Abstract
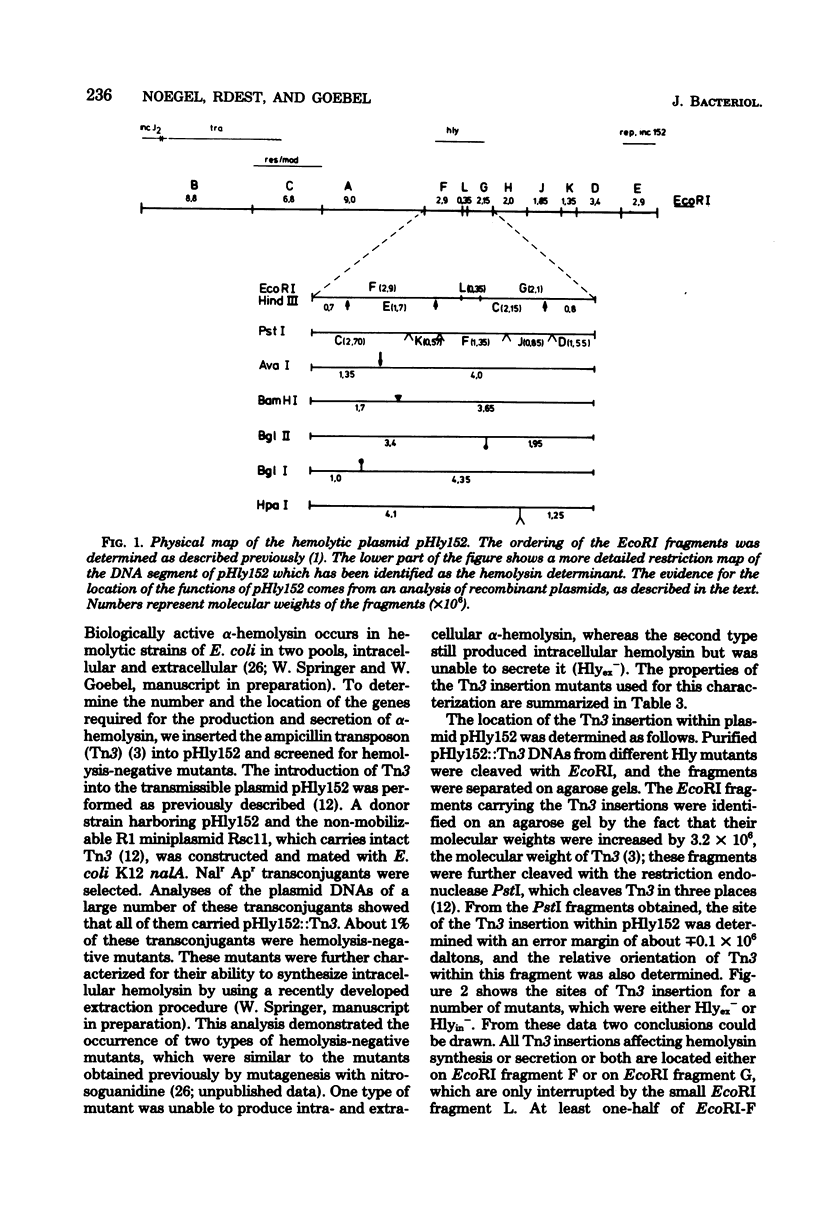
The alpha-hemolytic Escherichia coli strain PM152 harbors three transmissible plasmids, which have molecular weights of 65 X 10(6) (pA152), 41 X 10(6) pHly152), and 32 X 10(6) (pC152). Plasmids pHly152 and pC152 belong to incompatibility groups J2 and N, respectively. By transforming E. coli K-12 with isolated plasmids, we showed that the genetic determinant required for hemolysis was located entirely on plasmid pHly152, and a physical map of this plasmid was constructed. By transposon mutagenesis, a deoxyribonucleic acid segment of about 3.5 X 10(6) daltons was identified as being essential for hemolysis. Most of the EcoRI and HindIII fragments of the hemolytic plasmid pHly152 were cloned by using pACYC184 and RSF2124 as vectors. Two classes of Tn3-induced hemolysis-negative mutants could be complemented by recombinant plasmids carrying fragments from the hemolysis region of pHly152, whereas a third class could be restored to hemolytic activity only by recombination between the mutant plasmids and a suitable recombinant deoxyribonucleic acid. These data suggest that there are at least three clustered cistrons which are required for hemolysis. Other EcoRI and HindIII fragments of pHly152 were identified as being essential for replication, incompatibility, transfer, and restriction.
