Abstract
The Escherichia coli YidC protein belongs to the Oxa1 family of membrane proteins that have been suggested to facilitate the insertion and assembly of membrane proteins either in cooperation with the Sec translocase or as a separate entity. Recently, we have shown that depletion of YidC causes a specific defect in the functional assembly of F1F0 ATP synthase and cytochrome o oxidase. We now demonstrate that the insertion of in vitro–synthesized F1F0 ATP synthase subunit c (F0c) into inner membrane vesicles requires YidC. Insertion is independent of the proton motive force, and proteoliposomes containing only YidC catalyze the membrane insertion of F0c in its native transmembrane topology whereupon it assembles into large oligomers. Co-reconstituted SecYEG has no significant effect on the insertion efficiency. Remarkably, signal recognition particle and its membrane-bound receptor FtsY are not required for the membrane insertion of F0c. In conclusion, a novel membrane protein insertion pathway in E. coli is described in which YidC plays an exclusive role.
Keywords: YidC; F1F0 ATP synthase; membrane insertion; membrane targeting; complex assembly
Introduction
All major energy-transducing cellular membranes contain one or more members of the cytochrome oxidase biogenesis (Oxa) family of membrane proteins (Kuhn et al., 2003). In the inner membrane of yeast mitochondria, Oxa1 is required for the insertion of several membrane proteins among which are subunits of the F1F0 ATP synthase and cytochrome c oxidase (Stuart, 2002). These include mitochondrion-encoded proteins, like Cox2 (Bauer et al., 1994; He and Fox, 1997), as well as nucleus-encoded proteins, like Oxa1 itself (Herrmann et al., 1997). The latter are first imported into the mitochondrial matrix and subsequently inserted into the inner membrane. The chloroplast homologue Alb3 is required for the insertion of the light-harvesting complex protein into the thylakoid membrane (Moore et al., 2000). In the Gram-negative bacterium Escherichia coli, an Oxa1 homologue termed YidC was found that could be cross-linked to the transmembrane segments of nascent membrane proteins, like FtsQ (Scotti et al., 2000), leader peptidase (Houben et al., 2000), and MtlA (Beck et al., 2001). This process involves targeting via the signal recognition particle (SRP) pathway (Herskovits et al., 2000) and the initial membrane insertion by the SecYEG translocase (De Gier and Luirink, 2001). However, YidC is not essential for the insertion of the Sec-dependent model protein FtsQ, as the Δψ-dependent membrane integration of this protein can be reconstituted with proteoliposomes containing only SecYEG (van der Laan et al., 2004). Initially, only the Sec-independent phage proteins M13 procoat (Samuelson et al., 2000) and Pf3 coat (Chen et al., 2002) were found to depend strictly on YidC for membrane insertion. Therefore, it was not clear why YidC is essential for cell viability. Recently, we have shown that depletion of YidC causes severe defects in the functional assembly of both cytochrome o oxidase and F1F0 ATP synthase (van der Laan et al., 2003), which is reminiscent of the effects of mutations in OXA1 (Altamura et al., 1996). In particular, the amounts of cytochrome o oxidase subunit a and the small, ring-forming F0 subunit c (F0c) were exceptionally sensitive to YidC depletion. Consequently, YidC-depleted cells and inner membrane vesicles (IMVs) are severely impaired in their ability to generate a proton motive force (PMF) with oxidizable substrates or ATP (van der Laan et al., 2003). Although the loss of cytochrome o oxidase or F1F0 ATPase individually may not be detrimental for the cells, loss of both enzymes will severely impair growth. Cells are no longer able to generate a PMF with either oxidizable substrates, or under fermentative conditions, with ATP. Thus, these cells will be impaired in maintaining the intracellular pH, a process that is essential for viability. Yi et al. (2003) have shown that in addition to F0c, membrane insertion of F0 subunit a (F0a) is also inhibited by depletion of YidC in vivo.
E. coli F1F0 ATP synthase consists of a membrane-integral F0 part (subunit composition a1b2c10) and a peripherally bound, catalytic F1 subcomplex (α3β3γδɛ; Capaldi and Aggeler, 2002). During the catalytic cycle, the reversible protonation of F0c at residue Asp61 induces a rotation of γ, ɛ, and the c ring relative to the a3b3 hexagon. Subunits F0b and F1δ form a so-called “stator” that ensures that F0a and the α3β3 hexagon do not rotate together with γɛcring. This causes a rotor torque that is believed to be translated into conformational changes of the catalytic residues by elastic power transmission finally leading the synthesis of ATP from ADP and phosphate (Weber and Senior, 2003). Although the mechanisms of energy transduction have been studied in great detail, remarkably little is known about the assembly of large energy-transducing membrane protein complexes like F1F0 ATP synthases or cytochrome oxidases. In yeast, proteins have been identified that are required for their biogenesis, but their precise function is not understood (Ackerman and Tzagoloff, 1990; Ackerman, 2002; Carr and Winge, 2003). The E. coli F1F0 ATP synthase represents the simplest form of this enzyme, containing only the core subunits described above. The observed YidC requirement for the assembly of functional F1F0 ATP synthase in vivo, and particularly of the membrane insertion of the F0c rotor ring subunit, indicates that the biogenesis of this key enzyme is more complex than anticipated so far (Arechaga et al., 2002). To understand the role of YidC in the membrane insertion and oligomeric assembly of F0c, we have used an in vitro translation/insertion assay that has been successfully used to determine the minimal requirements for the Sec-dependent membrane insertion of FtsQ (van der Laan et al., 2004). Here, we demonstrate that in vitro membrane insertion of F0c is blocked by YidC depletion and can be reconstituted with proteoliposomes containing only YidC, whereas SecYEG, the PMF, and the SRP pathway are not required.
Recently, Serek et al. (2004) reported that YidC alone reconstituted into proteoliposomes stimulates the in vitro membrane insertion of Pf3 coat, indicating that YidC can function as a separate membrane protein insertase. Our data demonstrate an essential role of the YidC insertase in the assembly of a Sec-independent, endogenous E. coli membrane protein. F0c is the first described natural substrate of this novel membrane protein biogenesis pathway, which appears to be used by bacteriophages to assemble their coat proteins in the host cell membrane.
