions with Prrp (Fig. 5 B) and Vg1RBP/vera (Fig. 5 C). In cytoplasmic fractions prepared from oocytes expressing Prrp-myc, XStau was found in Prrp-containing complexes (Fig. 5 B, lane 4). The interaction between Prrp and XStau appears indirect, as the association was disrupted by RNase A treatment (Fig. 5 B, lane 5). To test whether XStau can associate with the core localization RNP in the nucleus, Vg1RBP/vera-FLAG was expressed in oocytes, and nuclear and cytoplasmic fractions were isolated. As shown in Fig. 5 C, Vg1RBP/vera-FLAG complexes contained XStau only in the cytoplasm (lane 8), and not in the nucleus (lane 5). A similar interaction was observed between XStau and hnRNP I (unpublished data). Together, these data suggest that Prrp, Vg1RBP/vera, hnRNP I, and XStau are present in a cytoplasmic RNP containing vegetally localized RNAs.
Discussion
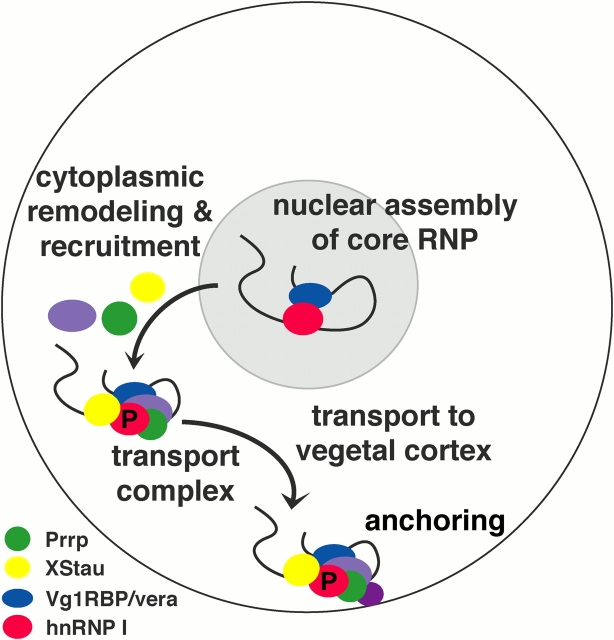
We have analyzed the Vg1 and VegT RNP complexes in both the nucleus and cytoplasm of Xenopus oocytes, and found that an early step in the localization pathway involves recognition of the localized RNAs by RNA-binding proteins in the nucleus. Our data support a model in which a core localization RNP is initially assembled in the nucleus, where factors such as hnRNP I and Vg1RBP/vera associate with the RNA (Fig. 6). Upon transport into the cytoplasm, the RNP complex is remodeled and additional transport factors are recruited, including XStau and Prrp. The association of XStau with the cytoplasmic vegetal RNP, combined with recent evidence that XStau interacts with both Vg1 RNA and a molecular motor (Yoon and Mowry, 2004), suggests that the vegetal localization RNP is actively transported along microtubules to the vegetal cortex during the later stages of localization. Ultimately, the vegetal RNP is anchored to the vegetal cortex, perhaps due to interactions between Prrp (Zhao et al., 2001) and proteins that could promote local remodeling of the cortical cytoskeleton.
Figure 6.
Model for the vegetal RNA localization pathway. Vegetal RNA localization begins in the nucleus (gray) where hnRNP I (red) and Vg1RBP/vera (blue) bind to the RNA, forming a core nuclear RNP. Upon export to the cytoplasm, the RNP complex is remodeled such that hnRNP I and Vg1RBP/vera no longer directly interact. In the cytoplasm, Prrp (green) and XStau (yellow) associate with the RNP, creating the cytoplasmic localization RNP. In this model, a motor protein promotes transport to the vegetal cortex, where anchoring could occur via an interaction of Prrp with factors that promote actin dynamics.