Full text
PDF













Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blohm D., Goebel W. Restriction map of the antibiotic resistance plasmid R1drd-19 and its derivatives pKN102 (R1drd-19B2) and R1drd-16 for the enzymes BamHI, HindIII, EcoRI and SalI. Mol Gen Genet. 1978 Nov 29;167(2):119–127. doi: 10.1007/BF00266905. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Boyer H. W., Helling R. B. Construction of biologically functional bacterial plasmids in vitro. Proc Natl Acad Sci U S A. 1973 Nov;70(11):3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N. Transposable genetic elements and plasmid evolution. Nature. 1976 Oct 28;263(5580):731–738. doi: 10.1038/263731a0. [DOI] [PubMed] [Google Scholar]
- Crosa J. H., Luttropp L. K., Falkow S. Molecular cloning of replication and incompatibility regions from the R-plasmid R6K. J Mol Biol. 1978 Sep 25;124(3):443–468. doi: 10.1016/0022-2836(78)90181-x. [DOI] [PubMed] [Google Scholar]
- Davis B. D., Tai P. C. The mechanism of protein secretion across membranes. Nature. 1980 Jan 31;283(5746):433–438. doi: 10.1038/283433a0. [DOI] [PubMed] [Google Scholar]
- Goebel W., Lindenmaier W., Pfeifer F., Schrempf H., Schelle B. Transposition and insertion of intact, deleted and enlarged ampicillin transposon Tn3 from mini-R1 (Rsc) plasmids into transfer factors. Mol Gen Genet. 1977 Nov 29;157(2):119–129. doi: 10.1007/BF00267389. [DOI] [PubMed] [Google Scholar]
- Goebel W., Royer-Pokora B., Lindenmaier W., Bujard H. Plasmids controlling synthesis of hemolysin in Escherichia coli: molecular properties. J Bacteriol. 1974 Jun;118(3):964–973. doi: 10.1128/jb.118.3.964-973.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel W., Schrempf H. Isolation and characterization of supercoiled circular deoxyribonucleic acid from beta-hemolytic strains of Escherichia coli. J Bacteriol. 1971 May;106(2):311–317. doi: 10.1128/jb.106.2.311-317.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heffron F., McCarthy B. J., Ohtsubo H., Ohtsubo E. DNA sequence analysis of the transposon Tn3: three genes and three sites involved in transposition of Tn3. Cell. 1979 Dec;18(4):1153–1163. doi: 10.1016/0092-8674(79)90228-9. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Chan R. K., Tye B. K., Botstein D. Mutagenesis by insertion of a drug-resistance element carrying an inverted repetition. J Mol Biol. 1975 Oct 5;97(4):561–575. doi: 10.1016/s0022-2836(75)80059-3. [DOI] [PubMed] [Google Scholar]
- Kollek R., Oertel W., Goebel W. Isolation and characterization of the minimal fragment required for autonomous replication ("basic replicon") of a copy mutant (pKN102) of the antibiotic resistance factor R1. Mol Gen Genet. 1978 Jun 1;162(1):51–57. doi: 10.1007/BF00333850. [DOI] [PubMed] [Google Scholar]
- Manis J. J., Kline B. C. F plasmid incompatibility and copy number genes: their map locations and interactions. Plasmid. 1978 Sep;1(4):492–507. doi: 10.1016/0147-619x(78)90007-0. [DOI] [PubMed] [Google Scholar]
- Mickel S., Ohtsubo E., Bauer W. Heteroduplex mapping of small plasmids derived from R-factor R12: in vivo recombination occurs at IS1 insertion sequences. Gene. 1977;2(3-4):193–210. doi: 10.1016/0378-1119(77)90017-8. [DOI] [PubMed] [Google Scholar]
- Minshew B. H., Jorgensen J., Counts G. W., Falkow S. Association of hemolysin production, hemagglutination of human erythrocytes, and virulence for chicken embryos of extraintestinal Escherichia coli isolates. Infect Immun. 1978 Apr;20(1):50–54. doi: 10.1128/iai.20.1.50-54.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molin S., Stougaard P., Uhlin B. E., Gustafsson P., Nordström K. Clustering of genes involved in replication, copy number control, incompatibility, and stable maintenance of the resistance plasmid R1drd-19. J Bacteriol. 1979 Apr;138(1):70–79. doi: 10.1128/jb.138.1.70-79.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti-Bragadin C., Samer L., Rottini G. D., Pani B. The compatibility of Hly factor, a transmissible element which controls alpha-haemolysin production in Escherichia coli. J Gen Microbiol. 1975 Feb;86(2):367–369. doi: 10.1099/00221287-86-2-367. [DOI] [PubMed] [Google Scholar]