Results
Insertion of F0c into IMVs requires YidC, but is independent of the PMF
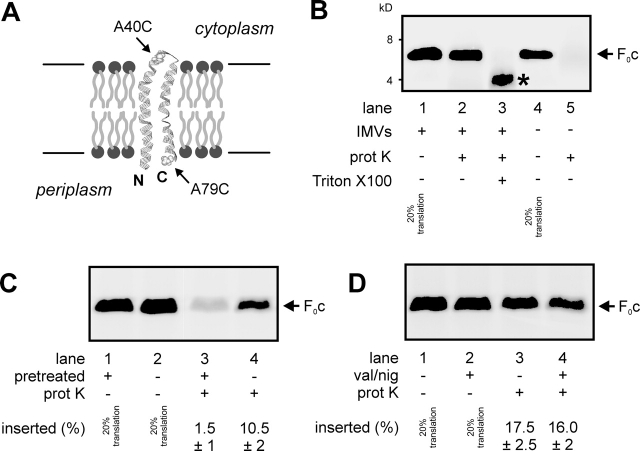
E. coli F0c is a very hydrophobic polypeptide of 79 amino acids in length. It consists of two transmembrane helices connected by a small polar loop that is exposed to the cytoplasm (Fig. 1 A; for review see Girvin et al., 1998). [35S]methionine-labeled F0c was synthesized in vitro in an E. coli S135 lysate (Fig. 1 B, lane 1). As the predicted molecular mass of F0c is 8.3 kD, the translation product showed an aberrant running behavior on SDS-PAGE typically observed for hydrophobic membrane proteins. Western blot analysis of wild-type E. coli IMVs revealed that endogenous F0c migrates at an identical position (unpublished data), indicating that the in vitro translation product is full-length F0c. When E. coli IMVs were present during translation, a significant fraction of the F0c was found to be resistant to added proteinase K (Fig. 1 B, lane 2), trypsin, and pronase (unpublished data). Furthermore, endogenous F0c in IMVs appeared protease resistant as well. Solubilization of IMVs with detergent before protease treatment led to partial cleavage of F0c (Fig. 1 B, lane 3), probably due to protection of the hydrophobic domains by the detergent micelles. In the absence of membranes and detergent, F0c was completely degraded (Fig. 1 B, lanes 4 and 5). Together, this indicates that the protease-protected conformation of F0c represents its membrane-inserted state. Insertion efficiency in the presence of saturating amounts of IMVs was 18–25%. When IMVs were pretreated with proteinase K before being added to the translation reaction, membrane insertion of F0c was almost completely blocked (Fig. 1 C), indicating that a membrane protein is needed to facilitate this process. However, the PMF is not required, as uncoupling of IMVs by the addition of the ionophores valinomycin and nigericin had no effect on the membrane insertion of F0c (Fig. 1 D), whereas it severely inhibited the PMF-dependent membrane insertion of the Sec-dependent model protein FtsQ (Fig. 2 C; van der Laan et al., 2004).
Figure 1.
In vitro–synthesized F 0 c inserts into E . coli inner membrane vesicles. (A) Membrane topology model of F0c based on the NMR solution structure (1A91.pdb). Arrows indicate the positions of the introduced cysteine mutations for topology analysis. (B) In vitro–synthesized F0c inserts into E. coli wild-type IMVs. Translation reactions were performed in the presence (lanes 1–3) or absence (lanes 4 and 5) of 5-μg IMVs. Subsequently, the translation reaction samples were treated with 0.4 mg/ml proteinase K (lanes 2, 3, and 5) in the presence (lane 3) or absence (lanes 1, 2, 4, and 5) of 1% (vol/vol) Triton X-100. In the presence of Triton X-100, a proteolytic fragment of F0c is formed (asterisk). Where indicated, 20% standards of the translation reactions are shown. (C) A membrane protein is required for the membrane insertion of F0c. IMVs were pretreated in the presence (lanes 1 and 3) or absence (lanes 2 and 4) of proteinase K as described in the Materials and methods section. (D) The PMF is not required for the membrane insertion of F0c. Insertion reactions were performed in the absence (lanes 1 and 3) or in the presence (lanes 2 and 4) of 1 μM valinomycin (val) and 1 μM nigericin (nig). Insertion efficiencies were calculated from three independent experiments.
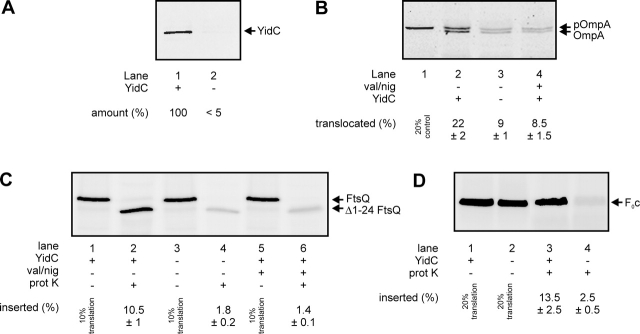
Figure 2.
YidC is required for the membrane insertion of F 0 c. (A) YidC can be efficiently depleted from E. coli JS7131 IMVs. Cells were grown as described in the Materials and methods section in the presence of 0.2% arabinose (lane 1) or 0.2% glucose (lane 2), and the amounts of YidC in IMVs were monitored by Western blotting. YidC-depleted IMVs exhibit reduced proOmpA translocation (B, lanes 1–3) and FtsQ insertion activities (C, lanes 1–4). The same extents of reduction are observed when the reactions are performed with wild-type IMVs in the presence of 1 μM valinomycin (val) and 1 μM nigericin (nig) to uncouple the PMF (B, lane 4; C, lanes 5 and 6). Post-translational proOmpA translocation as well as cotranslational FtsQ insertion reactions contained 25-μg IMVs. Efficiencies given in B and C were calculated from two independent experiments. (D) YidC depletion blocks membrane insertion of F0c. F0c insertion reactions were performed as described in the legend to Fig. 1. Efficiencies given in D were calculated from three independent experiments.
Previously, we have shown that the amount of F0c in IMVs depleted from YidC is strongly reduced (van der Laan et al., 2003). To determine if YidC plays a direct role in the membrane insertion of F0c, in vitro insertion assays were performed with YidC-depleted IMVs prepared from E. coli strain JS7131 (Samuelson et al., 2000) grown in the presence of 0.2% glucose. To achieve optimal depletion (Fig. 2 A) and to avoid pleiotrophic effects related to the stationary phase, cells were diluted 1:1 after every generation. This procedure led to a very efficient YidC depletion (Fig. 2 A) and an almost complete inability to generate a PMF with ATP (unpublished data). Consequently, PMF-dependent processes such as the translocation of proOmpA (Fig. 2 B, lanes 2 and 3) and the membrane insertion of FtsQ (Fig. 2 C, lanes 1–4) were strongly affected in these YidC-depleted IMVs. The extents of inhibition caused by the depletion of YidC were comparable to that observed with wild-type IMVs in the presence of uncouplers (Fig. 2 B, lane 4; Fig. 2 C, lanes 5 and 6). Remarkably, the membrane insertion of F0c into IMVs was almost completely blocked upon depletion of YidC (Fig. 2 D). As we have shown that F0c inserts independently of the PMF (Fig. 1 D), it can be concluded that this effect is directly caused by the loss of YidC.
YidC proteoliposomes catalyze the SecYEG-independent membrane insertion of F0c
The observation that YidC is required for the insertion of F0c into IMVs did not exclude that other membrane proteins, like the Sec translocase or other unknown components, are involved as well. Only the reconstitution of F0c insertion into proteoliposomes could clearly define the minimal requirements for this process. Therefore, we prepared (proteo-)liposomes containing either YidC together with SecYEG, YidC, or SecYEG alone, or containing no protein, and titrated them into the translation mixture. Upon addition of protein-free liposomes, small amounts of protease-protected F0c were detected (Fig. 3 A), indicating a low level of spontaneous membrane insertion. However, proteoliposomes containing YidC supported highly efficient membrane insertion of F0c (Fig. 3, A and B). Insertion efficiency achieved in the presence of saturating amounts of proteoliposomes was ∼40%. Co-reconstitution of SecYEG did not significantly increase the amount of membrane-inserted F0c (Fig. 3, A and B). Moreover, the presence of SecYEG alone did not stimulate F0c insertion compared with protein-free liposomes (Fig. 3, A and B). In contrast, proteoliposomes containing SecYEG mediated the translocation of proOmpA independently of YidC, showing that the reconstituted Sec translocase is functional (Fig. 3 A). These data indicate that YidC alone is able to catalyze membrane integration of F0c independently of the Sec translocase. To further examine a possible role of SecYEG in YidC-mediated insertion of F0c, experiments were performed using limiting YidC concentrations. YidC was reconstituted at different protein-to-lipid ratios in the presence or absence of a fixed amount of SecYEG. However, in no case was a significant stimulation of F0c insertion by SecYEG observed (Fig. 4, A and B).
