Abstract
In Saccharomyces cerevisiae, spindle orientation is controlled by a temporal and spatial program of microtubule (MT)–cortex interactions. This program requires Bud6p/Aip3p to direct the old pole to the bud and confine the new pole to the mother cell. Bud6p function has been linked to Kar9p, a protein guiding MTs along actin cables. Here, we show that Kar9p does not mediate Bud6p functions in spindle orientation. Based on live microscopy analysis, kar9Δ cells maintained Bud6p-dependent MT capture. Conversely, bud6Δ cells supported Kar9p-associated MT delivery to the bud. Moreover, additive phenotypes in bud6Δ kar9Δ or bud6Δ dyn1Δ mutants underscored the separate contributions of Bud6p, Kar9p, and dynein to spindle positioning. Finally, tub2 C354S, a mutation decreasing MT dynamics, suppressed a kar9Δ mutation in a BUD6-dependent manner. Thus, Kar9p-independent capture at Bud6p sites can effect spindle orientation provided MT turnover is reduced. Together, these results demonstrate Bud6p function in MT capture at the cell cortex, independent of Kar9p-mediated MT delivery along actin cables.
Introduction
The fidelity of cell division rests on a precise spatial and temporal relationship between chromosomal segregation and the division plane. The axis for chromosomal segregation is defined by the position of the spindle poles, which generate both the mitotic spindle and astral microtubules (MTs) that interact with the cell cortex. Cell division occurs perpendicular to this axis, once chromosomal segregation is complete (Rappaport, 1996; Glotzer, 2001).
The budding yeast Saccharomyces cerevisiae is a powerful model for understanding the multiple layers of control operating to orient the mitotic spindle in a cell dividing asymmetrically. Yeast cells position the preanaphase spindle along the mother-bud axis near the bud neck. As the spindle elongates during anaphase, one pole translocates into the bud and chromosomal segregation between the mother and daughter cell is accomplished (Segal and Bloom, 2001).
Yeast cells control spindle position through the functional counterpart of the centrosome, the spindle pole body (SPB), which interacts with the cell cortex via astral MTs (Carminati and Stearns, 1997; Shaw et al., 1997; Segal et al., 2000a). These interactions are confined to specific areas of the cell cortex according to a program that appears to follow the cortical distribution of the actin-interactor Bud6p/Aip3p throughout the cell cycle (Amberg et al., 1997; Segal et al., 2002).
Current models assign a key role to Kar9p in enforcing daughter-bound fate of a single SPB after spindle assembly (Kusch et al., 2003). Kar9p interacts with the MT-binding protein Bim1p, the yeast EB1 homologue (Tirnauer et al., 1999; Korinek et al., 2000; Lee et al., 2000; Miller et al., 2000), and is preferentially recruited at the daughter-bound SPB (SPBd) to travel toward the MT plus end (Liakopoulos et al., 2003). In turn, Kar9p-bound MTs interact with the cargo domain of Myo2p to be guided along actin cables to the bud (Beach et al., 2000; Yin et al., 2000; Hwang et al., 2003). This model explains the dependency of spindle orientation on the actin cytoskeleton (Palmer et al., 1992; Theesfeld et al., 1999). However, it does not address the issue of how SPB asymmetry is established or the nature of the relationship between Kar9p and Bud6p in MT capture. A more integral model must also incorporate the underlying asymmetric behavior between the old and new SPB with regard to astral MT organization (Shaw et al., 1997, Segal et al., 2000a). This intrinsic asymmetry requires Cdk activity (Segal et al., 2000a) but is independent of cortical determinants or Kar9p (Yeh et al., 2000).
In light of refined proposals suggesting that Kar9p provides a bridge between MTs and actin transport to be delivered to the bud while bound to MTs exclusively (Hwang et al., 2003; Kusch et al., 2003; Liakopoulos et al., 2003), the role of Bud6p in capture at the cell cortex must be revisited and incorporated into these models. Alternatively, Bud6p has been implicated in organization of actin cables (Amberg et al., 1997; Moseley et al., 2004). According to this view, Bud6p role in MT capture is limited to supporting Kar9p function (Heil-Chapdelaine et al., 1999).
Here, we have explored the relationship between Kar9p and Bud6p. We used time-lapse analysis to examine Bud6p–MT interactions in kar9Δ cells and, conversely, the impact of a bud6Δ mutation on Kar9p-mediated MT delivery to the bud. We also analyzed kar9Δ bud6Δ cells to test for synergism between the two mutations. Finally, we showed the ability of a tub2 allele that decreases MT turnover (Gupta et al., 2002), to suppress a kar9Δ mutation in a Bud6p-dependent manner. Our data suggest that Bud6p role in MT capture at the cell cortex is separable from Kar9p-mediated delivery of MTs to the bud along actin cables.
Results
MT–Bud6p dynamic interactions in kar9Δ cells
We have previously studied the dynamic behavior of astral MT–cortex interactions in wild-type cells expressing GFP-Bud6 and GFP-Tub1. This analysis showed a high incidence of MT interactions at GFP-Bud6 cortical sites throughout the cell cycle. Moreover, shrinkage of MTs at the cortex occurs at Bud6p sites. This mode of interaction is abolished in bud6Δ cells and is therefore Bud6p-dependent (Segal et al., 2002).
To evaluate the involvement of Kar9p in Bud6p-associated MT capture, we undertook a comparative analysis of wild-type versus kar9Δ cells coexpressing GFP-Bud6 and GFP-α tubulin (Tub1p) fusions. Interactions were studied along the cell cycle divided arbitrarily into three stages based on spindle pathway landmarks and the program of Bud6p localization (Fig. 1 A).
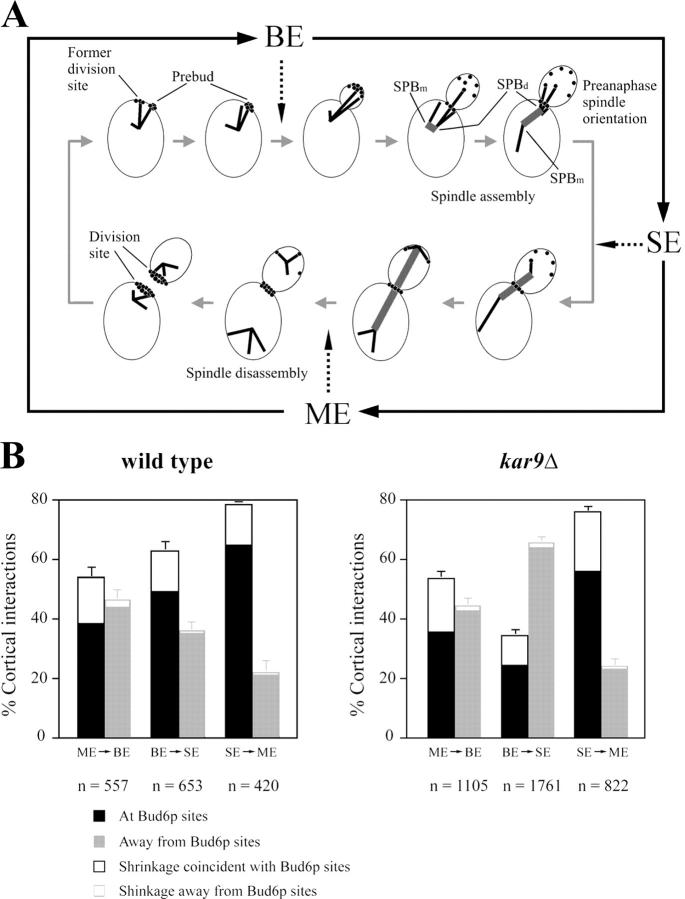
Figure 1.
Dynamic behavior of cortical Bud6p–astral MT interactions in kar9Δ cells. (A) Landmark events in Bud6p-mediated spindle orientation in wild-type cells. Solid black arrows show operational definition of cell cycle stages used in this work. In an unbudded cell (top left), astral MTs interact with Bud6p (black circles) at the recent division site. Accumulation of Bud6p at the prebud drives MT interactions toward the new budding site with minimal probing of the mother cortex. After bud emergence (BE), interactions continue with the bud cortex. As the spindle assembles, Bud6p accumulates at the bud neck. This causes the daughter-bound pole (SPBd) to interact with the bud and bud neck cortex whereas the SPBm is prevented from interacting with the bud. As a result, the preanaphase spindle orients along the mother-bud axis. During spindle elongation (SE), the SPBd translocates into the bud and MT interactions with Bud6p continue until spindle disassembly at mitotic exit (ME). After spindle disassembly, MT interactions focus on the Bud6p ring directing SPBs toward the recent division site. (B) Distribution of astral MT–cortex interactions in wild-type versus kar9Δ cells expressing GFP-Bud6 and GFP-Tub1. MT–cortex interactions were scored in time-lapse recordings of 54 wild-type and 172 kar9Δ cells as described in Materials and methods. The distribution shows interactions coinciding with cortical GFP-Bud6 (black bars) or away from GFP-Bud6 (gray bars). Open boxes in each category indicate percentage of shrinkage at the cell cortex as an indicator of a Bud6p-dependent mode of interaction (Segal et al., 2002). n = total number of cortical interactions scored. At least data from 320 (wild type) or 690 (kar9Δ) MTs were collected. Error bars indicate 95% confidence limits.