Both hnRNP I and Vg1RBP/vera have been shown to be critical for cytoplasmic localization of Vg1 and VegT RNA (Deshler et al., 1997, 1998; Havin et al., 1998; Cote et al., 1999; Bubunenko et al., 2002; Kwon et al., 2002; Lewis et al., 2004), but interactions with these RNAs in the nucleus were not revealed. We have demonstrated that both hnRNP I and Vg1RBP/vera associate with Vg1 and VegT RNAs in the nucleus (Fig. 2), suggesting that these proteins are recruited early in the transport pathway, during assembly of a nuclear, core RNP complex (Fig. 6). Both hnRNP I and Vg1RBP/vera have been implicated with nuclear functions in other cell types. In mammalian cells, hnRNPI/PTB functions in various aspects of nuclear RNA biogenesis, including splicing and intranuclear transport (for review see Valcarcel and Gebauer, 1997), and shuttles between the nucleus and the cytoplasm (Michael et al., 1995b; Kamath et al., 2001; Li and Yen, 2002; Xie et al., 2003). Vg1RBP/vera is an orthologue of ZBP1, which was identified through its role in cytoplasmic localization of β-actin RNA in chick fibroblasts (Ross et al., 1997), and has recently been shown to colocalize with β-actin RNA in the nucleus as well (Oleynikov and Singer, 2003). In Xenopus oocytes, hnRNP I and Vg1RBP/vera colocalize with Vg1 RNA in the vegetal cortical cytoplasm (Cote et al., 1999; Zhang et al., 1999), but both proteins can also be detected in the nucleus (Fig. 1; Zhang et al., 1999). Our results now provide direct biochemical evidence that hnRNP I and Vg1RBP/vera associate with vegetal RNAs in the nucleus, perhaps functioning to initiate cytoplasmic localization through formation of a core nuclear RNP localization complex.
Previously, we suggested that the clustering of the binding sites for hnRNP I and Vg1RBP/vera may demarcate a consensus LE RNA and serve to facilitate interactions (Bubunenko et al., 2002; Lewis et al., 2004). Here, we show that these proteins are capable of interacting in vitro and in vivo (Fig. 3 and Fig. 4). However, the association in the nucleus and the cytoplasm is distinct; the interaction may be direct in the nucleus, but is dependent on RNA binding in the cytoplasm (Fig. 4). The possibility that hnRNP I and Vg1RBP/vera can interact directly in the nucleus may suggest that the two proteins associate with one another before binding the vegetally localized RNA. Alternatively, one of the proteins may bind to the vegetally localized RNA first and recruit the second protein to the RNA, thereby nucleating assembly of the nuclear localization RNP.
The dynamic nature of the Vg1RBP/vera–hnRNP I interaction between the nucleus and the cytoplasm (Fig. 4) suggests a remodeling of the localization RNP complex upon export to the cytoplasm (Fig. 6). Our observations that distinct sets of factors comprise the nuclear and cytoplasmic vegetal RNPs lend support to this idea as well. Remodeling of the localization RNP may be linked to the phosphorylation state of hnRNP I, as phosphorylation is required for nuclear export of the mammalian homologue (Xie et al., 2003). The phosphorylation state of the Xenopus protein similarly reflects its nucleocytoplasmic distribution (Xie et al., 2003) and could alter its interactions with other proteins. Although the role of hnRNP I phosphorylation in localization is still unknown, it will be important to determine if it influences the composition of Vg1 or VegT RNPs.
In contrast to Vg1RBP/vera and hnRNP I, both Prrp and XStau associate with the vegetal RNP only in the cytoplasm, indicating that these factors may function in the cytoplasm to direct vegetal RNA localization at later stages during the transport pathway. Neither a nuclear localization signal nor a nuclear export signal has been identified in Prrp; however, Zhao et al. (2001) noted a COOH-terminal domain of the protein that has a similar amino acid composition to the M9 nucleocytoplasmic shuttling domain of hnRNP A1 (Michael et al., 1995a; Siomi and Dreyfuss, 1995). Although our results do not provide evidence for Prrp function in the nucleus, a nuclear role for Prrp cannot be excluded. Although XStau may be present in both the nucleus and the cytoplasm (Fig. 5 C), the protein is most abundant in the cytoplasm, and we observed an interaction of XStau with the localization RNP proteins only in the cytoplasm (Fig. 5, B and C). Hence, our data indicate that both Prrp and XStau join the core localization RNP once it is exported to the cytoplasm (Fig. 6).
