Abstract
Somatic DNA rearrangements in B lymphocytes, including V(D)J gene rearrangements and isotype switching, generally occur in cis, i. e., intrachromosomally. We showed previously, however, that 3 to 7% of IgA heavy chains have the VH and Cα regions encoded in trans. To determine whether the trans-association of VH and Cα occurred by trans-chromosomal recombination, by trans-splicing, or by trans-chromosomal gene conversion, we generated and analyzed eight IgA-secreting rabbit hybridomas with trans-associated VH and Cα heavy chains. By ELISA and by nucleotide sequence analysis we found that the VH and Cα regions were encoded by genes that were in trans in the germline. We cloned the rearranged VDJ-Cα gene from a fosmid library of one hybridoma and found that the expressed VH and Cα genes were juxtaposed. Moreover, the juxtaposed VH and Cα genes originated from different IgH alleles. From the same hybridoma, we also identified a fosmid clone with the other expected product of a trans-chromosomal recombination. The recombination breakpoint occurred within the Sμ/Sα region, indicating that the trans-association of VH and Cα genes occurred by trans-chromosomal recombination during isotype switching. We conclude that trans-chromosomal recombination occurs at an unexpectedly high frequency (7%) within the IgH locus of B lymphocytes in normal animals, which may explain the high incidence of B-cell tumors that arise from oncogene translocation into the IgH locus.
Ig genes undergo somatic recombination both during early B-cell development and again during antigen-induced immune responses. The V, D, and J gene segments recombine in proB and preB cells leading to the expression of IgM and IgD on the surface of mature B cells (1). Later, during isotype switching, the VDJ genes of these B lymphocytes rearrange to downstream CH genes leading to the expression of the IgG, IgA, or IgE isotypes (2–4). Although the mechanism for class switching has not been elucidated, we know that most switch rearrangements occur in or around switch regions that are characterized by a series of tandem repeat structures found 5′ of Cμ, Cγ, Cα, and Cɛ genes (2, 5).
Although switch recombination occurs generally by intrachromosomal DNA recombination between VH and CH genes in cis, Landucci-Tosi et al. (6) and Pernis et al. (7) identified trans-associated rabbit IgG molecules in which VH and CH are derived from genes in trans. These investigators used antibodies specific for VH and Cγ allotypes and showed that, in IgH heterozygous rabbits, some of the IgG molecules had the VH allotype encoded by one IgH allele and the Cγ allotype encoded by the other IgH allele. Subsequently, Knight et al. (8) identified trans-associated secretory IgA molecules from colostrum and showed that they represented as many as 8% of total IgA molecules.
The trans-association of VH and Cα could result from trans-chromosomal recombination, from trans-splicing of RNA, or from trans-chromosomal gene conversion. Support for each of these mechanisms has been reported. Kipps and Herzenberg (9) initially obtained evidence for trans-chromosomal recombination by identifying isotype switch variants that apparently resulted from in vitro recombination between VH and CH genes in trans. Subsequently, Gerstein et al. (10) and Umar and Gearhart (11) showed that VDJ transgenes could recombine interchromosomally with the endogenous IgH locus. Further, Giusti and colleagues (12, 13) showed that recombination between VDJ transgenes and the endogenous IgH locus could occur by gene conversion, a nonreciprocal homologous recombination.
Trans mRNA has been proposed as a mechanism to explain the simultaneous expression of multiple Ig isotypes in individual B lymphocytes (14–17). Recently, Fujieda et al. (17) identified chimeric I-CH transcripts in interleukin 4-stimulated B cells and suggested that these chimeric transcripts resulted from trans-splicing of two Ig pre-mRNA transcripts.
Studies of trans Ig molecules in rabbit are facilitated by the presence of allotypic markers in both the V and C regions of the heavy chains (reviewed in ref. 18). The VH allotypes a1, a2, and a3 are encoded by allelic genes and are found on 80 to 90% of Ig heavy-chains. The a1 and a2 allotype Ig can be readily identified by differences in the amino acid sequence in FR1 and FR3 (19). Allotypic markers also are found on the C region of rabbit IgM, IgG, and IgA heavy chains. Cα allotypic markers are found on each of the 13 IgA subclasses resulting from 13 nonallelic germline Cα genes (20–21). The anti-allotype antibodies that react with the Cα allotypes are designated anti-f, anti-g, or anti-f, g (18). The VH and Cα genes are linked closely and the number of IgH haplotypes, designated A through N, is limited (18).