The kar9Δ mutation did not affect MT–Bud6p contacts from mitotic exit to generation of a new budding site (ME to BE: 54.7%, n = 1105 in kar9Δ vs. 53.8%, n = 557 in wild type, Fig. 1 B). Once the Bud6p ring at the previous division site disassembled and accumulation began at the prebud site, MTs reoriented to this new area of capture (Fig. 2 A). In addition, interactions at Bud6p sites occurred at wild-type frequencies from onset of anaphase to mitotic exit in kar9Δ cells (Fig. 1 B, SE to ME: 76.1%, n = 822 in kar9Δ vs. 78.1%, n = 420 in wild type) and MT shrinkage at Bud6p sites was unperturbed (Fig. 1 B, open boxes within black bars).
Figure 2.
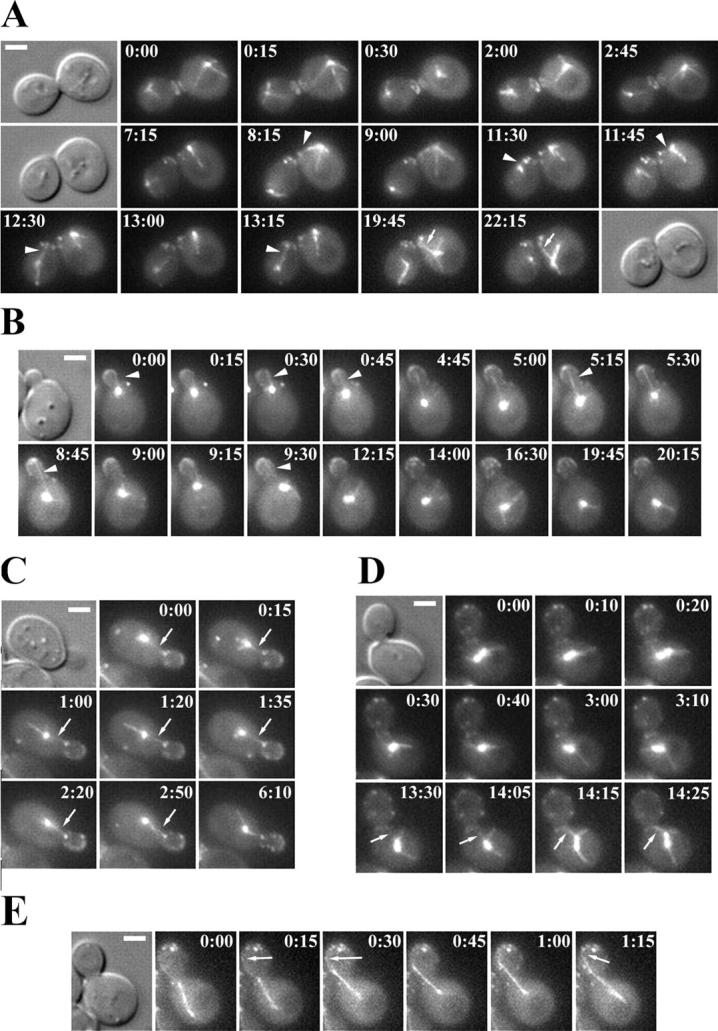
Bud6p–astral MT interactions in kar9Δ cells. Selected frames from representative time-lapse series analyzed in Fig. 1 B, illustrating dynamic Bud6p-astral MT interactions in kar9Δ GFP:BUD6 GFP:TUB1 cells. (A) After mitotic exit and cytokinesis (0:00), astral MTs from each SPB progressively reached for the recent division site decorated by a GFP-Bud6 ring (arrowheads). In the mother cell, these interactions shifted to the prebud site newly marked by GFP-Bud6 (19:45–22:15, arrow). (B) MT interactions with the cortex of a small bud (0:00–9:30, arrowheads) were progressively lost as the spindle began to assemble. MT contacts within the mother cell became then randomized (12:15–20:15). (C) MT interactions with cortical Bud6p within the proximal region of the bud were coupled to MT growth and shrinkage (arrows). (D) MT–Bud6p interactions at the bud neck cortex after spindle assembly. These interactions led to movement of one SPB toward the bud neck (13:30–14:25, arrows). (E) MT–Bud6p interactions during anaphase. A budded cell initiating spindle elongation within the mother exhibited MT interactions with cortical Bud6p within the bud (arrows), which accompanied orientation of the spindle in mid-anaphase. Time elapsed is indicated in min:s. Bars, 2 μm.
In contrast, kar9Δ cells exhibited a marked decrease in interactions with Bud6p decorated areas after bud emergence (Fig. 1 B, BE to SE). Cells showed repeated MT interactions with the bud tip in small-budded cells (100% cells within 10 min from bud emergence, n = 32). Yet, as the bud continued to grow, cells failed to maintain MTs oriented in the bud (Fig. 2 B, arrowheads). As a result, MTs spent significant time probing the mother cell cortex away from Bud6p marked regions. However, once GFP-Bud6 decorated the bud neck, MTs resumed interactions with this discrete area (Fig. 2, C and D, arrows). In spite of the reduction in Bud6p–MT contacts in S phase, kar9Δ cells still showed MT shrinkage at Bud6p sites (Fig. 1 B).
Bud6p–astral MT interactions during anaphase in kar9Δ cells occurred at wild-type levels. These interactions contributed to spindle positioning even if anaphase began within the mother cell (Fig. 2 E).
The correlation between MT–Bud6p interactions and the duration of cortical contacts previously reported in wild-type cells (Segal et al., 2002) was still apparent in kar9Δ cells. On average, interactions at Bud6p sites lasted 2.2 ± 1.2 min (n = 75) from mitotic exit to bud emergence, 2.0 ± 0.3 min (n = 42) from bud emergence to spindle assembly and increased to 3.0 ± 1.8 min (n = 92) from spindle elongation to mitotic exit. In contrast, interactions away from Bud6p sites lasted 0.6 ± 0.2 min (n = 112). These values were in agreement with those in wild-type cells (for review see Segal et al., 2002).
Together, these results confirmed that a kar9Δ mutation did not alter the dynamic characteristics of cortical Bud6p–MT interactions, instead, it perturbed the maintenance of astral MT orientation toward the bud from late bud emergence through spindle assembly and thus, indirectly, decreased the incidence of capture at Bud6p sites.
Analysis of Kar9p-driven MT delivery to the bud in wild-type or bud6Δ cells
We then examined wild-type or bud6Δ cells expressing CFP-Tub1 and Kar9-GFP to determine the dynamic behavior of Kar9p bound to MTs, the effect on astral MT orientation to cell cortex areas and the relationship to MT-driven SPB movement toward the bud.
Decoration of MTs by Kar9-GFP was initiated by recruitment at the SPB in most cases, both in wild-type or bud6Δ cells (93.5%, n = 113 MTs and 96.2%, n = 104, respectively). Kar9p was detected at both SPBs at onset of spindle assembly (14 of 16 time-lapse series spanning spindle assembly) but was clearly asymmetric in spindles longer than 1.2 ± 0.2 μm (Fig. 3). Kar9p traveled along MTs toward the plus or minus end. In addition, Kar9p moved while fixed at the plus end of a growing or shrinking MT (Fig. 4 A). These modes of dynamic behavior occurred significantly in bud6Δ cells. However, the bud6Δ mutation slightly reduced Kar9p translocation along persistent MTs (Fig. 4 A, black and gray bars in wild type vs. bud6Δ).
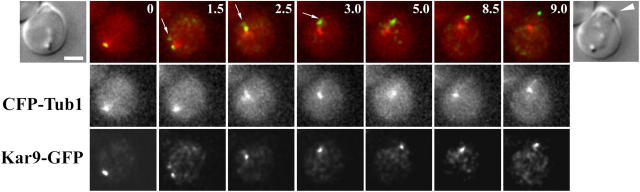
Figure 3.

Association of Kar9-GFP to SPBs during spindle assembly in wild-type cells. Selected frames of time-lapse series of wild-type cells expressing Kar9-GFP (overlaid in green) and CFP-Tub1 (red) showing Kar9p recruitment at both SPBs during the assembly of the mitotic spindle. The DIC image corresponds to the first frame. (A) Kar9-GFP associated with MTs from both SPBs. In the cell on the left, the label decorated one pole by 90 s (arrow). The cell on the right exhibited Kar9-GFP label on both poles during SPB separation (0–105 s). The label was present on a single pole by 150 s (arrow). (B) Kar9-GFP decorated both poles during the early part of spindle assembly (0–46 s). The time lapse also shows that SPB movement toward the bud did not coincide with the shrinkage of an MT decorated by Kar9-GFP (72–108 s, arrows). Instead, movement toward the bud occurred as an MT grew (168–192 s, arrows) and then maintained a constant length (192–216 s, arrows). (C) Kar9-GFP initially localized to MTs emanating from both SPBs (0 s) but only one pole established interactions with the bud (60–120 s, arrows), whereas label on the second pole progressively disappeared. Numbers indicate time elapsed in seconds. Bars, 2 μm.