Figure 3.
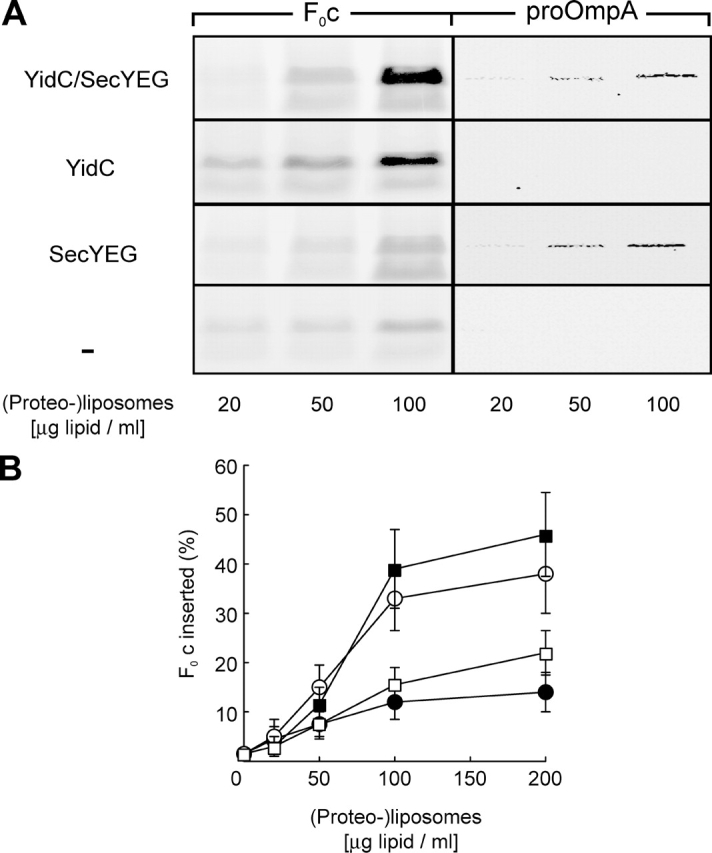
YidC alone is sufficient to catalyze the membrane insertion of F0c into proteoliposomes. (A) Co-translational F0c insertion as well as post-translational proOmpA translocation reactions were performed in the presence of the indicated amounts of (proteo-)liposomes reconstituted with YidC and SecYEG (YidC/SecYEG), YidC alone, SecYEG alone, or without any proteins (−). (B) F0c insertion efficiency at different (proteo-)liposome concentrations was quantified from the amount of protease-protected material in the presence of SecYEG/YidC (▪), YidC (○), SecYEG (□), or protein-free (•) liposomes. The amount of F0c synthesized was comparable in all reactions (not depicted) and was set as 100%. All data points shown in B are averages from five independent experiments in which proteoliposomes from three independent reconstitutions were used. Purified SecYEG and YidC were both used at a protein/lipid ratio of 1:100 (wt/wt).
Figure 4.
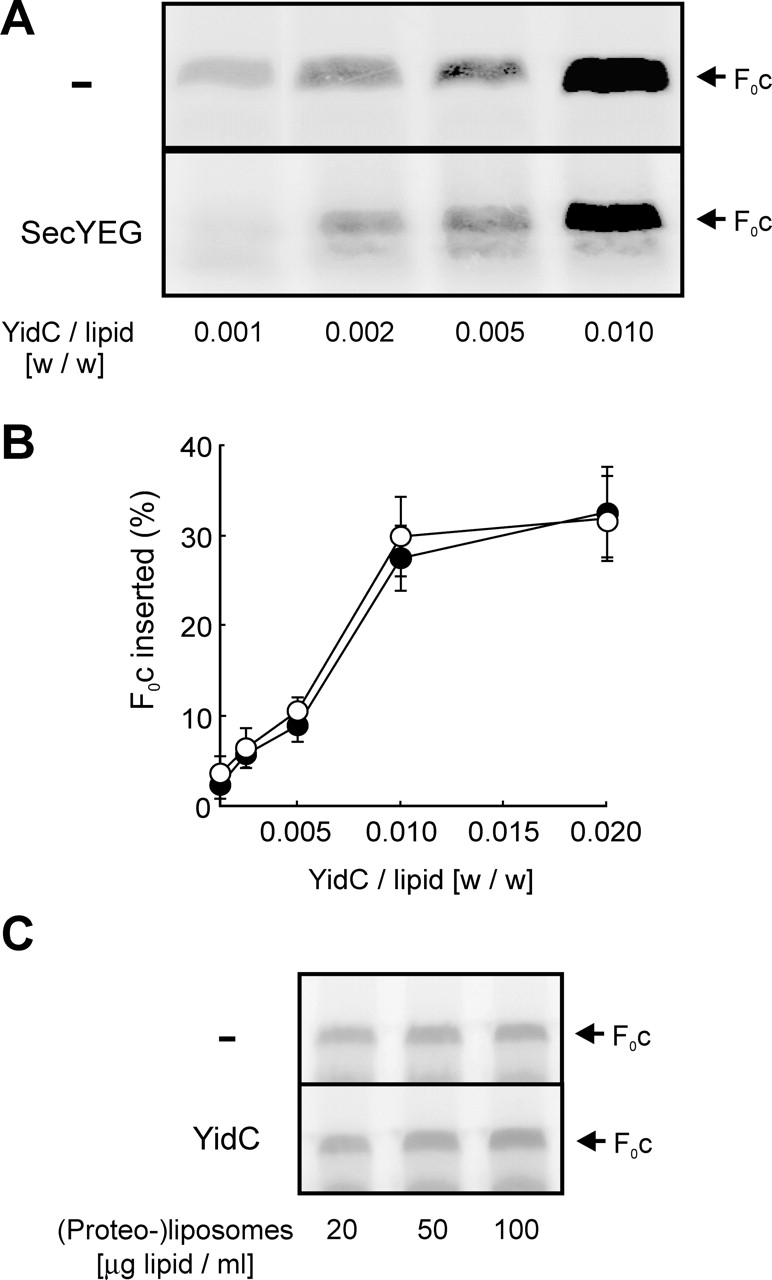
SecYEG is not required for F0c membrane insertion. (A) Reactions were performed as described in the legend of Fig. 3 in the presence of 100 μg lipid/ml proteoliposomes. For the reconstitutions, YidC was used at the indicated protein-to-lipid ratios together with (SecYEG) or without (−) a fixed amount of SecYEG (protein/lipid ratio of 1:100 [wt/wt]). (B) F0c insertion efficiencies were quantified from the amount of protease-protected material in the presence (•) or absence (○) of SecYEG. Synthesis of F0c was comparable in all reactions (not depicted). All data points shown in B are averages from three independent experiments. Proteoliposomes from two independent reconstitutions were used. When added after translation has been stopped with chloramphenicol, YidC proteoliposomes do not stimulate membrane insertion of F0c (C). Translation reactions were performed in the absence of membranes. Reactions were stopped by the addition of 25 μg/ml chloramphenicol. After 5 min, indicated amounts of YidC proteoliposomes or protein-free liposomes were added, and reaction mixtures were incubated for 20 min at 37°C.