- Noegel A., Rdest U., Springer W., Goebel W. Plasmid cistrons controlling synthesis and excretion of the exotoxin alpha-haemolysin of Escherichia coli. Mol Gen Genet. 1979 Oct 1;175(3):343–350. doi: 10.1007/BF00397234. [DOI] [PubMed] [Google Scholar]
- Nordström K., Ingram L. C., Lundbäck A. Mutations in R factors of Escherichia coli causing an increased number of R-factor copies per chromosome. J Bacteriol. 1972 May;110(2):562–569. doi: 10.1128/jb.110.2.562-569.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Hoppensteadt F. C. On plasmid incompatibility. Plasmid. 1978 Sep;1(4):421–434. doi: 10.1016/0147-619x(78)90001-x. [DOI] [PubMed] [Google Scholar]
- Oertel W., Kollek R., Beck E., Goebel W. The nucleotide sequence of a DNA fragment from the replication origin of the antibiotic resistance factor R1drd19. Mol Gen Genet. 1979 Mar 27;171(3):277–285. doi: 10.1007/BF00267582. [DOI] [PubMed] [Google Scholar]
- Oliver D. R., Brown B. L., Clewell D. B. Characterization of plasmids determining hemolysin and bacteriocin production in Streptococcus faecalis 5952. J Bacteriol. 1977 May;130(2):948–950. doi: 10.1128/jb.130.2.948-950.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts R. J. Restriction and modification enzymes and their recognition sequences. Gene. 1980 Mar;8(4):329–343. doi: 10.1016/0378-1119(80)90040-2. [DOI] [PubMed] [Google Scholar]
- Rogolsky M. Nonenteric toxins of Staphylococcus aureus. Microbiol Rev. 1979 Sep;43(3):320–360. doi: 10.1128/mr.43.3.320-360.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royer-Pokora B., Goebel W. Plasmids controlling synthesis of hemolysin in Escherichia coli. II. Polynucleotide sequence relationship among hemolytic plasmids. Mol Gen Genet. 1976 Mar 22;144(2):177–183. doi: 10.1007/BF02428106. [DOI] [PubMed] [Google Scholar]
- Smith H. W., Halls S. The transmissible nature of the genetic factor in Escherichia coli that controls haemolysin production. J Gen Microbiol. 1967 Apr;47(1):153–161. doi: 10.1099/00221287-47-1-153. [DOI] [PubMed] [Google Scholar]
- So M., Gill R., Falkow S. The generation of a ColE1-Apr cloning vehicle which allows detection of inserted DNA. Mol Gen Genet. 1975 Dec 30;142(3):239–249. doi: 10.1007/BF00425649. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Timmis K. N., Andrés I., Slocombe P. M. Plasmid incompatibility: cloning analysis of an incFII determinant of R6-5. Nature. 1978 May 4;273(5657):27–32. doi: 10.1038/273027a0. [DOI] [PubMed] [Google Scholar]
- Timmis K., Cabello F., Cohen S. N. Cloning, isolation, and characterization of replication regions of complex plasmid genomes. Proc Natl Acad Sci U S A. 1975 Jun;72(6):2242–2246. doi: 10.1073/pnas.72.6.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton J. R., Smith D. H. New hemolysin (gamma) produced by Escherichia coli. J Bacteriol. 1969 Apr;98(1):304–305. doi: 10.1128/jb.98.1.304-305.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz F., Müller D., Ortiz J. M., Goebel W. Hemolysis determinant common to Escherichia coli hemolytic plasmids of different incompatibility groups. J Bacteriol. 1980 Aug;143(2):825–833. doi: 10.1128/jb.143.2.825-833.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz F., Zabala J. C., Ortiz J. M. Incompatibility among alpha-hemolytic plasmids studied after inactivation of the alpha-hemolysin gene by transposition of Tn802. Plasmid. 1979 Oct;2(4):507–519. doi: 10.1016/0147-619x(79)90050-7. [DOI] [PubMed] [Google Scholar]