Figure 4.
Kar9p dynamic behavior in wild-type versus bud6Δ cells. (A) Kar9p dynamic behavior in association with MTs in wild type (107 cells recorded) or bud6Δ (99 cells recorded). Transits along MTs or movement at the MT plus end were as described in Materials and methods. Open boxes in each category indicate percentage of events confined to the bud neck. Error bars indicate 95% confidence limits. (B–D) Representative time-lapse series of Kar9p dynamic behavior in association with MTs oriented toward the bud in wild-type cells. Overlays of CFP-Tub1 (red) and Kar9-GFP (green) images and the DIC image corresponding to the first frame are shown. (B) After bud emergence, a cell exhibited Kar9-GFP transits along MTs oriented toward the emerging bud. Arrows at 4 and 5 min point to an MT reaching the cortex in the absence of Kar9-GFP at the MT plus end. A second MT is decorated by Kar9-GFP at the plus end (4–5 min). (C) Kar9-GFP transits along MTs in a cell before spindle assembly. In the absence of Kar9-GFP association, MTs interacted with the cell cortex. Of two MTs directed toward the bud at 4 min (arrowheads), only one is decorated at the plus end by Kar9-GFP (arrows). A line scan for fluorescence intensity along the overlapping MTs (4 min) is shown below (arrows show position of MT plus ends). (D) After spindle assembly, Kar9-GFP was bound to the MT plus end (0 min) of a shrinking MT (0.5 min) followed by localization at the SPB in the presence of an MT interacting with the cell cortex (3 min, arrows). Numbers indicate time elapsed in minutes. Bars, 2 μm.
The distribution of cortical interactions involving Kar9p-bound MTs by cell compartment was altered in bud6Δ cells. In general, MTs decorated by Kar9p were already directed toward the bud or became oriented when Kar9p occupied the plus end (Fig. 4, B–D). A bud6Δ mutation markedly decreased interactions confined to the bud neck (Fig. 4 A, open portion of bars for each mode of dynamic behavior). Thus, Kar9p directed MTs to the bud, yet, Bud6p appeared to dictate bud neck capture.
Of all Kar9p-bound MTs in wild-type cells, 31% (n = 113 MTs) were associated with SPB movement toward the bud (24% in bud6Δ cells, n = 104 MTs).
To correlate the direction of movement with Kar9p dynamic behavior, all events involving Kar9p return to the spindle pole were scored for coupled SPB movement toward the cortex. As shown in Fig. 5 A, there was no correlation between the direction of SPB movement and shrinkage of Kar9p-bound MTs in wild-type or bud6Δ cells (< 7%, n > 120). In fact, the SPB remained stationary relative to the cortex in the majority of Kar9p returns to the pole.
Figure 5.
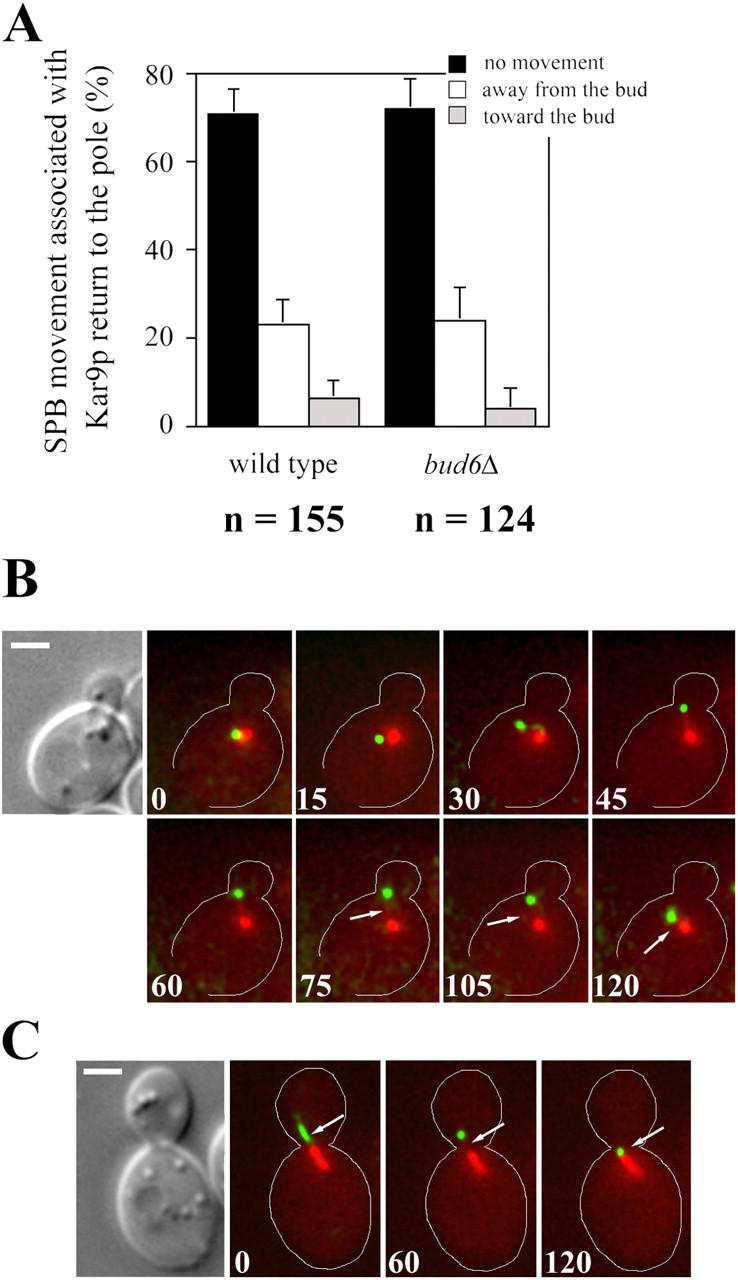
Analysis of SPB movement coupled to Kar9p-bound MTs. (A) SPB movement associated with Kar9p return to the pole in wild-type or bud6Δ cells was categorized as described in Materials and methods. Movement coupled to all Kar9p returns to the pole by either MT shrinkage or translocation was scored in time-lapse recordings described in Fig. 4. Error bars indicate 95% confidence limits. (B and C) Selected frames from a time-lapse series showing orientation of an MT decorated by Kar9-GFP at the plus end in a wild-type cell. Overlays of fluorescence images of CFP-Tub1 (red) and Kar9-GFP (green), and the DIC image corresponding to the first frame are shown. (B) A small-budded cell with the SPB already positioned near the bud neck showed Kar9-GFP localization at the plus end of a growing MT (15 s). After orientation toward the bud (30–75 s) the MT shrunk (arrows), bringing Kar9-GFP toward the SPB. Shrinkage was not coupled to SPB movement toward the bud (105–120 s). (C) Absence of coupling between shrinkage of an MT bound to Kar9-GFP (arrows) and SPB movement in a cell exhibiting an oriented preanaphase spindle. Numbers indicate time elapsed in seconds. Bars, 2 μm.
In wild-type cells, Kar9p dynamic behavior in the absence of SPB movement was confined to the vicinity of the bud neck within the mother (Fig. 5 B). This was associated with angular movement of MTs of constant length similar to previously reported Kar9p or Myo2p-dependent transports (Hwang et al., 2003; Liakopoulos et al., 2003). Alternatively, Kar9p-bound MTs shrunk away from the cortex without causing SPB movement (Fig. 5 C, arrow). In conclusion, Kar9p did not mediate SPB movement via changes in MT length. This was in contrast with, spindle orientation associated with cortical Bud6p relying primarily on MT shrinkage.
In bud6Δ cells SPBs were initially present away from the bud and became quickly repositioned as a Kar9p-bound MT moved toward the bud without observable shrinkage, presumably, along an actin cable (Fig. 6, arrows). This type of processive MT movements toward the bud were absent in bud6Δ kar9Δ cells (n > 150 MTs). Thus, a bud6Δ mutant supported Kar9p-dependent MT orientation and relied on long-range Kar9p-bound transport to compensate for lack of early MT–cortex interactions with the new bud and the inability to mobilize SPBs by MT shrinkage (Segal et al., 2002).
Figure 6.
Kar9p-mediated MT transport in a bud6Δ cell. Selected frames from a time-lapse series showing Kar9p-mediated orientation of the SPB in a bud6Δ cell expressing CFP-Tub1 and Kar9-GFP. Overlays of CFP-Tub1 (red) and Kar9-GFP (green) images are shown. The DIC images correspond to 0 and 9 min (arrowhead indicates the bud). Initially, the SPB was positioned far from the prebud site (0–1.5 min). As Kar9-GFP occupied the plus end of an MT (1.5–3.0 min, arrows), the SPB became rapidly oriented toward the budding site (5 min). Kar9-GFP then continued to direct MTs into the growing bud (5–9 min). Numbers indicate time elapsed in minutes. Bar, 2 μm.