Recently, we have shown that the insertion of the Sec-dependent membrane protein FtsQ is a strictly cotranslational process. In vitro, no insertion is observed when the membranes are added after translation has been terminated by the addition of chloramphenicol (Houben et al., 2000; van der Laan et al., 2004). Under identical conditions, post-translational insertion of F0c into YidC proteoliposomes also could not be detected (Fig. 4 C).
YidC-mediated membrane insertion of F0c does not require the SRP pathway
Co-translational targeting to the Sec translocase occurs via the SRP pathway (Herskovits et al., 2000). SRP binds to particularly hydrophobic signal sequences or transmembrane segments as they emerge from the ribosome. Upon interaction of SRP with its membrane-bound receptor FtsY, GTP hydrolysis drives the release of SRP from the nascent chain and the transfer of the translating ribosome to the Sec translocase. It is not clear whether the SRP pathway also delivers proteins directly to YidC. The YidC-dependent phage proteins M13 procoat (De Gier et al., 1998) and Pf3 coat (Chen et al., 2002) do not require SRP. However, Fröderberg et al. (2003) have constructed a fusion protein that does not depend on SecYEG, but requires YidC as well as the SRP pathway in vivo. In addition, the chloroplast YidC homologue Alb3 forms a complex with cpSRP and cpFtsY that can be stabilized by the addition of the nonhydrolysable GTP analogue GMP-PNP (Moore et al., 2003). As F0c represents the first “native” substrate of a novel membrane protein insertion pathway in which YidC seems to play a key role, we analyzed the involvement of the SRP pathway as well as the SecA motor protein. SecA is strictly required for the SecYEG-dependent translocation of the large periplasmic domain of FtsQ (van der Laan et al., 2004). As expected, immunodepletion of SecA from the translation lysate had no effect on the insertion of F0c into YidC proteoliposomes (Fig. 5 A, lane 2), whereas under identical conditions it completely blocks FtsQ insertion (van der Laan et al., 2004). Remarkably, immunodepletion of the SRP receptor FtsY also did not significantly affect YidC-mediated F0c insertion (Fig. 5 A, lane 3). Efficient FtsY depletion was demonstrated by Western blotting (Fig. 5 B) and by the inhibitory effect on membrane insertion of FtsQ (Fig. 5 C) as described before (van der Laan et al., 2004). To confirm this observation, we applied a second, independent experimental approach using a translation lysate that had been depleted from SRP in vivo. E. coli SRP consists of a 4.5S RNA and a 48-kD protein called Ffh. Strain HDB51 (Lee and Bernstein, 2001) carries the ffh gene under control of the arabinose promoter. Therefore, cells can be depleted from Ffh by growing them in the presence of glucose. Translation lysates were prepared from cells grown under Ffh depletion conditions as well as from Ffh-containing control cells grown in the presence of arabinose. Efficiency of depletion was monitored by Western blotting (Fig. 5 D). In agreement with the results obtained with FtsY-depleted lysate, we did not observe any major effect of Ffh depletion on the insertion of F0c into IMVs, YidC proteoliposomes, or YidC/SecYEG proteoliposomes (Fig. 5 E). In contrast, the insertion of FtsQ into either IMVs or SecYEG proteoliposomes was strongly inhibited upon depletion of Ffh (Fig. 5 F). Together, these data demonstrate that membrane targeting and YidC-mediated membrane insertion of F0c does not require the SRP pathway.
Figure 5.
Membrane insertion of F0c does not require the SRP pathway or SecA. (A) Translation lysate was immunodepleted from either FtsY or SecA, and F0c insertion reactions were performed in the presence of YidC proteoliposomes (YidC PL, 100 μg lipid/ml; protein/lipid ratio 1:100). (B) A Western blot with α-FtsY antiserum using 20 μg wild-type (lane 1) or FtsY-depleted (lane 2) lysate. (C) Depletion of FtsY from the lysate inhibits the FtsQ insertion into SecYEG proteoliposomes. Reactions were performed with FtsY-depleted (lanes 1 and 2) or wild-type (lanes 3 and 4) lysate in the presence of Na+-loaded SecYEG proteoliposomes (200 μg lipid/ml) and 1 μM valinomycin to generate a transmembrane electrical potential. (D) Ffh can be efficiently depleted in vivo in E. coli strain HDB51. A Western blot performed with 25 μg HDB51 wild-type (lane 1) or Ffh-depleted (lane 2) lysate is shown. (E) Ffh depletion has no effect on F0c insertion into IMVs (lane 1) or proteoliposomes containing YidC (lane 2) or YidC together with SecYEG (lane 3). Reactions were performed with wild-type or Ffh-depleted HDB51 lysate in the presence of 5-μg IMVs or 100 μg lipid/ml proteoliposome. Purified YidC and SecYEG were reconstituted at a protein/lipid ratio of 1:100. (F) Ffh depletion inhibits membrane insertion of FtsQ. Reactions were performed with Ffh-depleted (lanes 1 and 2) and wild-type (lanes 3 and 4) HDB51 lysate in the presence of 25-μg IMVs or 200 μg lipid/ml Na+-loaded SecYEG proteoliposome. FtsQ and F0c insertion efficiencies were calculated from two and three independent experiments, respectively.
F0c inserted into YidC proteoliposomes has acquired the native transmembrane topology
Next, we addressed the question of whether F0c is inserted into YidC proteoliposomes in the correct transmembrane topology. F0c exposes both the NH2 and the COOH terminus to the periplasm while the small loop connecting the two hydrophobic helices is located in the cytoplasm (Fig. 1 A). In our in vitro system, the lumen of the proteoliposomes corresponds to the periplasmic side of the membrane. If F0c has acquired the correct transmembrane topology, the COOH terminus should be inside the proteoliposomes, and therefore not accessible for membrane-impermeable chemical reagents. In contrast, it should be possible to modify the cytoplasmic loop that remains on the outside of the proteoliposomes with such probes. We introduced unique cysteine residues into the protein at position 79 at the COOH terminus (Fig. 1 A, F0c A79C) and at position 40 in the cytoplasmic loop (Fig. 1 A, F0c A40C). Both modified proteins inserted into proteoliposomes in a YidC-dependent manner comparable to wild-type F0c (Fig. 6 A). For the determination of the transmembrane topology of proteoliposome-inserted F0c, the protein was synthesized without radioactive tracers in the presence of YidC proteoliposomes. After the translation–insertion reaction and subsequent proteinase K treatment, proteoliposomes were isolated from the reaction mixture and incubated with the membrane-permeable, fluorescent cysteine-modifying reagent fluorescein maleimide. As expected, both F0c A79C and F0c A40C were labeled, whereas the cysteine-less wild-type F0c was not (Fig. 6 B, lanes 2 and 3). Pretreatment of the proteoliposomes with the membrane-impermeable cysteine-reactive reagent 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMdiS) completely prevented subsequent fluorescein maleimide labeling of F0c A40C (Fig. 6 B, lane 4), whereas F0c A79C could still be labeled after AMdiS pretreatment (Fig. 6 B, lane 4). Five independent experiments yielded an averaged labeling efficiency of ∼60%. When the proteoliposomes were solubilized in detergent before incubation with AMdiS, subsequent labeling of F0c A79C with fluorescein maleimide was not possible (unpublished data). These results indicate that the cytoplasmic loop of F0c is indeed localized on the outside, whereas the COOH terminus has been translocated into lumen of the proteoliposomes. These data show that the F0c has acquired the correct membrane topology after YidC-mediated insertion into proteoliposomes.