We have uncovered distinct nuclear and cytoplasmic steps in the vegetal RNA localization pathway through biochemical analysis of the Vg1 and VegT RNPs. Moreover, we suggest that vegetal RNP assembly in the nucleus may be required for the formation of a cytoplasmic localization-competent RNP, thus linking nuclear and cytoplasmic steps in the vegetal RNA transport pathway. Connections between steps in RNA biogenesis are increasingly appreciated (for review see Reed, 2003). For example, recent analyses on mRNA localization in Drosophila have revealed a requirement for components of the exon junction complex (Hachet and Ephrussi, 2001; Mohr et al., 2001). We have shown that Xenopus hnRNP I and Vg1RBP/vera first bind to vegetally localized RNA in the nucleus, but roles for these factors in other post-transcriptional functions in the oocytes remain to be determined. Nonetheless, it is tempting to speculate that such proteins could link cytoplasmic RNA localization to earlier nuclear events in RNA biogenesis such as transcription and splicing. The challenge now is to determine how the nuclear and cytoplasmic steps in the vegetal RNA localization pathway are interlinked to ensure proper regulation and coordination of RNA localization, from initiation in the nucleus to its final destination the cytoplasm.
Materials and methods
Cloning and recombinant protein production
The hnRNP I–coding region (AF091370) was amplified by PCR from pBShnRNP I (Cote et al., 1999) and cloned into pSP64T (Krieg and Melton, 1984) with a FLAG epitope (pSP64TSNFLAG) to create pSP64ThnRNPIFLAG. The Vg1RBP/vera-coding region (AF064634; Havin et al., 1998) was amplified by PCR from pVg1RBP (a gift of A. Git and N. Standart, University of Cambridge, Cambridge, UK) and cloned into pSP64TFLAG to generate pSP64TVg1RBP/veraFLAG. Prrp-myc (pCS3+MT-prrp) was a gift of P. Huber (Zhao et al., 2001).
Vg1RBP/vera-FLAG and hnRNPI-FLAG were in vitro translated in wheat germ extract (Promega) according to the manufacturer's instructions, and were used directly for immunoprecipitation. For antibody production, the NH2-terminal 136 amino acids of hnRNP I (including RRM1) were amplified from pBShnRNP I and cloned into pGEX4T1 (Amersham Biosciences) to generate pGST-N+RRM1. GST-N+RRM1 was expressed and purified by GST chromatography as in Coligan et al. (1995), followed by Mono Q™ chromatography.
Immunoblot analysis
Immunoblotting was performed as described in Denegre et al. (1997) with primary antibodies as follows: anti-hnRNP I, anti-XStau (Yoon and Mowry, 2004), anti-Vg1RBP/vera (supplied by J. Yisraeli, Hebrew University, Jerusalem, Israel, and by A. Git and N. Standart, University of Cambridge; Zhang et al., 1999; Git and Standart, 2002), or anti-tubulin (Sigma-Aldrich). Secondary antibodies were peroxidase-conjugated goat anti–rabbit IgG or rabbit anti–mouse IgG (Sigma-Aldrich). All incubations were performed in S-Blotto (50 mM Tris, pH 7.5, 250 mM NaCl, 5% powdered milk, and 1% Tween 20) except for the final wash (S-Blotto minus milk) before detection.
Immunolocalization
Immunofluorescence was performed as in Denegre et al. (1997), except stage I/II and III/IV oocytes were fixed for 1–2 h, and stage V/VI were fixed overnight in MEMFA (0.1 mM MOPS, pH 7.4, 2 mM EGTA, 1 mM MgSO4, and 3.7% formaldehyde). The primary antibody was anti-hnRNP I at 1:250 for stage I–IV and 1:100 for stage V–VI, and the secondary antibody was Alexa® 568–conjugated goat anti–rabbit (Molecular Probes, Inc.) at 1:100. After fixation, oocytes were mounted in 2:1 benzyl benzoate/benzyl alcohol and analyzed on a confocal microscope (LSM 410; Carl Zeiss MicroImaging, Inc.) using a 10× Plan-Apochromat objective with 0.45 NA (Carl Zeiss MicroImaging, Inc.) at RT. Images were obtained using Renaissance software v.2.17 (Microcosm) and prepared using Adobe Photoshop® 7.0.