Spindle orientation in bud6Δ kar9Δ or bud6Δ dyn1Δ mutants
Genetic analysis of spindle orientation has assigned motor activities and cortical determinants to putative early and late pathways required for spindle position (Heil-Chapdelaine et al., 1999). Kar9p and dynein are regarded as key components of the “early” and “late” pathways, respectively. To further assess the contribution of Bud6p in these pathways, the phenotypes resulting from double mutant combinations bud6Δ kar9Δ or bud6Δ dyn1Δ (DYN1/DHC1, dynein heavy chain; Eshel et al., 1993; Li et al., 1993) were characterized.
Astral MT behavior and spindle orientation defects observed in bud6Δ kar9Δ cells demonstrated the additive impact of deleting BUD6 over a single kar9Δ mutation. Early orientation of astral MTs toward the emerging bud was abolished as in single bud6Δ mutants (not depicted; Segal et al., 2002), whereas spindle positioning in the double mutant was markedly impaired relative to either single mutant (Table I and Fig. 7).
Table I.
Time-lapse analysis of spindle behavior at anaphase onset in bud6 , kar9 , and bud6kar9 mutants
| Strain | % lack of retention at the bud neck |
% correct alignmenta |
% mid-anaphase loss of positioning |
n b |
|---|---|---|---|---|
| wild type | 0 | 100 | 0 | 28 |
| bud6Δ | 27 | 87 | 13 | 15 |
| kar9Δ | 0 | 60 | 0 | 25 |
| kar9Δ bud6Δ | 26 | 33 | 13 | 39 |
Results are expressed as percentage relative to the total number of cells recorded.
Alignment of the spindle at anaphase onset was considered correct if an imaginary line drawn through the long axis of the spindle transversed the mother-bud neck (Theesfeld et al., 1999) as separation of the poles began at the “fast phase” of anaphase B (Yeh et al., 1995).
n = total number of anaphase cells recorded.
Figure 7.

Spindle orientation phenotypes in bud6Δ kar9Δ cells. Selected frames from representative time-lapse series showing spindle orientation defects in bud6Δ kar9Δ GFP:TUB1 cells. For quantitative information see Table I. (A) A misoriented preanaphase spindle (0–11.0 min) became aligned as an astral MT interacted with the bud cortex (12.0 min). The interaction, however, resulted in the translocation of the spindle past the bud neck (17.0–18.0 min). At onset of anaphase (18.5 min) an MT interacting with the mother cell cortex repositioned one pole past the bud neck within the mother cell as the spindle elongated. (B) A misaligned spindle began elongation within the mother cell in the absence of MT interactions with the bud neck (2.5–20.5 min). The spindle became oriented by dynein-driven sliding that pulled the mid-anaphase spindle into the bud (27.5–29.0 min). (C) A spindle that oriented at onset of anaphase along the mother-bud axis (4.5 min) and initiated spindle elongation across the bud neck (6.0–7.0 min) became misaligned part way through anaphase (8.0–8.5 min) but rapidly repositioned (9.0–10.0 min) to proceed with anaphase. Numbers indicate time elapsed in minutes. Bars, 2 μm.
A range of phenotypes in bud6Δ kar9Δ cells highlighted the contribution of Bud6p to cortical capture in the absence of Kar9p.
First, retention of the spindle at the bud neck was compromised causing transient positioning within the bud (Fig. 7 A, 17.0–18.5 min). This phenotype was as prevalent (26%, n = 39 cells recorded) as in bud6Δ cells and never observed in kar9Δ cells (Table I).
Second, onset of spindle elongation along the mother-bud axis was markedly reduced (33%, n = 39), possibly due to the additional absence of MT interactions with the bud neck (Fig. 7 B and see last section of Results describing Fig. 9 A). This contrasted with kar9Δ cells (Table I), in which MT interactions with the bud neck still contribute toward orientation (Segal et al., 2000b; and see last section of Results describing Fig. 9 A). Initial spindle elongation within the mother cell in kar9Δ bud6Δ cells was followed by dynein-driven positioning of the spindle part way through anaphase (Fig. 7 B) as in kar9Δ cells (Segal et al., 2000b; Yeh et al., 2000).
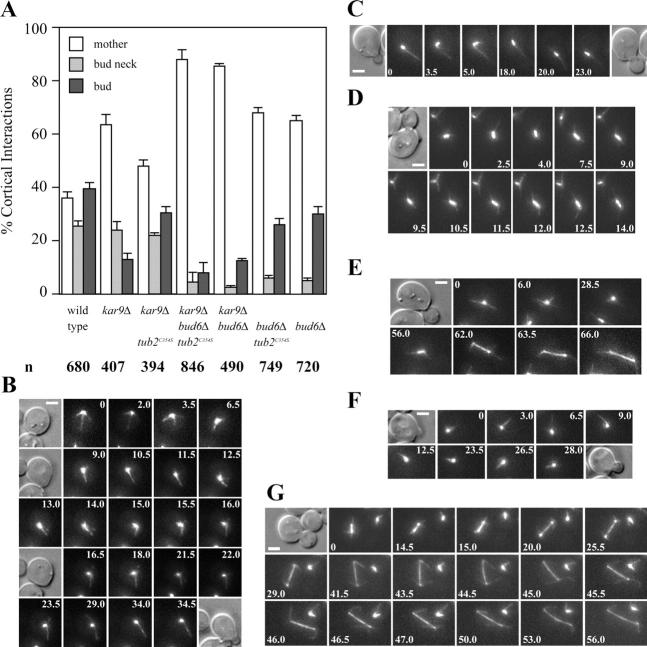
Figure 9.
Effect of decreased MT turnover on orientation of MT–cortex interactions in kar9Δ cells. (A) Astral MT–cortex interactions in the indicated strains expressing GFP-Tub1 (number of cells recorded: 81 wild type, 77 kar9Δ, 64 kar9Δ tub2 C354S, 52 kar9Δ tub2 C354S bud6Δ, 65 kar9Δ bud6Δ, 51 bud6Δ tub2 C354S, and 45 bud6Δ) were scored by cell compartment (mother, bud neck, or bud) from bud emergence to preanaphase spindle assembly. n = total number of interactions. Error bars indicate 95% confidence limits. (B–E) Selected frames from representative time-lapse series showing orientation of MT–cortex interactions in kar9Δ tub2 C354S GFP:TUB1 cells. (B) Early orientation of astral MT–cortex interactions toward the prebud site occurred after mitotic exit (9.0 min). Orientation was maintained throughout bud emergence (16.0–34.5 min). DIC images corresponding to 0, 9.0, 16.5, and 34.5 min are shown. (C) A small budded cell showing an astral MT oriented toward the bud (0 min) maintained its orientation throughout spindle assembly and alignment (18.0–23.0 min) of the preanaphase spindle along the mother-bud axis. DIC images correspond to the first and last frame of the series. (D) Selected frames from a time-lapse series showing preanaphase spindle orientation. A cell completed spindle assembly (0–7.5 min), maintaining MT–bud cortex interactions during spindle alignment. (E) Selected frames from a time-lapse series showing correct SPB orientation before spindle assembly (0–6 min). MT–cortex interactions within the bud continued throughout spindle assembly and orientation (56 min). The spindle was correctly aligned as the cell proceeded through anaphase (62–66 min). (F and G) Time-lapse series showing failure to orient MTs toward the emerging bud in a kar9Δ bud6Δ tub2 C354S GFP:TUB1 cell. (F) Astral MTs interacted with the cell cortex away from the bud. DIC images correspond to the first and last frame. (G) Defective preanaphase spindle orientation (0–15.0 min) and misaligned spindle elongation (20.0–29.0 min) followed by spindle orientation through dynein-dependent interactions (44.5–53.0 min) in mid-anaphase. Numbers indicate time elapsed in minutes. Bars, 2 μm.
Third, aberrant loss of spindle orientation in mid-anaphase of bud6Δ kar9Δ mutants (Fig. 7 C, 7.5–9.5 min), as observed in 13% (n = 39) of cells recorded (Table I), underscored Bud6p-dependent capture in anaphase. This indicated that dynein-driven events might be impaired yet sufficient to sustain viability of bud6Δ kar9Δ cells.
We also determined the effects of deleting BUD6 on spindle phenotypes of dyn1Δ cells, with particular focus on anaphase. In wild-type cells, the “fast phase” of spindle elongation (Fig. 8 A, 0–2.5 min and 6.0–10 min) is typically coupled to translocation of the SPBd into the bud (Yeh et al., 1995). SPB translocation is delayed in dyn1Δ mutants, although spindle elongation still begins along the mother-bud axis (Table II and Fig. 8 B). Spindles only became overtly misaligned during anaphase in 9% of cells (n = 35) after initial elongation along the mother-bud axis (Fig. 8 C).
Figure 8.