Figure 6.

F0c inserted into YidC proteoliposomes has acquired its native transmembrane topology. (A) F0c A79C and F0c A40C insert into YidC proteoliposomes comparable to the wild-type protein. Reactions were performed as described in the legend to Fig. 3 in the presence of the indicated amounts of (proteo-)liposomes. (B) For cysteine accessibility experiments, wild-type F0c and both single-cysteine derivatives were synthesized and inserted into YidC proteoliposomes in nonradioactive translation reactions. Subsequent labeling was performed with 1 mM fluorescein maleimide (lanes 1–4) as described in the Materials and methods section. A preincubation with 0.5 mM AMdiS was performed where indicated (lane 4). To control specificity of labeling, the same reactions were performed in the absence of mRNA (lane 1) or with the cysteine-free wild-type F0c (lane 2). (C) In vitro–synthesized F0c assembles into large oligomeric complexes after insertion into YidC proteoliposomes. Reactions were performed in the presence of protein-free liposomes (lane 1) or YidC proteoliposomes (lane 2) as described in the Materials and methods section, and the high molecular mass oligomers were analyzed by Blue Native PAGE. On this gel system, monomeric F0c (in the presence of 1% SDS) migrates as a very diffuse polypeptide band (not depicted).
F0c inserted into YidC proteoliposomes oligomerizes into large complexes
In vivo, F0c subunits assemble into a ring-like structure that is part of the rotor domain of F1F0 ATP synthase (Capaldi and Aggeler, 2002). Therefore, we asked if in vitro–synthesized F0c inserted into YidC proteoliposomes forms large oligomeric complexes as well. First, an insertion reaction, as described earlier in this paper, was performed. Subsequently, the proteoliposomes were reisolated from the reaction mixture, solubilized under mild conditions, and subjected to Blue Native PAGE (Fig. 6 D). Radioactively labeled F0c was indeed detected almost exclusively in a distinct high mol wt complex with a molecular mass of ∼80–100 kD after insertion into YidC proteoliposomes (Fig. 6 D, lane 2). These data demonstrate that membrane-inserted F0c is competent for oligomerization.
Discussion
About 30% of all structural genes in E. coli encode membrane proteins. These proteins have to be targeted to the membrane, inserted into the lipid bilayer, and correctly folded, and some of them have to be assembled into sophisticated supramolecular complexes. Examples of such complexes are the major energy-transducing membrane protein complexes such as the redox-driven proton pump cytochrome o oxidase or the PMF-driven mechanoenzyme F1F0 ATP synthase. Little is known about folding of membrane proteins and complex assembly, but it has been well established that the majority of membrane proteins is targeted via the SRP pathway to the SecYEG translocase, where membrane insertion takes place (De Gier and Luirink, 2001). In contrast, two special membrane proteins turned out to have completely different requirements for their insertion. The small bacteriophage proteins M13 procoat and Pf3 coat do not need the SRP pathway (De Gier et al., 1998; Chen et al., 2002) or SecYEG (Geller and Wickner, 1985; Kiefer and Kuhn, 1999) to become integrated into the membrane, but they depend on YidC in vivo (Samuelson et al., 2000; Chen et al., 2002). These two proteins do not stably integrate into the membrane, but in a second step assemble into the phage coat at the extracellular surface of the membrane (Russel et al., 1997). In vitro, they insert into protein-free liposomes with low efficiency (Geller and Wickner, 1985; Ridder et al., 2000; Serek et al., 2004), and their topologically correct insertion is strongly stimulated by the PMF (Date et al., 1980; Kiefer and Kuhn, 1999; Serek et al., 2004). Moreover, it has been shown that nascent chains of Pf3 coat can be cross-linked to YidC, but not SecY (Chen et al., 2002). These data indicate that in E. coli, an alternative membrane protein insertion pathway exists in which YidC plays a key role. Recently, Serek et al. (2004) demonstrated that Pf3 coat can be incorporated into YidC proteoliposomes in vitro, indicating that YidC functions as an individual membrane protein insertase. However, the YidC requirement of the phage proteins M13 procoat and Pf3 coat does not explain why YidC is an essential protein for E. coli. Other proteins that have been shown to strictly require YidC in vivo are artificial fusion proteins (Fröderberg et al., 2003; Yi et al., 2003) or membrane proteins that have been modified by epitope tags that can alter their membrane insertion pathways (Facey and Kuhn, 2003).
The first step toward the discovery of the native substrates of YidC was the observation that YidC-depleted IMVs contain strongly reduced amounts of cytochrome o oxidase and F1F0 ATP synthase (van der Laan et al., 2003), two major energy-transducing membrane protein complexes. In the case of F1F0 ATP synthase, the amount of the small ring-forming F0c protein was especially affected. Here, we show by means of in vitro insertion experiments that YidC alone mediates the membrane integration of F0c. Proteoliposomes containing only YidC catalyze efficient F0c insertion. Although F0c shows some spontaneous insertion into liposomes, reconstituted YidC dramatically stimulates F0c integration into proteoliposomes. At all YidC concentrations tested, co-reconstitution of the SecYEG complex has no effect on the insertion efficiency, indicating that the Sec translocase is not required for membrane integration of F0c.
It has been shown that membrane partitioning of M13 and Pf3 coat occurs in the absence of a PMF, but that the proteins do not acquire their native topology under these conditions (Date et al., 1980; Chen et al., 2002). Insertion of F0c does not depend on the PMF. We demonstrate by cysteine accessibility experiments that the COOH terminus of the protein, which is located in the periplasm in vivo, is translocated into YidC proteoliposomes, as it is protected from labeling with a membrane-impermeable probe. In contrast, the cytoplasmic loop connecting the two transmembrane helices remains accessible on the outer surface of the proteoliposomes. Thus, the final topology of F0c differs from M13 procoat and Pf3 coat. Although with M13 procoat the hydrophilic loop must be translocated across the membrane, the corresponding domain of F0c remains cytosolic. The first step in the membrane insertion of both M13 procoat and F0c might be the binding to the membrane surface mediated by electrostatic interactions with charged phospholipid headgroups. Presumably, the three positive charges in the cytosolic loop of F0c then prevent membrane translocation, and partitioning of the hydrophobic transmembrane domains into the lipid bilayer is assisted by YidC. The short NH2-terminal, periplasmic tail of F0c contains two negative charges. However, a possible electrophoretic contribution to their translocation is obviously not required. Interestingly, stepwise readdition of charged amino acids into an uncharged Pf3 variant (Pf3-4N) demonstrated that negatively charged residues in the periplasmic NH2-terminal domain show a clear electrophoretic response only when the hydrophobicity of the transmembrane segment is limiting (Kiefer and Kuhn, 1999). However, F0c is a very hydrophobic protein. This could explain why YidC-mediated hydrophobic interactions seem to be the sole driving force for F0c membrane insertion.