Oocyte microinjection
Stage III/IV oocytes (as in Dumont, 1972) were obtained surgically from X. laevis females (Nasco), and were defoliculated by incubation in 2 mg/ml collagenase (Sigma-Aldrich). RNA for injection was transcribed from pSP64ThnRNPIFLAG, pSP64TVg1RBP/veraFLAG, or pCS3+MT-prrp using the mMESSAGE mMACHINE® kit (Ambion). After microinjection with ∼2 nl of in vitro–transcribed RNA at 500 nM, oocytes were cultured for 18 h as in Gautreau et al. (1997).
Preparation of lysates
Defolliculate oocytes were placed in YSS buffer (50 mM Tris, pH 8.0, 50 mM NaCl, 0.01% Ipegal [Sigma-Aldrich], 1 U/ml RNasin [Promega], 0.1 μg/ml leupeptin, 0.1 μg/ml aprotonin, 0.1 μg/ml trypsin inhibitor, 0.4 mM Pefabloc®, 1.0 mM DTT, and 100 mM sucrose). Oocytes were lanced with a 26-gauge needle and the nucleus was squeezed from the oocyte, leaving the cytoplasm intact within the oocyte membrane. For preparation of S10 lysates, nuclei or enucleated oocytes (cytoplasm) were homogenized in YSS (1 nucleus/μl or 1 cytoplasm/μl) and were centrifuged for 10 min at 10,000 g. S10 supernatants were collected and used immediately for analysis. For RNase treatment of lysates, RNase A (Sigma-Aldrich) was added at 0.1 μg/μl and incubated for 20 min at 37°C; mock treatment was with RNasin (Promega) at 0.12 U/μl, incubated for 20 min at 37°C.
Immunoprecipitation and RT-PCR
For immunoprecipitations, 5-μl anti-hnRNP I, anti-Vg1RBP/vera (Zhang et al., 1999; Git and Standart, 2002), anti-myc (9E10; Sigma-Aldrich), or control (anti-VP67; Volodina et al., 2003) antibodies were incubated overnight at 4°C with 10 μl protein G–Sepharose beads (Amersham Biosciences) in 1 ml YSS. FLAG immunoprecipitations were performed using 10 μl anti-FLAG beads (Sigma-Aldrich). After four washes with YSS, 20 μl of nuclear or cytoplasmic S10 lysates or 4 μl of in vitro translation mixture was added to the beads in 1 ml YSS. When indicated, RNase treatment was performed as above. After overnight incubation at 4°C, beads were washed 4× with 1 ml YSS, resuspended in sample buffer, and separated by SDS-PAGE. For RT-PCR, samples were resuspended after immunoprecipitation in 100 μl YSS. Isolation of RNA and RT-PCR were performed as in LaBonne and Whitman (1994). Vg1 primers were 5′-CGATGACATCCACCCAACAC-3′ and 5′-GAGGGTCACAGTCAGCAAGG-3′, and the VegT primers were as in Zhang and King (1996). PCR products were analyzed by agarose gel electrophoresis along with an amplification control from total cDNA generated from nuclear or cytoplasmic lysates.
Acknowledgments
We thank P. Huber, N. Standart, and J. Yisraeli for supplying reagents. We also thank D. Black, D. Cameron, J. Clifton, R. Lewis, T. Messitt, and T. Serio for helpful comments on the manuscript.
T.L. Kress was a predoctoral trainee, supported in part by a grant (5-T32-GM07601) from the National Institutes of Health. This work was supported by a grant (R01 HD30699) from the National Institutes of Health to K.L. Mowry.
Abbreviation used in this paper: LE, localization element.