Spindle defects in bud6Δ dyn1Δ cells. (A) Spindle elongation and translocation of the SPBd into the bud are coupled in early anaphase of wild-type cells. Representative time-lapse series showing correct insertion of the spindle at the bud neck (0 min, 6.0 min) and initiation of spindle elongation across the bud neck (2.5 min, 8.5 min). For quantitative information see Table II. (B and C) Initiation of spindle elongation in dyn1Δ GFP:TUB1 cells occurred along the mother-bud axis but translocation of the SPBd into the bud was delayed. (B) SPBd translocation into the bud followed elongation of the spindle within the mother cell (9.0–12.0 min) without significant delay. (C) At anaphase the spindle rotated to become misaligned (25.5–43.0 min). (D–F) Spindle defects in bud6Δ dyn1Δ GFP:TUB1 cells. (D) After initiation of spindle elongation (1.5–2.5 min) the spindle became oriented along the mother-bud axis and continued elongation past the bud neck (5.0–9.0 min). (E) Spindle morphologies underscoring the genetic interaction between dyn1Δ and bud6Δ observed in asynchronous cells. Pairs of DIC and fluorescence images are shown. (a) Initiation of anaphase in the mother cell as in dyn1Δ cells. (b–d) Disrupted spindle polarity in anaphase. (d) Spindle elongation continued within the mother cell causing the spindle to curve. (e) Spindle disassembly in a mother cell followed by rebudding. (f) Cells containing multiple SPBs reached 10% in asynchronous populations. (F) Time-lapse series showing spindle disassembly (7.5–10.0 min) within the mother cell. Numbers indicate time elapsed in minutes. Bars, 2 μm.
Table II.
Spindle behavior in bud6 , dyn1 , and bud6dyn1 mutants
| Strain | % correctly aligned spindles at onset of anaphasea |
% late anaphase spindles exhibiting correct polarityb |
|---|---|---|
| wild type | 100 (n = 28) | 100 (n = 200) |
| bud6Δ | 87 (n = 15) | 92 (n = 300) |
| dyn1Δ | 94c (n = 35) | 90 (n = 210) |
| bud6Δ dyn1Δ | 72 (n = 18) | 60 (n = 302) |
Alignment of the spindle at anaphase onset was scored as described in Table I. Results are expressed as percentage of total anaphase cells recorded (n).
Polarity in late anaphase spindles was considered correct if a single pole was still connected to the bud cortex, irrespective of spindle alignment. Results are expressed as percentage of total anaphase cells counted (n) in asynchronous populations of the indicated strains.
In 9% of anaphase cells recorded, the spindle became overtly misaligned during anaphase, after initial elongation along the mother-bud axis (Fig. 8 C).
In contrast, bud6Δ dyn1Δ mutants already showed a mild preanaphase spindle orientation defect and concomitant reduction of spindle elongation along the mother-bud axis (Table II). Spindle alignment could be rectified during early anaphase, presumably, through Kar9p-directed astral MT delivery to the bud (Fig. 8 D).
Impaired MT capture at the bud neck in dyn1Δ bud6Δ cells led to loss of polarity of mid-anaphase spindles held within the mother cell, as astral MTs emerging from both poles entered the bud (Fig. 8 E; for review see Yeh et al., 2000). In asynchronous populations, only 60% (n = 302) of late anaphase spindles in dyn1Δ bud6Δ cells retained apparent polarity (only one SPB connected to the bud) in contrast to 90% (n = 210) in dyn1Δ cells (Table II).
Finally, bud6Δ dyn1Δ cells containing misaligned anaphase spindles within the mother failed to restrain mitotic exit. Progression of the spindle pathway past spindle disassembly was sufficiently frequent to be recorded by real-time microscopy (Fig. 8 F, 6.0–10.0 min). Accordingly, asynchronous bud6Δ dyn1Δ cultures contained 10% of cells exhibiting supernumerary SPBs (Fig. 8, E and F). Such cells were not present in bud6Δ kar9Δ or single bud6Δ, kar9Δ, or dyn1Δ mutant populations (n = 1,000 cells).
The observed interactions between bud6Δ and kar9Δ or dyn1Δ mutations stress the separate contributions of Kar9p and Bud6p to spindle orientation. In addition, the impact of Bud6p-dependent MT capture at the bud neck for correct spindle dynamics and progression (Segal et al., 2000b) was further demonstrated by the more complex spindle defects of bud6Δ kar9Δ or bud6Δ dyn1Δ mutants during anaphase.
Effect of reduced MT turnover on spatial distribution of MT–cortex interactions in kar9Δ cells
The observed dynamic behavior of Kar9p was difficult to reconcile with the proposed role for Kar9p in cortical capture of MTs. Kar9p kept a fixed distance from the SPB during MT tracking along actin cables, and frequently decorated MTs already reaching within the bud. Moreover, the phenotype of kar9Δ cells after bud emergence (Fig. 2 B), suggested that Kar9p might additionally control MT dynamic behavior to promote persistence of MTs in the bud. This raised the possibility that the requirement for Kar9p could be bypassed by a decrease in MT instability. Under these conditions, MT capture coincident with cortical Bud6p in the bud might prevail and suppress the spindle orientation defect of kar9Δ mutants.
To address this question, time-lapse analysis was performed to compare wild-type, kar9Δ, and kar9Δ tub2 C354S cells for the distribution of MT–cortex interactions by cell compartment from bud emergence to assembly of ∼2-μm-long spindles (Fig. 9 A). The tub2 C354S allele was introduced to reduce MT dynamics (Gupta et al., 2002).
Wild-type cells exhibited a characteristic distribution of MT–cortex interactions in the mother, the bud and at the bud neck. These interactions (Fig. 9 A) led to correctly oriented preanaphase spindles, which initiated elongation along the mother-bud axis (Table III). In kar9Δ cells, interactions within the bud were selectively decreased (Fig. 9 A). As a result, correct alignment at anaphase onset was reduced to 60% (n = 25; Table III).
Table III.
Effect of tub2 C354S on spindle alignment in kar9 Δ cells
| Strain | % spindle alignmenta
at anaphase onset in time-lapse series |
% correct preanaphaseb
spindle orientation in asynchronous cells |
|---|---|---|
| wild type | 100 (n = 28) | 98 ± 2 |
| kar9Δ | 60 (n = 25) | 63 ± 4 |
| kar9Δ tub2 C354S | 86 (n = 22) | 90 ± 3 |
| kar9Δ tub C354S bud6Δ | 25 (n = 40) | 29 ± 4 |
| kar9Δ bud6Δ | 33 (n = 39) | 32 ± 3 |
| bud6Δ tub2 C354S | 80 (n = 20) | 72 ± 3 |
| bud6Δ | 87 (n = 15) | 79 ± 4 |
Scoring was carried out in time-lapse series spanning early anaphase as indicated in Table I. n = total anaphase cells recorded.
Spindle orientation was scored in 500 preanaphase cells in asynchronous cultures of the indicated strains expressing GFP-Tub1. Results were expressed as mean ± SD for triplicate experiments. Alignment of the preanaphase spindle was considered correct if an imaginary line drawn through the long axis of the spindle transversed the mother-bud neck (Theesfeld et al., 1999).
Introduction of the tub2 C354S allele in the kar9Δ mutant restored astral MT–cortex interactions with the bud (Fig. 9, A and B) and significantly improved preanaphase spindle orientation (Fig. 9, C–E; Table III). Astral MTs became oriented toward the bud (Fig. 9 B) and continued to interact with the bud cortex until anaphase (Fig. 9, B–E). Elongation of the spindle along the mother-bud axis was increased to 86% of anaphase cells recorded (n = 22; Table III).
The tub2 C354S allele could not correct spindle orientation in kar9Δ bud6Δ cells (Fig. 9, A, F, and G; Table III). The kar9Δ bud6Δ tub2 C354S mutant initiated anaphase along the mother-bud axis in only 25% of cells recorded (n = 40). This highlighted the importance of Bud6p in directing MT capture at the incipient bud cortex. Accordingly, tub2 C354S failed to change bud6Δ mutant phenotypes (Fig. 9 A and Table III).
These data were validated by separately scoring preanaphase spindle orientation in asynchronous cultures of all strains studied by time-lapse analysis (Table III).
Thus, decreased MT turnover suppressed spindle orientation defects of kar9Δ cells, in a Bud6p-dependent manner. This result confirmed the participation of Bud6p in MT capture at the cell cortex and pointed to an additional role for Kar9p in control of MT dynamic behavior. Indeed, Kar9p-bound MTs entering the bud underwent repeated cycles of recovery (unpublished data). Such cycles were confirmed by time-lapse analysis of wild-type cells expressing GFP-Tub1 (at least 2 cycles in 60% of MTs entering the bud, n = 135) but were markedly decreased in a kar9Δ mutant in which 97% of MTs entering the bud (n = 105) underwent a single cycle of growth and shrinkage past the bud neck (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200407167/DC1).
Discussion
Kar9p is not directly involved in cortical capture at Bud6p sites
The presumed involvement of Bud6p in actin organization has prompted models in which the role of Bud6p in spindle orientation is based on supporting Kar9p function (Heil-Chapdelaine, et al., 1999; Miller et al., 1999). These models assumed that either Kar9p localization at the bud cortex requires actin or, alternatively, Kar9p bound to MT plus ends interfaces with Myo2p-dependent transport. Accordingly, by perturbing the actin cytoskeleton, a bud6Δ mutation disrupts Kar9p-mediated spindle orientation.