In the present study, we examined the molecular basis for trans-recombinant IgA heavy chains in rabbits heterozygous for the C and E heavy chain haplotypes. The VH and Cα allotypes encoded by rabbits of the C haplotype are a1f72g74, and those of the E haplotype are a2f71g75. We generated IgA-secreting hybridomas and identified those hybridomas that secreted trans-associated IgA heavy chains. By determining the haplotype origin of the VH and Cα genes through restriction fragment length polymorphisms (RFLPs) and nucleotide sequence analyses, we found that the trans-association of VH and Cα resulted from both trans-chromosomal recombination and gene conversion.
MATERIALS AND METHODS
Rabbits and IgA Heavy Chain Genes.
Rabbits of the C haplotype a1f72/a1f72 (allotypes VHa1 and Cαf72) and of the E haplotype a2f71/a2f71 (allotypes VHa2 and Cαf71) were maintained in our colony at Loyola University of Chicago. Hybridomas were derived from heterozygous C/E (a1f72/a2f71) rabbits (22) 295J2, 83K, and 229L2.
The 13 nonallelic Cα genes in the rabbit germline (21) are numbered Cα1 to Cα13, in the order in which they were discovered. The Cα genes with known locations relative to other CH genes are in the order: 5′-Cμ–Cγ–Cɛ–Cα4–Cα5–Cα6–3′. The Cα5 and Cα6 genes were cloned from C haplotype rabbits. We do not know whether the number and organization of Cα genes in the C and E haplotypes are identical. Cα5 of a G haplotype rabbit was cloned from a 6- to 8-kilobase (kb) BamHI size-selected DNA library as described (21), and the nucleotide sequence of the Cα gene was determined (accession no. AFO90366). For this study, this gene is referred to as Cα5 of the E haplotype rather than the G haplotype because the E and G haplotypes have the same Cα allotypes (f71g75) (18) and because, by Southern blot analysis, these haplotypes have identical Cα gene RFLPs (20). Accordingly, we assume Cα5 of these two haplotypes is identical. The nucleotide sequence for Cα5 from the C haplotype is accession no. X82111 (21).
IgA-Secreting Hybridomas.
IgA-secreting hybridomas were generated by the fusion of the rabbit plasmacytoma cell line 240E-1 with mesenteric lymph node and Peyer’s patches lymphocytes from rabbits, 295J2, 83K, and 229L2 as described (23). Hybridoma supernatant fluids were tested by ELISA by using 96-well plates coated with goat anti-rabbit Cα antibody. IgA-secreting hybridomas were identified with biotinylated goat anti-rabbit Cα antibody and HRP-avidin (Vector Laboratories). Supernatant fluids containing IgA were tested further with the anti-allotype antisera anti-a1, anti-a2, anti-f71g75, anti-f72g74, and anti-g74 followed by biotinylated goat anti-rabbit Fcγ and HRP-avidin. In some cases, we used biotinylated anti-a1 and anti-a2 followed by HRP-avidin.
We obtained a total of 756 hybridomas, of which, 183 were IgA-secreting, and 106 of these reacted with anti-Cα allotype antibodies. Although we had expected that the polyclonal anti-Cα allotype antisera would react with all of the IgA-secreting hybridomas (24), we believe that the polyclonal antisera used in the ELISA probably have insufficient antibody against some of the 13 IgA subclasses such that they were not detected in the ELISA.
Reverse Transcription–PCR and Nucleotide Sequence Analysis.
Heavy chain genes of trans-associated IgA-secreting hybridomas were cloned by reverse transcription–PCR (22). The cDNA was synthesized by using oligo(dT) as primer and Superscript as the reverse transcriptase (ProMega). For PCR, we used as 5′ primer VHprB 5′CTGCAGCTCTGGCACAGGAGCTC3′ (22) and as 3′ primer OAEx2α 5′CTTCAAGCTTCTCAGGGTGCAGGTGAGGCT3′, based on the nucleotide sequence in exon 2 that is conserved among the 13 germline Cα genes (21). The PCR products were cloned into M13 mp18/19, and the nucleotide sequences were determined in a single orientation by using Sequenase (U.S. Biochemical, Cleveland, OH).
Fosmid Library.
A partial MboI genomic DNA library from hybridoma M19 cells was prepared in the fosmid vector pFOS1 as described by Kim et al. (25). Fragments of 50 to 100 kb were separated by pulsed-field gel electrophoresis and were ligated to fosmid vector arms as described (25). Approximately 200,000 colonies were screened by hybridization with a JH probe. DNA from selected clones was subcloned as XbaI fragments into pUC19 or pGem3 for restriction mapping. Southern blot analysis was performed with probes for JH, Eμ, Sμ, Sα, Cα, and VH genes (20, 21, 26, 27).