References
- Arn, E.A., B.J. Cha, W.E. Theurkauf, and P.M. Macdonald. 2003. Recognition of a bicoid mRNA localization signal by a protein complex containing Swallow, Nod, and RNA binding proteins. Dev. Cell. 4:41–51. [DOI] [PubMed] [Google Scholar]
- Barbarese, E., D.E. Koppel, M.P. Deutscher, C.L. Smith, K. Ainger, F. Morgan, and J.H. Carson. 1995. Protein translation components are colocalized in granules in oligodendrocytes. J. Cell Sci. 108:2781–2790. [DOI] [PubMed] [Google Scholar]
- Bertrand, E., P. Chartrand, M. Schaefer, S.M. Shenoy, R.H. Singer, and R.M. Long. 1998. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 2:437–445. [DOI] [PubMed] [Google Scholar]
- Betley, J.N., M.C. Frith, J.H. Graber, S. Choo, and J.O. Deshler. 2002. A ubiquitous and conserved signal for RNA localization in chordates. Curr. Biol. 12:1756–1761. [DOI] [PubMed] [Google Scholar]
- Bilodeau, P.S., J.K. Domsic, A. Mayeda, A.R. Krainer, and C.M. Stoltzfus. 2001. RNA splicing at human immunodeficiency virus type 1 3′ splice site A2 is regulated by binding of hnRNP A/B proteins to an exonic splicing silencer element. J. Virol. 75:8487–8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubunenko, M., T.L. Kress, U.D. Vempati, K.L. Mowry, and M.L. King. 2002. A consensus RNA signal that directs germ layer determinants to the vegetal cortex of Xenopus oocytes. Dev. Biol. 248:82–92. [DOI] [PubMed] [Google Scholar]
- Chan, A.P., and L.D. Etkin. 2001. Patterning and lineage specification in the amphibian embryo. Curr. Top. Dev. Biol. 51:1–67. [DOI] [PubMed] [Google Scholar]
- Coligan, J.E., B.M. Dunn, H.L. Ploegh, D.W. Speicher, and P.T. Wingfield. 1995. Purification of recombinant proteins. Current Protocols in Protein Science. Vol. 1. John Wiley & Sons, Inc., New York. 6.6.1–6.6.26.
- Cote, C.A., D. Gautreau, J.M. Denegre, T.L. Kress, N.A. Terry, and K.L. Mowry. 1999. A Xenopus protein related to hnRNP I has a role in cytoplasmic RNA localization. Mol. Cell. 4:431–437. [DOI] [PubMed] [Google Scholar]
- Dale, L., G. Matthews, and A. Colman. 1993. Secretion and mesoderm-inducing activity of the TGF-β-related domain of Xenopus Vg1. EMBO J. 12:4471–4480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denegre, J.M., E.R. Ludwig, and K.L. Mowry. 1997. Localized maternal proteins in Xenopus revealed by subtractive immunization. Dev. Biol. 192:446–454. [DOI] [PubMed] [Google Scholar]
- Deshler, J.O., M.I. Highett, and B.J. Schnapp. 1997. Localization of Xenopus Vg1 mRNA by Vera protein and the endoplasmic reticulum. Science. 276:1128–1131. [DOI] [PubMed] [Google Scholar]
- Deshler, J.O., M.I. Highett, T. Abramson, and B.J. Schnapp. 1998. A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr. Biol. 8:489–496. [DOI] [PubMed] [Google Scholar]
- Dumont, J.N. 1972. Oogenesis in Xenopus laevis (Daudin). I. Stages of oocyte development in laboratory maintained animals. J. Morphol. 136:153–179. [DOI] [PubMed] [Google Scholar]
- Farina, K.L., and R.H. Singer. 2002. The nuclear connection in RNA transport and localization. Trends Cell Biol. 12:466–472. [DOI] [PubMed] [Google Scholar]
- Gautreau, D., C.A. Cote, and K.L. Mowry. 1997. Two copies of a subelement from the Vg1 RNA localization sequence are sufficient to direct vegetal localization in Xenopus oocytes. Development. 124:5013–5020. [DOI] [PubMed] [Google Scholar]
- Git, A., and N. Standart. 2002. The KH domains of Xenopus Vg1RBP mediate RNA binding and self-association. RNA. 8:1319–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, W., F. Pan, H. Zhang, G.J. Bassell, and R.H. Singer. 2002. A predominantly nuclear protein affecting cytoplasmic localization of β-actin mRNA in fibroblasts and neurons. J. Cell Biol. 156:41–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachet, O., and A. Ephrussi. 2001. Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr. Biol. 11:1666–1674. [DOI] [PubMed] [Google Scholar]