It has been established that the YidC-dependent proteins M13 procoat and Pf3 do not require the SRP-targeting pathway to become inserted into the membrane (De Gier et al., 1998; Chen et al., 2002). Consistently, we now demonstrate by two independent approaches, i.e., the in vitro depletion of FtsY and in vivo depletion of Ffh, that inactivation of the SRP pathway has no effect on the insertion of the physiological YidC substrate F0c into both IMVs and YidC proteoliposomes. Although an SRP requirement was not observed, proteoliposomes have to be present cotranslationally. Recently, a direct interaction between mitochondrial ribosomes and the YidC homologue Oxa1p has been reported (Jia et al., 2003; Szyrach et al., 2003). YidC lacks the COOH-terminal cytosolic domain that is required for ribosome binding to Oxa1p (Jia et al., 2003). So far, there is no evidence for a direct coupling between translation and membrane insertion of F0c. However, it seems likely that a very hydrophobic protein like F0c quickly aggregates in the absence of membranes, and this may complicate the post-translational insertion reaction, as the time window between translation and membrane insertion of such small membrane proteins is probably very short in vivo. In this respect, F0c does not seem to differ much from M13 procoat, a protein of similar length that also does not require SRP for membrane targeting.
Our data represent the first direct demonstration of a physiologically important catalytic activity of YidC, which casts a light on the essential role of YidC in E. coli. We have functionally reconstituted a novel membrane protein insertion pathway, and we have shown that the F0c protein is a substrate of it. Viruses and phages generally make use of key biogenesis pathways of their host cells in order to assure their own propagation. For the assembly of filamentous phages like M13 and Pf3, the accumulation of coat proteins in the cytoplasmic membrane of the host is an essential step. It now appears that M13 and Pf3 make use of a cellular machinery that plays a central role the biogenesis of major energy-transducing membrane protein complexes like F1F0 ATP synthase. The mechanism of YidC-mediated membrane protein integration remains to be elucidated. Classical protein translocation and insertion machineries like Sec or Tat translocases use ATP and/or the PMF as energy sources to actively transport proteins or domains of proteins across the membrane. In contrast, no PMF requirement was found for the YidC-mediated insertion of F0c, and YidC contains no obvious conserved nucleotide-binding domain.
It has been suggested that the function of YidC might be analogous to a chaperone that stabilizes folding and assembly intermediates of membrane proteins or membrane protein complexes. Remarkably, all three substrates of the YidC pathway are small and very hydrophobic proteins that assemble into large oligomeric structures like the phage coat or the rotor ring of the F1F0 ATP synthase. The mechanisms of assembly may be vastly different. However, phage coat assembly takes place at the outer surface of the inner membrane, and the final oligomer is not a membrane-inserted complex. Also, substrates of Oxa1, like Cox2p (He and Fox, 1997), and Alb3, like light-harvesting complex protein (Moore et al., 2000), are part of energy-transducing membrane protein complexes. YidC could play an important role as a chaperone in the assembly of these complexes. It might stabilize the transmembrane topology of a single F0c or act as a platform on which subunits can accumulate and organize into a ring structure. This ring is a very rigid complex (Arechaga et al., 2002) However, some intermediates in the process of complex formation might have to be stabilized by YidC. The role of YidC as well as the Sec translocase in the membrane insertion of other F0 subunits is unclear. Yi et al. (2003) have reported that the polytopic subunit F0a also requires YidC to become integrated into the membrane. However, F0a is known to be unstable in the absence of F0c, so that the F0a insertion defect might be at least partly indirect (Hermolin and Fillingame, 1995).
The reconstituted system described here will be a valuable tool in the detailed analysis of YidC-mediated membrane protein integration. Important questions are where the initial interaction of YidC with its substrates takes place, and how binding to and release from YidC are regulated. To understand the role of YidC in the formation of membrane protein complexes, the reconstituted assay described in this paper should be expanded to all subunits of the F0 complex, e.g., by combination with the reconstitution of Sec-dependent membrane protein insertion on which we have recently reported (van der Laan et al., 2004). This will be a challenge for future analyses.
Materials and methods
Strains and plasmids
Wild-type IMVs were prepared from E. coli SF100 (Baneyx and Georgiou, 1990), which was also used for overexpression of SecYEG (van der Does et al., 1996) and YidC (van der Laan et al., 2001). Strain JS7131 (Samuelson et al., 2000) was used to obtain YidC-depleted IMVs. Strain MC4100 was used to obtain S135 lysate. Strain HDB51 (Lee and Bernstein, 2001) was used to prepare Ffh-depleted and wild-type control lysate. Plasmid pBSKftsQ (van der Laan et al., 2004) was used for in vitro transcription of ftsQ. For in vitro transcription of atpE, the gene encoding the F0c protein, plasmid pET20atpE was constructed. atpE was PCR amplified from pBWU13 (Iwamoto et al., 1991) and cloned into pET20b (Novagen), yielding pET20atpE. pET20atpE-A40C and pET20atpE-A79C for the in vitro synthesis of F0c single-cysteine derivatives were constructed by replacing the GCG codon at position 40 or position 79 of atpE, respectively, with a TGC codon. Plasmid pBWU13 was provided by K. Altendorf and G. Deckers-Hebestreit (University of Osnabrück, Osnabrück, Germany). Strain HDB51 was obtained from J. Luirink (Free University Amsterdam, Amsterdam, Netherlands).
Depletion of YidC and Ffh
Depletion of YidC and Ffh was performed essentially as described by Nouwen et al. (2001) for SecDFyajC. For YidC depletion, an overnight culture E. coli JS7131 grown in Luria-Bertoni (LB) medium supplemented with 0.2% (wt/vol) arabinose and 25 μg/ml spectinomycin was diluted 1:100 into the same medium without the antibiotic and grown until an OD660 of 0.8. Wild-type IMVs were prepared from these cells. A part of the same culture was washed once with warm LB and resuspended in LB containing 0.2% (wt/vol) glucose at an OD660 of 0.4. After every generation, the culture was diluted with 1 vol of the same medium until the cells stopped growing. YidC-depleted IMVs were prepared from this culture. E. coli HDB51 was grown overnight in LB with 25 μg/ml kanamycin, 10 μg/ml tetracycline, 100 μg/ml ampicillin, and 0.2% (wt/vol) arabinose. Cells were diluted 1:200 into the same medium without antibiotic and grown until an OD660 of 0.2. HDB51 wild-type lysate was prepared from these cells. Depletion of Ffh was induced in the same way as described for YidC, with the exception that cells were initially resuspended at an OD660 of 0.1.
Proteinase K pretreatment of IMVs
To inactivate membrane proteins, IMVs were treated with 0.2 mg/ml proteinase K for 20 min on ice. Proteinase K activity was subsequently blocked by the addition of 0.5 mM PMSF, and IMVs were collected by centrifugation through a cushion consisting of 50 mM Hepes-KOH, pH 7.5, 0.5 mM PMSF, and 20% (wt/vol) sucrose. IMVs were washed with 50 mM Hepes-KOH, pH 7.5, and resuspended in 50 mM Hepes-KOH, pH 7.5, and 20% glycerol. Control IMVs were subjected to the same treatment, with the exception that proteinase K was left out.
In vitro transcription, translation, and insertion reaction
The RiboMAX™ in vitro transcription kit (Promega) was used for the synthesis of mRNA. Plasmids pBSKftsQ (FtsQ), pET20atpE (wild-type F0c), pET20atpE-A40C (F0c A40C, containing a unique cysteine residue at position 40), or pET20atpE-A79C (F0c A79C, containing a unique cysteine at position 79), respectively, served as DNA templates. In vitro translation–insertion reactions were performed for 20 min at 37°C in the presence of the indicated amounts of IMVs or (proteo-)liposomes as described previously (van der Laan et al., 2004).