PCR-Amplification of Cα5 and Cα6 from genomic DNA.
Primers for Cα5(E) were 5′CTGCAACCCCCCCGATCA3′ from exon 1 and 5′GGCCGGATGTGGTGGCT3′ from the hinge region of Cα5 of FOS6/FOS12. Primers for Cα6(C) were 5′GTGAACACTAGACCCATCCTCAT3′ from exon 1 and 5′GTGGAAGCTTGGCAGGTGGTTGTATCTGAA3′ from the hinge region of germline Cα6 (21). (Bold type indicates mismatches with the corresponding gene from the opposite allele). Hot-start PCR (28) was performed in a 30-μl volume at 0.1 μM primer with 27 cycles (45 s at 94°C, 45 s at 67°C, and 45 s at 72°C). The target template was either 200 ng of genomic DNA or 10 pg of plasmid DNA from cloned Cα5 (C or E haplotypes) or Cα6 (C haplotype) genes (21). The PCR products were separated by PAGE (5%) and were photographed after staining with ethidium bromide.
RESULTS
Generation of IgA-Secreting Hybridomas with Trans-Associated VH and Cα Heavy Chains.
To examine the molecular events leading to IgA with trans-associated VH and Cα heavy chains, we generated IgA-secreting hybridomas from Peyer’s patch and mesenteric lymph node cells from normal unimmunized heterozygous C/E haplotype rabbits. Of the 106 IgA-secreting hybridomas that reacted with the anti-Cα allotype antisera, most reacted with antibodies to the cis-associated VH and Cα allotypes (VHa1 and Cαf72,g74 or VHa2 and Cαf71,g75). However, seven of the hybridomas (7%) reacted with antibodies to the VH and Cα allotypes that were encoded in trans (VHa1 and Cαf71,g75 or VHa2 and Cαf72,g74) (Table 1). These hybridomas were cloned by limiting dilution and were analyzed further to determine the mechanism of the trans-association of the VH and Cα allotypes.
Table 1.
ELISA of IgA-secreting hybridomas from heterozygous C (a1f72)/E (a2f71) rabbits
| Experiment no. (Rabbit) | No. of
allotype+ wells*
|
No. of hybridomas with
trans- associated VH and Cα
|
||||
|---|---|---|---|---|---|---|
| a1 | f72g74 | a2 | f71g75 | a1f71 | a2f72 | |
| 1 (295J2) | 73 | 63 | 24 | 7 | 1 | 2 |
| 2 (83K) | 29 | 19 | 1 | 7 | 2 | 0 |
| 3 (229L2) | 9 | 7 | 2 | 3 | 1 | 1 |
| Total | f71 + f72 = 106 | 7 (7%)† | ||||
The allotypes a1, f72, and g74 are encoded by the C haplotype; allotypes a2, f71, and g75 are encoded by the E haplotype.
One clone (F41) that appears to result from a nonreciprocal trans-chromosomal recombination (gene conversion) is not included in this table.
Molecular Analysis of Trans-Associated IgA.
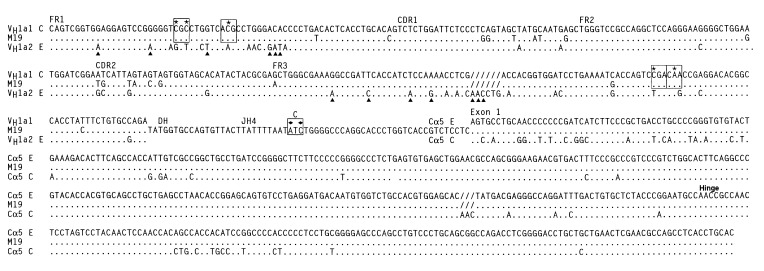
To confirm the trans phenotype of the VH and Cα allotypes of the seven hybridomas, we amplified the heavy chain genes by reverse transcription–PCR and determined their nucleotide sequences. Fig. 1 shows the sequence of the VDJ region, the hinge region, and Cα exon1 of hybridoma M19 (a1f71), which secretes trans-associated IgA molecules. The V region of the M19 VDJ gene is similar to the a1-encoding gene VH1-a1 of the C haplotype, and it encodes the four VH a1 allotype-associated amino acids and none of the 10 VH a2 allotype-associated amino acids in FR1 and FR3. For the other hybridomas, the deduced amino acid sequence of the VH region also correlated with the amino acid sequence characteristic of the VHa allotype identified by ELISA (Table 2). We found that two of the hybridomas, M9 and M19, have the C haplotype sequence polymorphism in JH4 (as indicated in Fig. 1) showing that, like the VH gene, the JH gene segment is also from the C haplotype (22). We conclude that the V, D, and J gene segments of hybridomas M9 and M19 are rearranged in cis. Presumably the V, D, and J gene segments of the other five hybridomas also rearranged in cis; however, no restrictions polymorphisms in the JH genes used in these clones were available to test directly this idea. The data confirm those of a previous study that found, by analyzing cDNA encoding trans-associated VH and Cα allotypes of heterozygous C/E rabbits, that the V, D, and J gene segments rearranged in cis (22).