Here, we show that the dynamic properties of Bud6p–MT interactions are unchanged by a kar9Δ mutation. MTs kinetically followed Bud6p from onset of anaphase until bud emergence in the next cell cycle as in wild-type cells (Figs. 1 and 2). In addition, Bud6p-dependent MT shrinkage coupled to SPB movement, was unperturbed. A kar9Δ mutation decreased the orientation and maintenance of MTs toward the bud in small-budded cells (Fig. 1 B and Fig. 2 B) thus, reducing Bud6p–MT interactions during S phase. Yet, kar9Δ cells were proficient in MT–Bud6p interactions at the bud neck after SPB separation (Fig. 2, C and D).
Together, these results indicated that MT capture at Bud6p sites is independent of Kar9p, yet, the efficiency by which MTs entered and were maintained in the bud depended on Kar9p. The role of Kar9p became critical during S phase until preanaphase orientation was accomplished, demarking the end of the actin-sensitive period of spindle orientation (Theesfeld et al., 1999). Bud6p functions in cortical capture beyond this period further support a role that is separable from its links with the actin cytoskeleton (Moseley et al., 2004).
Kar9p-mediated delivery of astral MTs to the bud occurs in bud6Δ cells
To correlate the behavior of Kar9p with parameters of astral MT function, we examined wild-type cells coexpressing CFP-Tub1 and Kar9-GFP. We also asked whether a bud6Δ mutation perturbed the behavior of Kar9p-bound MTs in support of a functional link between Bud6p and Kar9p.
Kar9p was present at both SPBs during spindle assembly but became asymmetric once the identity of the poles was defined (Fig. 3). This dependency on correctly polarized MT–cortex interactions for asymmetric recruitment to the SPBd resembled that reported for mitotic exit regulators (Pereira et al., 2001; Smeets and Segal, 2002). Accordingly, Maekawa and Schiebel (2004) demonstrated that Cdc28p-Clb4p is recruited, in fact, to the SPBd via asymmetrically localized Kar9p. This precludes the converse view that Cdc28p-Clb4p prevents recruitment of Kar9p at the mother-bound SPB (SPBm) to generate polarity.
Kar9p traveled along MTs or moved when fixed at the MT plus end during periods of MT shortening or growth. Yet, association of Kar9p with an MT plus end did not always coincide with its interaction with the cell cortex (Fig. 4, B–D) or necessarily precede orientation of MTs toward the bud. These findings are not compatible with Kar9p serving solely as a cortical anchor or a bridge with the actin cytoskeleton.
Kar9p-bound MTs were directed to the bud in bud6Δ cells (Figs. 4 and 6), whereas events ending at the bud neck were markedly reduced (Fig. 4 A). Thus, a bud6Δ mutation did not perturb Kar9p-based MT delivery but impaired interactions with the bud neck or bud cortex.
The role of septins in MT capture at the bud neck has been previously addressed but was inferred to reflect Kar9p-dependent capture (Kusch et al., 2002). Yet, a bud6Δ mutation was not evaluated in that work. Because MT capture at the bud neck is proficient in kar9Δ cells (Fig. 2), whereas defective in a bud6Δ mutant (Fig. 4; Segal et al., 2002), it is clear that Bud6p, rather than Kar9p, creates an area of specialized capture at the bud neck. Indeed, disruption of the septin ring after a temperature shift in cdc3 ts, cdc10 ts, or cdc11 ts strains impaired Bud6p localization to the bud neck in S phase (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200407167/DC1), explaining the disruption of astral MT–bud neck interactions in these mutants.
Finally, Kar9p-bound MTs did not support shrinkage coupled to SPB movement toward the cortex (Fig. 5 A). Interestingly, although the SPBd is associated with, on average, three MTs in wild-type cells only one is usually decorated by Kar9p at any given time. Thus, MT shrinkage coincident with Bud6p or Kar9p-associated transport may take place on different MT subpopulations (Fig. 10). Indeed, contrary to the high frequency of MT interactions with cortical Bud6p, Kar9-GFP rarely colocalized with CFP-Bud6 (unpublished data).
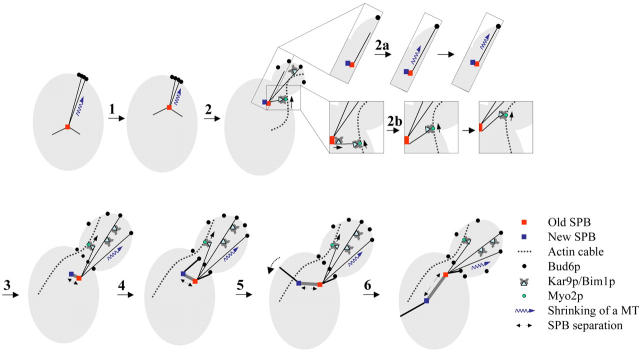
Figure 10.
The role of Bud6p and Kar9p in cortical capture. A model summarizing astral MT dynamic behavior associated with Bud6p or Kar9p during MT capture and establishment of spindle polarity. (1) As a cell proceeds through G1, the old SPB moves toward the incipient bud site by shrinking of astral MTs (thin black lines) at cortical Bud6p sites. (2) After bud emergence and SPB duplication, Kar9p or Bud6p-associated capture coexist and can separately direct astral MTs to the bud cortex. For example, in 2a, an MT reaching cortical Bud6p undergoes shrinkage coupled with SPB movement toward the cortex (without Kar9p being present at the MT plus end); in 2b, Kar9p translocates to an MT plus end and binds to Myo2p to guide the MT to the bud via an actin cable. In addition, Kar9p can travel along astral MTs already contacting the cortex (at Bud6p). Kar9p may also prevent MTs in the bud from undergoing catastrophe past the bud neck. As a result of these combined contributions of Bud6p and Kar9p, dynamic MT–cortex interactions are maintained as the bud grows and SPB separation begins. (3) During early spindle assembly, Bud6p associated capture and Kar9p-dependent delivery continue to focus MTs toward the bud. (4) As new astral MTs are organized by the respective SPB outer plaques (thick black lines) cortical capture begins at newly defined Bud6p sites at the bud neck. These interactions effectively restrict the new pole to the mother cell (irrespective of Kar9p initial presence on both SPBs; Fig. 4 C). (5) Progressively, Kar9p is recruited solely at the SPBd (spindle > 1 μm). (6) Coupled to continued SPB separation, MTs generated by the SPBm are no longer directed toward the bud and the spindle becomes aligned as it reaches final preanaphase length (∼2 μm).
The analysis presented here showed separable contributions of Bud6p and Kar9p toward spindle orientation summarized in Fig. 10. Earlier events would be dictated by the Bud6p cortical program of MT interactions and enforced by progressive asymmetric recruitment of Kar9p to the SPBd. Bud6p would couple SPB movement with cortical capture, whereas Kar9p would secure delivery of MTs from the SPBd to the bud. In addition, Kar9p transits along MTs within the bud may preserve these MTs from shrinking past the bud neck and instead redirect them to interact with actin cables via Myo2p.
Genetic analysis places Bud6p in the “early” and “late” pathways of spindle orientation
Genetic analysis has assigned motors, MT binding proteins and putative cortical anchors to “early” and “late” pathways participating in spindle orientation (Heil-Chapdelaine et al., 1999; Pearson and Bloom, 2004). Bud6p cortical program underlies the orientation of MT–cortex interactions throughout the spindle pathway (Segal et al., 2002). Yet, Bud6p relationship with actin organization (Moseley et al., 2004) pointed to links with the early Kar9p-dependent pathway.
Here, we show that kar9Δ bud6Δ cells exhibited phenotypes suggestive of synergism between the two mutations (Fig. 7 and Table I). These cells still relied on dynein-driven MT sliding to orient anaphase spindles. These interactions, however, were not completely proficient as evidenced by transient loss of orientation in the late portion of anaphase (Fig. 7 C).
Additive effects were also observed in bud6Δ dyn1Δ cells. Preanaphase spindle orientation was comparable to that of bud6Δ single mutants, yet, defective cortical capture at the bud neck also resulted in disruption of spindle polarity in anaphase cells (Fig. 8 and Table II).
Failure to translocate a spindle pole into the bud during anaphase blocks mitotic exit and cell division, under the control of the mitotic exit network (Bardin and Amon, 2001; McCollum and Gould, 2001). This surveillance mechanism is based on the separation between the upstream activator of the network Lte1p confined to the bud cortex, and its effector, the small GTPase Tem1p, localized to the SPBd. Once the SPBd translocates into the bud, these two regulators may interact to trigger mitotic exit. Adames et al. (2001) have proposed that cortical capture at the bud neck is also critical for this control. In agreement with this proposal, dyn1Δ bud6Δ cells failed to restrain spindle disassembly within the mother cell. This underscores the importance of Bud6p in cortical capture at the bud neck as bud6Δ cells still compartmentalize Lte1p to the bud (Jensen et al., 2002). Conversely, bud6Δ cells experience a delay in mitotic exit when astral MTs from the SPBd, already positioned in the bud, grow aberrantly past the bud neck and interact with the mother cell cortex (Segal et al., 2002).