Labeling of F0c A40C and F0c A79C
To determine the transmembrane topology of in vitro–synthesized and membrane-inserted F0c, translation reactions using F0c A40C or F0c A79C mRNAs and YidC proteoliposomes were performed in the absence of any radioactively labeled amino acid. Subsequently, 0.4 mg/ml protease K was added to degrade nonincorporated F0c. After 30 min on ice, 0.5 mM PMSF was added to block proteinase K activity. Proteoliposomes were isolated from the reaction mixture by centrifugation through a cushion consisting of 50 mM Hepes-KOH, pH 7.0, 50 mM KCl, and 20% (wt/vol) glycerol. Membranes were resuspended in 50 mM Hepes-KOH, pH 7.0, and 50 mM KCl, and were incubated for 10 min on ice in the presence or absence of 0.5 mM AMdiS (Molecular Probes, Inc.) as indicated. Subsequently, 1 mM fluorescein maleimide (Molecular Probes, Inc.) was added, and incubation was continued for another 10 min. Labeling reactions were quenched with 5 mM DTT and samples were subjected to precipitation with TCA. Samples were analyzed by 17.5% SDS-PAGE, and fluorescently labeled proteins were visualized using a Lumi-Imager F1 workstation (Roche).
Oligomerization of in vitro–synthesized F0c
F0c was synthesized as radioactively labeled protein in the presence of proteoliposomes containing either YidC, SecYEG together with YidC, or with no proteins. 0.4 mg/ml proteinase K was added and samples were incubated for 30 min on ice to degrade noninserted F0c. 0.5 mM PMSF was used to inactivate proteinase K, and F0c-loaded proteoliposomes were isolated by centrifugation through a cushion consisting of 50 mM Hepes-KOH, pH 8.0, 50 mM KCl, and 20% (vol/vol) sucrose. Pellets were resuspended in 50 μl solubilization buffer (50 mM Hepes, pH 8.0, 50 mM KCl, 20% glycerol, and 0.05% dodecyl maltoside) and were incubated for 15 min on ice. Subsequently, samples were mixed with gel-loading buffer. Blue Native PAGE analysis was performed on 8–18% gradient gels as described previously (Schägger and von Jagow, 1991).
Other methods
SecYEG (Manting et al., 2000) and YidC (van der Laan et al., 2001) were purified as described. Reconstitutions were performed as described previously (van der Laan et al., 2001). Where indicated, SecA or FtsY was removed from the lysate by immunodepletion (van der Laan et al., 2004); depletion was verified by immunoblotting. ProOmpA translocation experiments were performed for 20 min at 37°C using fluorescently labeled precursor protein (De Keyzer et al., 2002).
Acknowledgments
We would like to thank Eli van der Sluis for critically reading the manuscript.
This work was supported by the Earth and Life Sciences Foundation, which is subsidized by the Netherlands Organization for Scientific Research (program grant 809.65.012).
Abbreviations used in this paper: AMdiS, 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid; F0a, F0 subunit a; F0c, F0 subunit c; IMV, inner membrane vesicle; PMF, proton motive force; SRP, signal recognition particle.
References
- Ackerman, S.H. 2002. Atp11p and Atp12p are chaperones for F1-ATPase biogenesis in mitochondria. Biochim. Biophys. Acta. 1555:101–105. [DOI] [PubMed] [Google Scholar]
- Ackerman, S.H., and A. Tzagoloff. 1990. ATP10, a yeast nuclear gene required for the assembly of the mitochondrial F1F0 complex. J. Biol. Chem. 265:9952–9959. [PubMed] [Google Scholar]
- Altamura, N., N. Capitanio, N. Bonnefoy, S. Papa, and G. Dujardin. 1996. The Saccharomyces cerevisiae OXA1 gene is required for the correct assembly of cytochrome c oxidase and oligomycin-sensitive ATP synthase. FEBS Lett. 382:111–115. [DOI] [PubMed] [Google Scholar]
- Arechaga, I., P.J.G. Butler, and J.E. Walker. 2002. Self-assembly of ATP synthase subunit c rings. FEBS Lett. 515:189–193. [DOI] [PubMed] [Google Scholar]
- Baneyx, F., and G. Georgiou. 1990. In vivo degradation of secreted fusion proteins by the Escherichia coli outer membrane protease OmpT. J. Bacteriol. 172:491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer, M., M. Behrens, K. Esser, G. Michaelis, and E. Pratje. 1994. PET1402, a nuclear gene required for proteolytic processing of cytochrome oxidase subunit 2 in yeast. Mol. Gen. Genet. 245:272–278. [DOI] [PubMed] [Google Scholar]
- Beck, K., G. Eisner, D. Trescher, R.E. Dalbey, J. Brunner, and M. Müller. 2001. YidC, an assembly site for polytopic Escherichia coli membrane proteins located in immediate proximity to the SecYE translocon and lipids. EMBO Rep. 2:709–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capaldi, R.A., and R. Aggeler. 2002. Mechanism of the F1F0-type ATP synthase, a biological rotary motor. Trends Biochem. Sci. 27:154–160. [DOI] [PubMed] [Google Scholar]
- Carr, H.S., and D.R. Winge. 2003. Assembly of cytochrome c oxidase within the mitochondrion. Acc. Chem. Res. 36:309–316. [DOI] [PubMed] [Google Scholar]
- Chen, M., J.C. Samuelson, F. Jiang, M. Müller, A. Kuhn, and R.E. Dalbey. 2002. Direct interaction of YidC with the Sec-independent Pf3 coat protein during its membrane protein insertion. J. Biol. Chem. 277:7650–7675. [DOI] [PubMed] [Google Scholar]
- Date, T., J.M. Goodman, and W.T. Wickner. 1980. Procoat, the precursor of M13 coat protein, requires an electrochemical potential for membrane insertion. Proc. Natl. Acad. Sci. USA. 77:4669–4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Gier, J.W., and J. Luirink. 2001. Biogenesis of inner membrane proteins in Escherichia coli. Mol. Microbiol. 40:314–322. [DOI] [PubMed] [Google Scholar]
- De Gier, J.W., P.A. Scotti, A. Saaf, Q.A. Valent, A. Kuhn, J. Luirink, and G. von Heijne. 1998. Differential use of the signal recognition particle translocase targeting pathway for inner membrane protein assembly in Escherichia coli. Proc. Natl. Acad. Sci. USA. 95:14646–14651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Keyzer, J., C. van der Does, and A.J. Driessen. 2002. Kinetic analysis of the translocation of fluorescent precursor proteins into Escherichia coli membrane vesicles. J. Biol. Chem. 277:46059–46065. [DOI] [PubMed] [Google Scholar]
- Facey, S.J., and A. Kuhn. 2003. The sensor protein KdpD inserts into the Escherichia coli membrane independent of the Sec translocase and YidC. Eur. J. Biochem. 270:1724–1734. [DOI] [PubMed] [Google Scholar]
- Fröderberg, L., E. Houben, J.C. Samuelson, M. Chen, S.K. Park, G.J. Phillips, R. Dalbey, J. Luirink, and J.W. de Gier. 2003. Versatility of inner membrane protein biogenesis in Escherichia coli. Mol. Microbiol. 47:1015–1027. [DOI] [PubMed] [Google Scholar]
- Geller, B.L., and W. Wickner. 1985. M13 procoat inserts into liposomes in the absence of other membrane proteins. J. Biol. Chem. 260:13281–13285. [PubMed] [Google Scholar]