Figure 1.
Comparison of nucleotide sequence of PCR-amplified VDJ-Cα cDNA from trans-associated VHa1 Cαf71 IgA-secreting hybridoma M19 to the sequence of genomic VH1-a1 (C haplotype), VH1-a2 (E haplotype), Cα5 (E haplotype), and Cα5 (C haplotype). The four VHa1 allotype-associated amino acids in FR1 and FR3 are boxed, and the allotype-associated nucleotides are indicated by asterisks; polymorphic nucleotides specific for C haplotype JH are boxed and indicated by diamonds (22). VH a2 allotype amino acids in FR1 and FR3 encoded by VH1-a2 are indicated by triangles.
Table 2.
Characterization of VDJ-Cα genes from seven trans-associated hybridomas from C/E haplotype (a1f72/a2f71) heterozygous rabbits*
| Hybridoma | (allotype) | Haplotype
|
Cα gene used | |
|---|---|---|---|---|
| VH | Cα | |||
| H34 | (a2f72) | E | C | Cᆇ |
| H276 | (a2f72) | E | C | Cα4 |
| H315 | (a2f72) | E | C | Cα5 |
| P39 | (a2f72) | E | C | Cᆠ|
| M9 | (a1f71) | C | E | Cα5 |
| M19 | (a1f71) | C | E | Cα5 |
| P4 | (a1f71) | C | E | Cα10‡ |
Based on nucleotide sequence analyses.
Cα gene cannot be identified definitively because many Cα genes of the C haplotype have not been characterized.
Cα1 exon deleted.
The nucleotide sequence of the Cα region of each of the seven hybridomas was compared with germline Cα sequences to determine which of the 13 Cα genes was expressed. We identified the Cα gene used in five of the seven hybridomas; three of them, including M19 in Fig. 1, used Cα 5, one used Cα4, and another used Cα10. The sequence of the cDNA of these five hybridomas was identical to that of the germline Cα gene from the haplotype indicated by ELISA. The Cα gene used in two hybridomas, H34 and P39, could not be identified definitively because the used Cα gene, as shown by ELISA with anti-allotype antisera, was derived from the C haplotype (f72 allotype), and we do not yet have germline sequences for all 13 Cα genes of this haplotype. Surprisingly, exon 1 in each of two of the PCR-amplified heavy chains was deleted (H34 and P4; Table 2). Such a deletion may have occurred during RNA processing, during isotype switch, or as a result of PCR-amplification. Taken together, the serologic and the nucleotide sequence analyses of the seven hybridomas show clearly that these hybridomas secreted trans-associated IgA heavy chains.
Genomic Cloning of Trans-Associated IgA Heavy Chain from a Fosmid DNA Library.
If the trans-association of the VH and Cα allotypes results from trans-chromosomal recombination or from trans-chromosomal gene conversion, then the expressed VH and Cα genes derived from different haplotypes will be juxtaposed. In trans-chromosomal gene conversion, the region 5′ of VH and 3′ of Cα would originate from the same IgH haplotype whereas in trans-chromosomal recombination, those regions would originate from different IgH haplotypes. If the trans-association of VH and Cα results from trans-splicing, then the expressed VH and Cα genes would not be juxtaposed but instead would remain in trans.
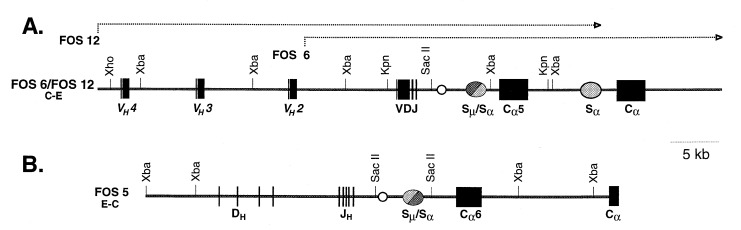
We cloned the rearranged VDJ and Cα genes from a fosmid DNA library from hybridoma M19 and screened the library with a JH probe. We isolated three clones, two of which, FOS6 and FOS12, also hybridized with both VH and Cα probes, and the third, FOS5, hybridized with a Cα probe but not with a VH probe. These three clones were restriction mapped and were examined by Southern blot analyses.