- Havin, L., A. Git, Z. Elisha, F. Oberman, K. Yaniv, S.P. Schwartz, N. Standart, and J.K. Yisraeli. 1998. RNA-binding protein conserved in both microtubule- and microfilament-based RNA localization. Genes Dev. 12:1593–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek, K.S., G.J. Kidd, J.H. Carson, and R. Smith. 1998. hnRNP A2 selectively binds the cytoplasmic transport sequence of myelin basic protein mRNA. Biochemistry. 37:7021–7029. [DOI] [PubMed] [Google Scholar]
- Horb, M.E., and G.H. Thomsen. 1997. A vegetally localized T-box transcription factor in Xenopus eggs specifies mesoderm and endoderm and is essential for embryonic mesoderm formation. Development. 124:1689–1698. [DOI] [PubMed] [Google Scholar]
- Jansen, R.P. 2001. mRNA localization: message on the move. Nat. Rev. Mol. Cell Biol. 2:247–256. [DOI] [PubMed] [Google Scholar]
- Joseph, E.M., and D.A. Melton. 1998. Mutant Vg1 ligands disrupt endoderm and mesoderm formation in Xenopus embryos. Development. 125:2677–2685. [DOI] [PubMed] [Google Scholar]
- Kamath, R.V., D.J. Leary, and S. Huang. 2001. Nucleocytoplasmic shuttling of polypyrimidine tract-binding protein is uncoupled from RNA export. Mol. Biol. Cell. 12:3808–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler, D.S., and D.A. Melton. 1995. Induction of dorsal mesoderm by soluble, mature Vg1 protein. Development. 121:2155–2164. [DOI] [PubMed] [Google Scholar]
- Kislauskis, E.H., X. Zhu, and R.H. Singer. 1994. Sequences responsible for intracellular localization of β-actin messenger RNA also affect cell phenotype. J. Cell Biol. 127:441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloc, M., N.R. Zearfoss, and L.D. Etkin. 2002. Mechanisms of subcellular mRNA localization. Cell. 108:533–544. [DOI] [PubMed] [Google Scholar]
- Krichevsky, A.M., and K.S. Kosik. 2001. Neuronal RNA granules: a link between RNA localization and stimulation-dependent translation. Neuron. 32:683–696. [DOI] [PubMed] [Google Scholar]
- Krieg, P.A., and D.A. Melton. 1984. Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 12:7057–7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll, T.T., W.M. Zhao, C. Jiang, and P.W. Huber. 2002. A homolog of FBP2/KSRP binds to localized mRNAs in Xenopus oocytes. Development. 129:5609–5619. [DOI] [PubMed] [Google Scholar]
- Kwon, S., T. Abramson, T.P. Munro, C.M. John, M. Kohrmann, and B.J. Schnapp. 2002. UUCAC- and vera-dependent localization of VegT RNA in Xenopus oocytes. Curr. Biol. 12:558–564. [DOI] [PubMed] [Google Scholar]
- LaBonne, C., and M. Whitman. 1994. Mesoderm induction by activin requires FGF-mediated intracellular signals. Development. 120:463–472. [DOI] [PubMed] [Google Scholar]
- Lewis, R.A., T.L. Kress, C.A. Cote, D. Gautreau, M.E. Rokop, and K.L. Mowry. 2004. Conserved and clustered RNA recognition sequences are a critical feature of signals directing RNA localization in Xenopus oocytes. Mech. Dev. 121:101–109. [DOI] [PubMed] [Google Scholar]
- Li, B., and T.S. Yen. 2002. Characterization of the nuclear export signal of polypyrimidine tract-binding protein. J. Biol. Chem. 277:10306–10314. [DOI] [PubMed] [Google Scholar]
- Long, R.M., R.H. Singer, X. Meng, I. Gonzalez, K. Nasmyth, and R.P. Jansen. 1997. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 277:383–387. [DOI] [PubMed] [Google Scholar]
- Long, R.M., W. Gu, X. Meng, G. Gonsalvez, R.H. Singer, and P. Chartrand. 2001. An exclusively nuclear RNA-binding protein affects asymmetric localization of ASH1 mRNA and Ash1p in yeast. J. Cell Biol. 153:307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lustig, K.D., K.L. Kroll, E.E. Sun, and M.W. Kirschner. 1996. Expression cloning of a Xenopus T-related gene (Xombi) involved in mesodermal patterning and blastopore lip formation. Development. 122:4001–4012. [DOI] [PubMed] [Google Scholar]