- Girvin, M.E., V.K. Rastogi, F. Abildgaard, J.L. Markley, and R.H. Fillingame. 1998. Solution structure of the transmembrane H+-transporting subunit c of the F1F0 ATP synthase. Biochemistry. 37:8817–8824. [DOI] [PubMed] [Google Scholar]
- He, S., and T.D. Fox. 1997. Membrane translocation of mitochondrially coded Cox2p: distinct requirements for export of N and C termini and dependence on the conserved protein Oxa1p. Mol. Biol. Cell. 8:1449–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermolin, J., and R.H. Fillingame. 1995. Assembly of F0 sector of Escherichia coli H+-ATP synthase. J. Biol. Chem. 270:2815–2817. [DOI] [PubMed] [Google Scholar]
- Herrmann, J.M., W. Neupert, and R.A. Stuart. 1997. Insertion into the mitochondrial inner membrane of a polytopic protein, the nuclear-encoded Oxa1p. EMBO J. 16:2217–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskovits, A.A., E.S. Bochkareva, and E. Bibi. 2000. New prospects in studying the bacterial signal recognition particle pathway. Mol. Microbiol. 38:927–939. [DOI] [PubMed] [Google Scholar]
- Houben, E.N., P.A. Scotti, Q.A. Valent, J. Brunner, J.L. de Gier, B. Oudega, and J. Luirink. 2000. Nascent Lep inserts into the Escherichia coli inner membrane in the vicinity of YidC, SecY and SecA. FEBS Lett. 476:229–233. [DOI] [PubMed] [Google Scholar]
- Iwamoto, A., H. Omote, H. Yanada, N. Tomioka, A. Itai, M. Maeda, and M. Futai. 1991. Mutations in Ser174 and the glycine-rich sequence (Gly149, Gly150, and Thr156) in the β subunit of Escherichia coli H+-ATPase. J. Biol. Chem. 266:16350–16355. [PubMed] [Google Scholar]
- Jia, L., M. Dienhart, M. Schramp, M. McCauley, K. Hell, and R.A. Stuart. 2003. Yeast Oxa1 interacts with mitochondrial ribosomes: the importance of the C-terminal region of Oxa1. EMBO J. 22:6438–6447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer, D., and A. Kuhn. 1999. Hydrophobic forces drive spontaneous membrane insertion of the bacteriophage Pf3 coat protein without topological control. EMBO J. 18:6299–6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn, A., R.A. Stuart, R. Henry, and R.E. Dalbey. 2003. The Alb3/Oxa1/YidC protein family: membrane-localized chaperones facilitating membrane protein insertion? Trends Cell Biol. 13:510–516. [DOI] [PubMed] [Google Scholar]
- Lee, H.C., and H.D. Bernstein. 2001. The targeting pathway of Escherichia coli presecretory and integral membrane proteins is specified by the hydrophobicity of the targeting signal. Proc. Natl. Acad. Sci. USA. 98:3471–3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manting, E.H., C. van Der Does, H. Remigy, A. Engel, and A.J. Driessen. 2000. SecYEG assembles into a tetramer to form the active protein translocation channel. EMBO J. 19:852–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, M., M.S. Harrison, E.C. Peterson, and R. Henry. 2000. Chloroplast Oxa1p homolog albino3 is required for post-translational integration of the light harvesting chlorophyll-binding protein into thylakoid membranes. J. Biol. Chem. 275:1529–1532. [DOI] [PubMed] [Google Scholar]
- Moore, M., R.L. Goforth, H. Mori, and R. Henry. 2003. Functional interaction of chloroplast SRP/FtsY with the ALB3 translocase in thylakoids: substrate not required. J. Cell Biol. 162:1245–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouwen, N., M. van der Laan, and A.J. Driessen. 2001. SecDFyajC is not required for the maintenance of the proton motive force. FEBS Lett. 508:103–106. [DOI] [PubMed] [Google Scholar]
- Ridder, A.N., S. Morein, J.G. Stam, A. Kuhn, B. de Kruijff, and J.A. Killian. 2000. Analysis of the role of interfacial tryptophan residues in controlling the topology of membrane proteins. Biochemistry. 39:6521–6528. [DOI] [PubMed] [Google Scholar]
- Russel, M., N.A. Linderoth, and A. Sali. 1997. Filamentous phage assembly: variations on a protein export theme. Gene. 192:23–32. [DOI] [PubMed] [Google Scholar]
- Samuelson, J.C., M. Chen, F. Jiang, I. Möller, M. Wiedmann, A. Kuhn, G.J. Phillips, and R.E. Dalbey. 2000. YidC mediates membrane protein insertion in bacteria. Nature. 406:637–641. [DOI] [PubMed] [Google Scholar]
- Schägger, H., and G. von Jagow. 1991. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal. Biochem. 199:223–231. [DOI] [PubMed] [Google Scholar]
- Scotti, P.A., M.L. Urbanus, J. Brunner, J.W. de Gier, G. von Heijne, C. van der Does, A.J. Driessen, B. Oudega, and J. Luirink. 2000. YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 19:542–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serek, J., G. Bauer-Manz, G. Struhalla, L. Van Den Berg, D. Kiefer, R.E. Dalbey, and A. Kuhn. 2004. Escherichia coli YidC is a membrane insertase for Sec- independent proteins. EMBO J. 23:294–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart, R.A. 2002. Insertion of proteins into the inner membrane of mitochondria: the role of the Oxa1 complex. Biochim. Biophys. Acta. 1592:79–87. [DOI] [PubMed] [Google Scholar]
- Szyrach, G., M. Ott, N. Bonnefoy, W. Neupert, and J.M. Herrmann. 2003. Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J. 22:6448–6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Does, C., T. den Blaauwen, J.G. de Wit, E.H. Manting, N.A. Groot, P. Fekkes, and A.J. Driessen. 1996. SecA is an intrinsic subunit of the Escherichia coli preprotein translocase and exposes its carboxyl terminus to the periplasm. Mol. Microbiol. 22:619–629. [DOI] [PubMed] [Google Scholar]
- van der Laan, M., E.N. Houben, N. Nouwen, J. Luirink, and A.J. Driessen. 2001. Reconstitution of Sec-dependent membrane protein insertion: nascent FtsQ interacts with YidC in a SecYEG-dependent manner. EMBO Rep. 2:519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan, M., M.L. Urbanus, C.M. ten Hagen-Jongman, N. Nouwen, B. Oudega, N. Harms, A.J. Driessen, and J. Luirink. 2003. A conserved function of YidC in the biogenesis of respiratory chain complexes. Proc. Natl. Acad. Sci. USA. 100:5801–5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan, M., N. Nouwen, and A.J. Driessen. 2004. SecYEG proteoliposomes catalyze the Δψ-dependent membrane insertion of FtsQ. J. Biol. Chem. 279:1659–1664. [DOI] [PubMed] [Google Scholar]
- Weber, J., and A.E. Senior. 2003. ATP synthesis driven by proton transport in F1F0-ATP synthase. FEBS Lett. 545:61–70. [DOI] [PubMed] [Google Scholar]
- Yi, L., F. Jiang, M. Chen, B. Cain, A. Bolhuis, and R.E. Dalbey. 2003. YidC is strictly required for membrane insertion of subunits a and c of the F1F0 ATP synthase and SecE of the SecYEG translocase. Biochemistry. 42:10537–10544. [DOI] [PubMed] [Google Scholar]