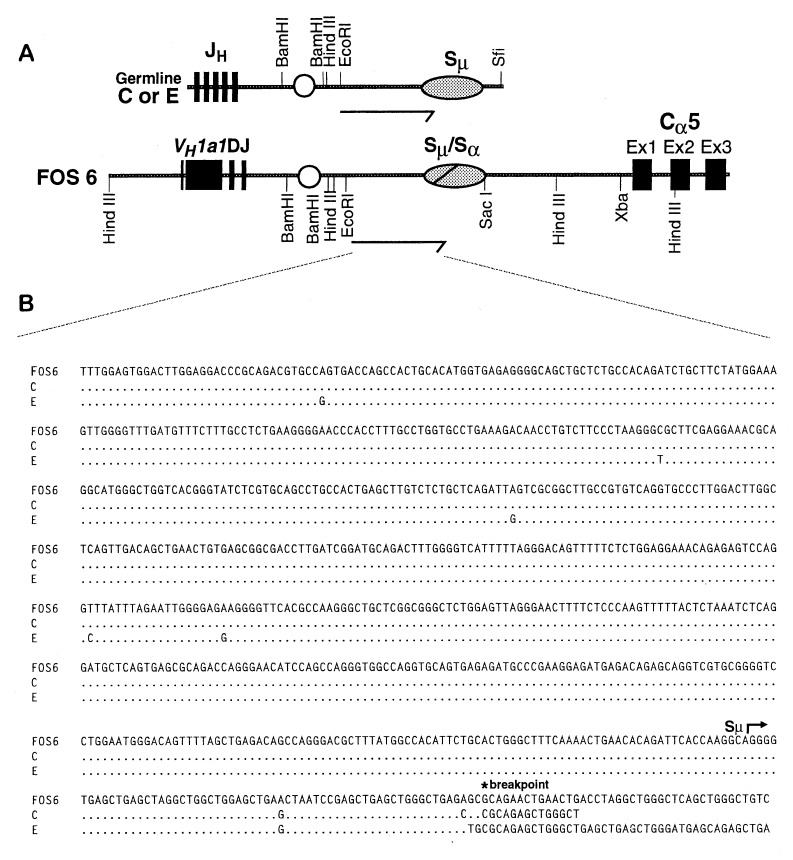
The Expressed IgH Allele.
Two of the fosmid clones, FOS6 and FOS12, were found to have overlapping restriction maps, and they spanned a total of 60 kb of DNA (Fig. 2A). This region contained the rearranged VDJ gene, three upstream VH genes, the heavy chain intron enhancer, and two Cα genes, each with a switch region ≈2 kb upstream. We determined the nucleotide sequence of the VDJ gene and proximal Cα gene and found that they were identical to that of the VDJ-Cα5 cDNA from the M19 hybridoma shown in Fig. 1. Thus, the fosmids contain, in juxtaposition, the expressed VH and Cα5 genes encoding the trans-associated α-chain of hybridoma M19. This finding rules out trans-splicing as the mechanism for trans-association of VH and Cα in clone M19.
Figure 2.
Partial restriction maps of fosmid clones from trans-IgA-secreting hybridoma M19. (A) Overlapping fosmid clones FOS6 and FOS12. (B) Clone FOS5. Solid boxes represent VH, D, JH, VDJ, and Cα exons; shaded ovals represent switch regions; unshaded circle represents intronic heavy chain enhancer.