- Matunis, E.L., M.J. Matunis, and G. Dreyfuss. 1992. Characterization of the major hnRNP proteins from Drosophila melanogaster. J. Cell Biol. 116:257–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayeda, A., S.H. Munroe, J.F. Caceres, and A.R. Krainer. 1994. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. EMBO J. 13:5483–5495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton, D.A. 1987. Translocation of a localized maternal mRNA to the vegetal pole of Xenopus oocytes. Nature. 328:80–82. [DOI] [PubMed] [Google Scholar]
- Michael, W.M., M. Choi, and G. Dreyfuss. 1995. a. A nuclear export signal in hnRNP A1: a signal-mediated, temperature-dependent nuclear protein export pathway. Cell. 83:415–422. [DOI] [PubMed] [Google Scholar]
- Michael, W.M., H. Siomi, M. Choi, S. Pinol-Roma, S. Nakielny, Q. Liu, and G. Dreyfuss. 1995. b. Signal sequences that target nuclear import and nuclear export of pre-mRNA-binding proteins. Cold Spring Harb. Symp. Quant. Biol. 60:663–668. [DOI] [PubMed] [Google Scholar]
- Min, H., C.W. Turck, J.M. Nikolic, and D.L. Black. 1997. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 11:1023–1036. [DOI] [PubMed] [Google Scholar]
- Mohr, S.E., S.T. Dillon, and R.E. Boswell. 2001. The RNA-binding protein Tsunagi interacts with Mago Nashi to establish polarity and localize oskar mRNA during Drosophila oogenesis. Genes Dev. 15:2886–2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry, K.L. 1996. Complex formation between stage-specific oocyte factors and a Xenopus mRNA localization element. Proc. Natl. Acad. Sci. USA. 93:14608–14613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry, K.L., and D.A. Melton. 1992. Vegetal messenger RNA localization directed by a 340-nt RNA sequence element in Xenopus oocytes. Science. 255:991–994. [DOI] [PubMed] [Google Scholar]
- Mowry, K.L., and C.A. Cote. 1999. RNA sorting in Xenopus oocytes and embryos. FASEB J. 13:435–445. [DOI] [PubMed] [Google Scholar]
- Norvell, A., R.L. Kelley, K. Wehr, and T. Schupbach. 1999. Specific isoforms of squid, a Drosophila hnRNP, perform distinct roles in Gurken localization during oogenesis. Genes Dev. 13:864–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleynikov, Y., and R.H. Singer. 2003. Real-time visualization of ZBP1 association with β-actin mRNA during transcription and localization. Curr. Biol. 13:199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios, I.M. 2002. RNA processing: splicing and the cytoplasmic localisation of mRNA. Curr. Biol. 12:R50–R52. [DOI] [PubMed] [Google Scholar]
- Palacios, I.M., and D. St Johnston. 2001. Getting the message across: the intracellular localization of mRNAs in higher eukaryotes. Annu. Rev. Cell Dev. Biol. 17:569–614. [DOI] [PubMed] [Google Scholar]
- Patton, J.G., S.A. Mayer, P. Tempst, and B. Nadal-Ginard. 1991. Characterization and molecular cloning of polypyrimidine tract-binding protein: a component of a complex necessary for pre-mRNA splicing. Genes Dev. 5:1237–1251. [DOI] [PubMed] [Google Scholar]
- Reed, R. 2003. Coupling transcription, splicing and mRNA export. Curr. Opin. Cell Biol. 15:326–331. [DOI] [PubMed] [Google Scholar]