To orient the spindle, MT-based motors must generate force relative to an attachment site to result in net displacement of the spindle pole (Pearson and Bloom, 2004). Num1p provides an anchor for dynein-driven sliding movements along the cortex as the spindle translocates into the bud (Heil-Chapdelaine et al., 2000). In contrast, the cortical anchor promoting dynein-dependent MT shrinkage (Carminati and Stearns, 1997) has not been assigned. Our studies point to the involvement of Bud6p in this mode of interaction. Indeed, Bud6p-dependent SPB repositioning after cytokinesis, which relies on MT shrinkage in association to the Bud6p ring (Segal et al., 2002), is abolished by a dyn1Δ mutation (unpublished data). Moreover, MT–Bud6p interactions during anaphase are preserved in the absence of Num1p (Segal et al., 2002). Thus, different modes of MT–cortex interactions might be modulated by association of MT-bound dynein–dynactin complex with alternative cortical anchors. This would add an additional layer of complexity to the control of competing modes of MT dynamic behavior accompanying the transitions from Kar9p to dynein-driven spindle positioning (Segal and Bloom, 2001).
Is MT plus end delivery to the bud the sole function of Kar9p?
Oriented MT capture at Bud6p sites in the bud still occurred in kar9Δ cells. Yet, orientation was not maintained as the bud continued to grow (Fig. 2). Therefore, we tested whether reduced MT instability could suppress kar9Δ phenotypes. Indeed, reduced MT turnover restored spindle orientation in kar9Δ cells dependent on Bud6p (Fig. 9), pointing to a role for Kar9p in maintaining MTs within the bud once Bud6p-dependent capture had targeted these MTs to the bud cortex. Consistent with this notion, Kar9p appeared to prevent MTs from undergoing shrinkage past the bud neck once inside the bud (Fig. S1).
We have previously proposed that Kar9p may affect MT dynamics in addition to its role as a bridge to the actin cytoskeleton (Segal and Bloom, 2001). Interestingly, the human adenomatous polyposis coli, the proposed counterpart of Kar9p, is critical for EB1-dependent MT polymerization. Adenomatous polyposis coli is subject to phosphorylation by Cdk controlling its binding to EB1 (Nakamura et al., 2001). Similarly, phosphorylation by Cdk may control Kar9p binding to Bim1p although the significance of this control is unclear (Liakopoulos et al., 2003). Finally, another work implicates Kar9p in modulation of MT dynamic behavior (Maekawa and Schiebel, 2004). According to this report, Kar9p targets Cdk to plus ends of MTs generated by the SPBd to control differential MT–cortex interactions at the bud tip or bud neck. The substrates mediating this control remain unknown.
Mechanisms of cortical capture are conserved between yeast and higher eukaryotes. Thus, the continued effort to dissect the regulation of dynamic aspects of MT–cortex interactions in yeast will yield new clues to understand the spatial control of MT capture, and its impact on developmental programs relying on regulated spindle orientation for the generation of cell diversity in metazoans.
Materials and methods
Yeast strains, plasmids, and genetic procedures
Yeast strains were isogenic to 15Dau (Segal et al., 2000a). The deletion alleles bud6Δ, kar9Δ, and dyn1Δ were generated using KAN R cassettes amplified by PCR (Segal et al., 2000b). Strains expressing a mutant β-tubulin conferring reduced MT dynamics were created by transforming yeast cells with a linear SacI–SphI fragment from pCS3-C354S (provided by R. Himes, University of Kansas, Lawrence, KS) encoding tub2 C354S (Gupta et al., 2002). A cdc3 ts strain and isogenic wild-type strains were a gift from M. Longtine (Oklahoma State University, Stillwater, OK). Strains expressing a GFP-Tub1 and GFP-Bud6 fusion were obtained by transformation with pAFS72 (Straight et al., 1997) and pRB2190 (Amberg et al., 1997), respectively. pCFP-TUB1 was used to express a CFP-α tubulin fusion (Jensen et al., 2001). pRS404KAR9tGFP contained a 495bp SacI–NotI fragment generated by PCR for 3′ in-frame fusion to GFP. Digestion with BamHI targeted the construct for integration at the endogenous KAR9. Standard yeast genetic procedures were used (Sherman et al., 1986). Cells were grown at 25°C unless indicated.
Microscopy methods
Cells were grown to ∼5 × 106 cells/ml in selective dextrose medium and then mounted in the same medium containing 25% gelatin to perform time-lapse recordings at RT (Maddox et al., 1999) using a Nikon Eclipse E800 with a CFI Plan Apochromat 100×, N.A. 1.4 objective, Chroma Technology Corp. filter sets and a Coolsnap-HQ CCD camera (Roper Scientific). Images were acquired using 2 × 2 binning. For cells expressing GFP fusions, five fluorescence images were acquired at a Z-distance of 0.8 μm between planes. A single differential interference contrast (DIC) image was taken in the middle focal plane. This acquisition regime was repeated at 10-, 15-, or 30-s intervals. Images were processed using Metamorph software (Universal Imaging Corp.; Maddox et al., 1999). The validity and considerations on the resolution of the analysis to assess dynamic interactions between cortical Bud6p and MTs have been addressed previously (Segal et al., 2002).
Interactions at or away from Bud6p were categorized as described previously (Segal et al., 2002). For simplicity, MT shrinkage at the cortex was the only category scored individually. This mode of interaction occurs at Bud6p cortical sites and is abolished in bud6Δ cells (Segal et al., 2002). For this analysis, the cell cycle was arbitrarily divided into three stages (Fig. 1): (1) from mitotic exit (ME) to generation of the new prebud site (from spindle disassembly and formation of a double ring of Bud6p at the division site till disappearance of the Bud6p ring and relocation to the new bud site); (2) from bud emergence (BE) to spindle assembly and orientation; and (3) from spindle elongation (SE) to ME. For the third stage, only interactions involving astral MTs from the SPBd were considered.
Duration of cortical interactions was determined by following the history of 50 MTs. Mean values correspond to the total time MTs contacted the cortex divided by the number of contact events (n). Results were expressed as mean ± SD.
Cells expressing CFP-Tub1 and Kar9-GFP fusions were recorded using a modification of the protocol that discriminates between CFP and GFP using a CFP/YFP filter set (Pearson et al., 2001). Three fluorescence images were acquired at a Z-distance of 0.8 μm. Z-stacks were acquired at 12-, 15-, 30-, or 60-s intervals.
Kar9-GFP dynamic behavior was categorized as follows: (1) Kar9p movement along an MT toward the plus end; (2) Kar9p movement toward the SPB along a persistent MT; (3) Kar9p movement at the plus end of a growing MT; (4) Kar9p movement at the plus end of a shrinking MT; and (5) Kar9p association to a plus end without recruitment via the SPB. Categories 1 and 2 included all events involving Kar9p presence along an MT, whereas categories 3 and 4 corresponded to all events in which Kar9p at the MT plus end moved as a consequence of MT growth and shrinkage.
To correlate Kar9p-bound MT shortening and SPB movement, Kar9p returns toward the spindle pole were scored for associated movement of the SPB as follows: (a) no movement (the SPB remained stationary); (b) movement away from the cortex (unrelated to the direction of the shortening MT); and (c) movement coupled to shortening (toward the cortex).
Quantitations described in Fig. 2 included data derived from 54 wild-type cells (total of 65.5 h recorded) and 172 kar9Δ cells (140 h). Analysis of cells coexpressing CFP-Tub1 and Kar9-GFP included recorded data of 107 wild-type cells (30 h) and 99 bud6Δ cells (31 h).
Quantitations described in Fig. 9 represent recorded data from the following strains expressing GFP-Tub1: 81 wild-type cells (89 h); 77 kar9Δ cells (76 h); 64 kar9Δ tub2 C354S cells (73 h); 51 bud6Δ tub2 C354S cells (50.5 h); 52 bud6Δ kar9Δ tub2 C354S (56.8 h); 65 kar9Δ bud6Δ cells (83 h); and 45 bud6Δ cells (70 h).
Comparative analysis of bud6Δ, kar9Δ, dyn1Δ, bud6Δ kar9Δ and dyn1Δ bud6Δ cells expressing GFP-Tub1 was performed using the same set of recordings described above. In addition, 31 dyn1Δ bud6Δ cells (26.2 h) and 36 dyn1Δ cells (50 h) were recorded.
Online supplemental material
Fig. S1 shows representative time-lapse series of wild-type or kar9Δ cells expressing GFP-Tub1 showing dynamic behavior of MTs entering the bud. Fig. S2 shows GFP-Bud6 localization in cdc3 ts cells. Online material is available at http://www.jcb.org/cgi/content/full/jcb.200407167/DC1.
Acknowledgments
We thank M. Longtine, T. Huffaker, and R. Himes for generous gift of plasmids and strains and members of Kerry Bloom's lab for their invaluable help in setting up our time-lapse microscope in Cambridge. We also thank E. Schiebel for sharing results before publication, F. Balloux for helpful discussions, and anonymous reviewers for critical reading of the manuscript.
This work was supported by the Marco Polo Fund (to S.M. Huisman); the Erasmus Exchange Program (to M. Bertrand); National Institutes of Health grant GM38328 (to S.I. Reed); Cancer Research UK, The Isaac Newton Trust, and The Wellcome Trust (to M. Segal).