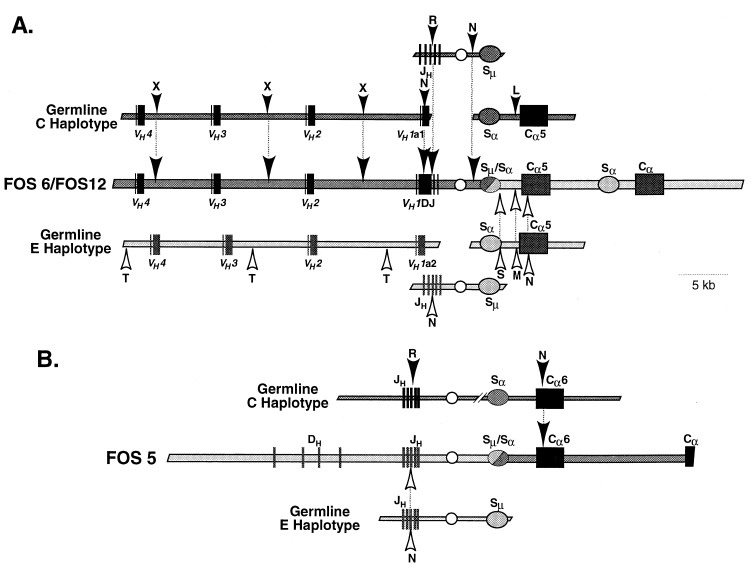
To distinguish between trans-recombination and trans-chromosomal gene conversion, we determined the haplotype origin of the regions surrounding the VH and Cα5 genes (Fig. 3A). The VDJ gene in FOS6 and FOS12 originated from the C haplotype, as shown by the identity of the restriction sites 5′ of the VDJ gene in the fosmid clones to the restriction sites in the region 5′ of VH1 of the C haplotype and not to those of the E haplotype. The restriction map and nucleotide sequence of Cα5 in the fosmids was identical to those of Cα5 cloned from the E haplotype and not those of the C haplotype, indicating that Cα5 originated from the E haplotype.
Figure 3.
Haplotype origin of the DNA in fosmids cloned from trans-IgA-secreting hybridoma M19. The restriction maps of FOS6 and FOS12 (A) and of FOS 5 (B) are compared with the restriction maps of germline VH, JH - Sμ, and Cα from C and E haplotypes. The polymorphisms indicated by arrowheads represent haplotype-specific restriction sites or nucleotide sequences. The letters associated with the arrowheads represent the following restriction sites: X, XbaI; R, EcoRI; L, SalI; T, SacII; S, SacI; M, SmaI; N, nucleotide sequence. Solid boxes, VH, VDJ, DH, JH and Cα exons; ovals, switch regions; circle, intronic heavy chain enhancer.
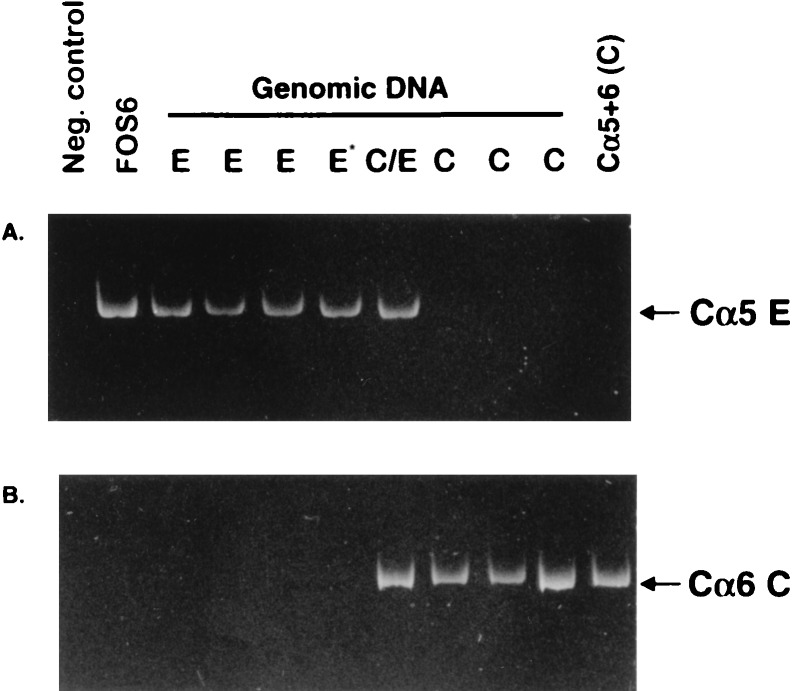
Because not all Cα genes of the C haplotype are cloned, we needed to test whether the Cα5 gene in the fosmids could have originated from the C haplotype rather than from the E haplotype. We searched for a gene such as Cα5(E) in genomic DNA of C haplotype rabbits by PCR by using as 5′ and 3′ primers oligomers specific for the Cα5 gene found in the fosmids and in Cα5 of the E haplotype. Although we obtained the expected PCR product from genomic DNA samples from E and C/E haplotype rabbits, we did not obtain a product from any of three samples from C haplotype rabbits (Fig. 4A). DNA from the C haplotype rabbits, however, could be PCR-amplified if primers for a known Cα gene of the C haplotype (Cα6C) were used (Fig. 4B). We conclude that the C haplotype does not contain a gene identical to the Cα5 gene in FOS6/FOS12. These data confirm our conclusion that the Cα gene in the fosmids did not originate from the C haplotype but instead were derived from the E haplotype. Taken together with the observation that the VDJ gene of FOS6/FOS12 originated from the C haplotype, we conclude that the trans-association of VH and Cα in hybridoma M19 resulted from a trans-chromosomal DNA recombination and not from trans-chromosomal gene conversion.