- Rook, M.S., M. Lu, and K.S. Kosik. 2000. CaMKIIα 3′ untranslated region-directed mRNA translocation in living neurons: visualization by GFP linkage. J. Neurosci. 20:6385–6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, A.F., Y. Oleynikov, E.H. Kislauskis, K.L. Taneja, and R.H. Singer. 1997. Characterization of a β-actin mRNA zipcode-binding protein. Mol. Cell. Biol. 17:2158–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S.P., L. Aisenthal, Z. Elisha, F. Oberman, and J.K. Yisraeli. 1992. A 69-kDa RNA-binding protein from Xenopus oocytes recognizes a common motif in two vegetally localized maternal mRNAs. Proc. Natl. Acad. Sci. USA. 89:11895–11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shestakova, E.A., R.H. Singer, and J. Condeelis. 2001. The physiological significance of β-actin mRNA localization in determining cell polarity and directional motility. Proc. Natl. Acad. Sci. USA. 98:7045–7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi, H., and G. Dreyfuss. 1995. A nuclear localization domain in the hnRNP A1 protein. J. Cell Biol. 129:551–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennard, F., G. Carnac, and J.B. Gurdon. 1996. The Xenopus T-box gene, Antipodean, encodes a vegetally localised maternal mRNA and can trigger mesoderm formation. Development. 122:4179–4188. [DOI] [PubMed] [Google Scholar]
- Takizawa, P.A., A. Sil, J.R. Swedlow, I. Herskowitz, and R.D. Vale. 1997. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 389:90–93. [DOI] [PubMed] [Google Scholar]
- Thomsen, G.H., and D.A. Melton. 1993. Processed Vg1 protein is an axial mesoderm inducer in Xenopus. Cell. 74:433–441. [DOI] [PubMed] [Google Scholar]
- Valcarcel, J., and F. Gebauer. 1997. Post-transcriptional regulation: the dawn of PTB. Curr. Biol. 7:R705–R708. [DOI] [PubMed] [Google Scholar]
- Volodina, N., J.M. Denegre, and K.L. Mowry. 2003. Apparent mitochondrial asymmetry in Xenopus eggs. Dev. Dyn. 226:654–662. [DOI] [PubMed] [Google Scholar]
- Weeks, D.L., and D.A. Melton. 1987. A maternal mRNA localized to the vegetal hemisphere in Xenopus eggs codes for a growth factor related to TGF-β. Cell. 51:861–867. [DOI] [PubMed] [Google Scholar]
- Wilkie, G.S., and I. Davis. 2001. Drosophila wingless and pair-rule transcripts localize apically by dynein-mediated transport of RNA particles. Cell. 105:209–219. [DOI] [PubMed] [Google Scholar]
- Xie, J., J.-A. Lee, T.L. Kress, K.M. Mowry, and D.L. Black. 2003. Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc. Natl. Acad. Sci. USA. 100:8776–8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yisraeli, J.K., S. Sokol, and D.A. Melton. 1990. A two-step model for the localization of maternal mRNA in Xenopus oocytes: involvement of microtubules and microfilaments in the translocation and anchoring of Vg1 mRNA. Development. 108:289–298. [DOI] [PubMed] [Google Scholar]
- Yoon, Y.J., and K.L. Mowry. 2004. Xenopus Staufen is a component of a ribonucleoprotein complex containing Vg1 RNA and kinesin. Development. In press. [DOI] [PubMed] [Google Scholar]
- Zhang, H.L., T. Eom, Y. Oleynikov, S.M. Shenoy, D.A. Liebelt, J.B. Dictenberg, R.H. Singer, and G.J. Bassell. 2001. Neurotrophin-induced transport of a β-actin mRNP complex increases β-actin levels and stimulates growth cone motility. Neuron. 31:261–275. [DOI] [PubMed] [Google Scholar]
- Zhang, J., and M.L. King. 1996. Xenopus VegT RNA is localized to the vegetal cortex during oogenesis and encodes a novel T-box transcription factor involved in mesodermal patterning. Development. 122:4119–4129. [DOI] [PubMed] [Google Scholar]
- Zhang, J., D.W. Houston, M.L. King, C. Payne, C. Wylie, and J. Heasman. 1998. The role of maternal VegT in establishing the primary germ layers in Xenopus embryos. Cell. 94:515–524. [DOI] [PubMed] [Google Scholar]
- Zhang, Q., K. Yaniv, F. Oberman, U. Wolke, A. Git, M. Fromer, W.L. Taylor, D. Meyer, N. Standart, E. Raz, and J.K. Yisraeli. 1999. Vg1 RBP intracellular distribution and evolutionarily conserved expression at multiple stages during development. Mech. Dev. 88:101–106. [DOI] [PubMed] [Google Scholar]
- Zhao, W.M., C. Jiang, T.T. Kroll, and P.W. Huber. 2001. A proline-rich protein binds to the localization element of Xenopus Vg1 mRNA and to ligands involved in actin polymerization. EMBO J. 20:2315–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]