Abbreviations used in this paper: DIC, differential interference contrast; MT, microtubule; SPB, spindle pole body; SPBd, daughter-bound SPB; SPBm, mother-bound SPB.
References
- Adames, N.R., J.R. Oberle, and J.A. Cooper. 2001. The surveillance mechanism of the spindle position checkpoint in yeast. J. Cell Biol. 153:159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg, D.C., J.E. Zahner, J.W. Mulholland, J.R. Pringle, and D. Botstein. 1997. Aip3p/Bud6p, a yeast actin-interacting protein that is involved in morphogenesis and the selection of bipolar budding sites. Mol. Biol. Cell. 8:729–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin, A.J., and A. Amon. 2001. Men and sin: what's the difference? Nat. Rev. Mol. Cell Biol. 2:815–826. [DOI] [PubMed] [Google Scholar]
- Beach, D.L., J. Thibodeaux, P. Maddox, E. Yeh, and K. Bloom. 2000. The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr. Biol. 10:1497–1506. [DOI] [PubMed] [Google Scholar]
- Carminati, J.L., and T. Stearns. 1997. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 138:629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshel, D., L.A. Urrestarazu, S. Vissers, J.C. Jauniaux, J.C. van Vliet-Reedijk, R.J. Planta, and I.R. Gibbons. 1993. Cytoplasmic dynein is required for normal nuclear segregation in yeast. Proc. Natl. Acad. Sci. USA. 90:11172–11176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer, M. 2001. Animal cell cytokinesis. Annu. Rev. Cell Dev. Biol. 17:351–386. [DOI] [PubMed] [Google Scholar]
- Gupta, M.L., Jr., C.J. Bode, D.A. Thrower, C.G. Pearson, K.A. Suprenant, K.S. Bloom, and R.H. Himes. 2002. β-Tubulin C354 mutations that severely decrease microtubule dynamics do not prevent nuclear migration in yeast. Mol. Biol. Cell. 13:2919–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil-Chapdelaine, R.A., N.R. Adames, and J.A. Cooper. 1999. Formin' the connection between microtubules and the cell cortex. J. Cell Biol. 144:809–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil-Chapdelaine, R.A., J.R. Oberle, and J.A. Cooper. 2000. The cortical protein Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J. Cell Biol. 151:1337–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, E., J. Kusch, Y. Barral, and T.C. Huffaker. 2003. Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J. Cell Biol. 161:483–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, S., M. Segal, D.J. Clarke, and S.I. Reed. 2001. A novel role of the budding yeast separin Esp1 in anaphase spindle elongation: evidence that proper spindle association of Esp1 is regulated by Pds1. J. Cell Biol. 152:27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen, S., M. Geymonat, A.L. Johnson, M. Segal, and L.H. Johnston. 2002. Spatial regulation of the guanine nucleotide exchange factor Lte1 in Saccharomyces cerevisiae. J. Cell Sci. 115:4977–4991. [DOI] [PubMed] [Google Scholar]
- Korinek, W.S., M.J. Copeland, A. Chaudhuri, and J. Chant. 2000. Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science. 287:2257–2259. [DOI] [PubMed] [Google Scholar]
- Kusch, J., A. Meyer, M.P. Snyder, and Y. Barral. 2002. Microtubule capture by the cleavage apparatus is required for proper spindle positioning in yeast. Genes Dev. 16:1627–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusch, J., D. Liakopoulos, and Y. Barral. 2003. Spindle asymmetry: a compass for the cell. Trends Cell Biol. 13:562–569. [DOI] [PubMed] [Google Scholar]
- Lee, L., J.S. Tirnauer, J. Li, S.C. Schuyler, J.Y. Liu, and D. Pellman. 2000. Positioning of the mitotic spindle by a cortical microtubule capture mechanism. Science. 287:2260–2262. [DOI] [PubMed] [Google Scholar]
- Li, Y.Y., E. Yeh, T. Hays, and K. Bloom. 1993. Disruption of mitotic spindle orientation in a yeast dynein mutant. Proc. Natl. Acad. Sci. USA. 90:10096–10100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liakopoulos, D., J. Kusch, S. Grava, J. Vogel, and Y. Barral. 2003. Asymmetric loading of Kar9 onto spindle poles and microtubules ensures proper spindle alignment. Cell. 112:561–574. [DOI] [PubMed] [Google Scholar]
- Maddox, P., E. Chin, A. Mallavarapu, E. Yeh, E.D. Salmon, and K. Bloom. 1999. Microtubule dynamics from mating through the first zygotic division in the budding yeast Saccharomyces cerevisiae. J. Cell Biol. 144:977–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa, H., and E. Schiebel. 2004. Cdk1-Clb4 controls the interaction of astral microtubule plus ends with subdomains of the daughter cell cortex. Genes Dev. 18:1709–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollum, D., and K.L. Gould. 2001. Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol. 11:89–95. [DOI] [PubMed] [Google Scholar]
- Miller, R.K., D. Matheos, and M.D. Rose. 1999. The cortical localization of the microtubule orientation protein, Kar9p, is dependent upon actin and proteins required for polarization. J. Cell Biol. 144:963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, R.K., S.C. Cheng, and M.D. Rose. 2000. Bim1p/Yeb1p mediates the Kar9p-dependent cortical attachment of cytoplasmic microtubules. Mol. Biol. Cell. 11:2949–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley, J.B., I. Sagot, A.L. Manning, Y. Xu, M.J. Eck, D. Pellman, and B.L. Goode. 2004. A conserved mechanism for Bni1- and mDia1-induced actin assembly and dual regulation of Bni1 by Bud6 and profilin. Mol. Biol. Cell. 15:896–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, M., X.Z. Zhou, and K.P. Lu. 2001. Critical role for the EB1 and APC interaction in the regulation of microtubule polymerization. Curr. Biol. 11:1062–1067. [DOI] [PubMed] [Google Scholar]
- Palmer, R.E., D.S. Sullivan, T. Huffaker, and D. Koshland. 1992. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 119:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, G., T.U. Tanaka, K. Nasmyth, and E. Schiebel. 2001. Modes of spindle pole body inheritance and segregation of the Bfa1p-Bub2p checkpoint protein complex. EMBO J. 20:6359–6370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, C.G., and K. Bloom. 2004. Dynamic microtubules lead the way for spindle positioning. Nat. Rev. Mol. Cell Biol. 5:481–492. [DOI] [PubMed] [Google Scholar]
- Pearson, C.G., P.S. Maddox, E.D. Salmon, and K. Bloom. 2001. Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 152:1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport, R. 1996. Cytokinesis in Animal Cells. Vol. 32. Cambridge University Press, Cambridge, UK. 386 pp.
- Segal, M., and K. Bloom. 2001. Control of spindle polarity and orientation in S. cerevisiae. Trends Cell Biol. 11:160–166. [DOI] [PubMed] [Google Scholar]
- Segal, M., D.J. Clarke, P. Maddox, E.D. Salmon, K. Bloom, and S.I. Reed. 2000. a. Coordinated spindle assembly and orientation requires Clb5-dependent kinase in budding yeast. J. Cell Biol. 148:441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal, M., K. Bloom, and S.I. Reed. 2000. b. Bud6 directs sequential microtubule interactions with the bud tip and bud neck during spindle morphogenesis in Saccharomyces cerevisiae. Mol. Biol. Cell. 11:3689–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal, M., K. Bloom, and S.I. Reed. 2002. Kar9p-independent microtubule capture at Bud6p cortical sites primes spindle polarity before bud emergence in Saccharomyces cerevisiae. Mol. Biol. Cell. 13:4141–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, S.L., E. Yeh, P. Maddox, E.D. Salmon, and K. Bloom. 1997. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J. Cell Biol. 139:985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., G. Fink, and J.B. Hicks. 1986. Methods in Yeast Genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- Smeets, M.F.M.A., and M. Segal. 2002. Spindle polarity in S. cerevisiae: MEN can tell. Cell Cycle. 1:308–311. [DOI] [PubMed] [Google Scholar]
- Straight, A.F., W.F. Marshall, J.W. Sedat, and A.W. Murray. 1997. Mitosis in living budding yeast: anaphase A but no metaphase plate. Science. 277:574–578. [DOI] [PubMed] [Google Scholar]
- Theesfeld, C.L., J.E. Irazoqui, K. Bloom, and D.J. Lew. 1999. The role of actin in spindle orientation changes during the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 146:1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer, J.S., E. O'Toole, L. Berrueta, B.E. Bierer, and D. Pellman. 1999. Yeast Bim1p promotes the G1-specific dynamics of microtubules. J. Cell Biol. 145:993–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, E., R.V. Skibbens, J.W. Cheng, E.D. Salmon, and K. Bloom. 1995. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 130:687–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, E., C. Yang, E. Chin, P. Maddox, E.D. Salmon, D.J. Lew, and K. Bloom. 2000. Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants. Mol. Biol. Cell. 11:3949–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, H., D. Pruyne, T.C. Huffaker, and A. Brestscher. 2000. Myosin V orientates the mitotic spindle in yeast. Nature. 406:1013–1015. [DOI] [PubMed] [Google Scholar]