Figure 4.
Ethidium bromide-stained PAGE of PCR-amplified exon 1—hinge region of Cα5 (E haplotype) and Cα6 (C haplotype) genes from genomic DNA of C, E and C/E haplotype rabbits. (A) Primers specific for Cα5E. (B) Primers specific for Cα6C. Cα5+6 (C), control plasmid DNA for Cα5 + Cα6 (C haplotype). The E* template DNA was from a G haplotype rabbit. The negative control contained plasmid DNA from 11 cloned Cα genes (excluding Cα5 and Cα6) (21). The position for the correct sized product is indicated.
The Unexpressed Allele.
The fosmid clone FOS5 hybridized to JH and Cα probes but not to a VH probe (Fig. 2B). Restriction mapping and Southern blot analysis of this clone showed that the JH gene segments were in germline configuration but that the proximal CH gene segment was a Cα gene. On the basis of this configuration, FOS5 likely represented the other expected product of the recombination found in FOS6/FOS12. If this were the case, the JH region would be from the unexpressed E haplotype and the Cα region from the C haplotype. We performed RFLP and nucleotide sequence analyses of FOS5 (Fig. 3B), which showed that the unrearranged germline JH region was derived from the E haplotype. In addition, nucleotide sequence analysis of the Cα region showed that it was identical to Cα6 of the C haplotype (data not shown). To exclude the possibility that the Cα6 gene was derived from the E haplotype, we attempted to PCR-amplify such a gene from genomic DNA of E haplotype rabbits by using, as 5′ and 3′ primers, oligomers specific for exon 1 and the hinge region, respectively, of Cα6 (C haplotype). Although we obtained a PCR product from genomic DNA samples from C and C/E haplotype rabbits, we did not obtain a product from any of four genomic DNA samples from E haplotype rabbits (Fig. 4B), even though these DNA samples readily PCR-amplified with E haplotype-specific Cα5 primers (Fig. 4A). These data confirm that Cα6 in FOS5 originated from the C haplotype. We conclude that FOS5 represents the unexpressed allele and further that it represents the other expected product of the trans-recombination observed in FOS6 and FOS12.
Breakpoint for Trans-Recombination.
The RFLP and nucleotide sequence analyses that we performed revealed that the breakpoint of the trans-recombination event between the VDJ and Cα5 genes (FOS6 and FOS12) occurred 3′ of JH on the C haplotype and 5′ of Cα5 on the E haplotype. Because isotype switch occurs by recombination within or adjacent to the switch regions, we hypothesized that the trans-recombination occurred within the switch region. To test this idea, we subcloned portions of the switch region from FOS6 as well as from germline Sμ from both the C and E haplotypes (Fig. 5A), and we determined the nucleotide sequence of a portion of this DNA (Fig. 5B). We found that, of five nucleotide polymorphisms in the 689-bp 5′ of Sμ, FOS6 had all five characteristic of the C haplotype and none characteristic of the E haplotype. Beginning 61 bp into the repeated elements of Sμ, the sequence of FOS6 still had switch-like repeated elements, but they diverged markedly from the Sμ region of the C and E haplotypes. We suggest that these divergent switch-like elements are derived from the Sα region associated with Cα5 and that both switch recombination and the trans-chromosomal recombination occurred at this juncture.
Figure 5.
Expanded restriction map and nucleotide sequence of the recombination breakpoint. (A) VDJ-Cα5 region of FOS6 compared with JH-Sμ region of C and E haplotypes. Arrows indicate regions from which nucleotide sequences were determined. (B) Comparison of the nucleotide sequence from the EcoRI site of a 2.8-kb EcoRI/SacI fragment cloned from FOS6 to that of a 2.8-kb EcoRI/SfiI fragment cloned from Sμ of haplotypes C and E. Dots indicate identical nucleotides. The beginning of Sμ and the proposed site of recombination (breakpoint) are indicated.
Trans-Chromosomal Gene Conversion.
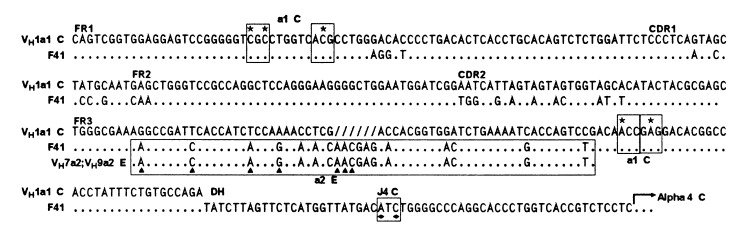
In the nucleotide sequence analysis of potential trans-associated IgA molecules, we found one hybridoma with an unusual sequence. This hybridoma, F41, secreted IgA that reacted with a2 and f72 g74 anti-allotype antibodies, suggesting that trans-recombination had occurred between VH of the E haplotype and Cα of the C haplotype. However, both the JH and most of the VH region were similar to genes of the C haplotype (Fig. 6), including nucleotides that encode all four a1 allotype-associated amino acids (two in FR1 and two in FR3). These data indicate that the V, D, and J gene rearrangements occurred on the C haplotype. The reaction with anti-a2 antibody probably resulted from the 65 bp in FR3 that were identical to the sequence of the a2-encoding VH genes, VH7-a2 and VH9-a2, from the E haplotype and encoded all five a2 allotype-associated amino acids of FR3. We suggest that this sequence in FR3 resulted from a gene conversion event in which a gene, such as VH7-a2 or VH9-a2 from the E haplotype, served as the donor.
Figure 6.
Trans-associated a2f72 α-chain cDNA derived from hybridoma F41. The nucleotide sequence of VDJ of F41 is compared with the sequence of the a1-encoding gene, VH1, (C haplotype). The area in FR3 identical to the a2-encoding genes VH7 and VH9 (accession nos. U51027 and U51029, respectively; E allele) is boxed; other markings are as in Fig. 1.
DISCUSSION
We developed IgA-secreting hybridomas that had trans-associated VH and Cα heavy chain allotypes and showed that the VH and Cα genes encoding these allotypes were derived from different parental haplotypes. From one of these hybridomas, M19, we cloned trans-derived, expressed VDJ and Cα genes and found them juxtaposed. We also cloned the other expected product of the trans-recombination, indicating that the trans-association of VH and Cα allotypes occurred by trans-chromosomal DNA recombination and not by trans-splicing of mRNA transcripts.
Trans-Chromosomal Recombination During Isotype Switching.
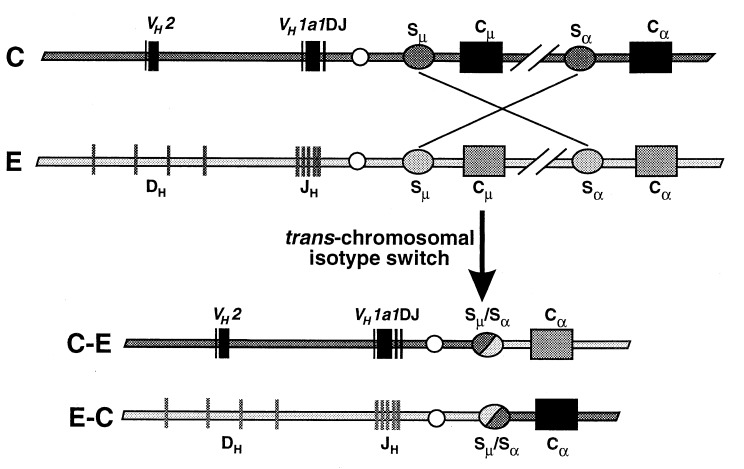
By RFLP and nucleotide sequence analyses of the fosmid clones containing the recombined VH and Cα genes (FOS6 and FOS12), we showed that the recombination breakpoint on the IgH allele with the VDJ gene occurred within Sμ. Although we did not search for the precise breakpoint of the recombination on the other IgH allele (FOS5), we showed that it occurred downstream of Eμ on the E haplotype and upstream of Cα6 on the C haplotype. We suggest that the trans-chromosomal DNA recombination likely occurred during isotype switch recombination between Sμ and Sα. We cannot, however, rule out the possibility that it occurred during an earlier isotype switch rearrangement, such as that between Sμ and Sγ, followed by an intrachromosomal isotype switch on both IgH chromosomes.
Switch recombination is known to occur frequently on both chromosomes (29–30), meaning that double-strand DNA breaks must occur on both chromosomes. If these double-strand breaks occur simultaneously, then the free ends of DNA from the two chromosomes must come into proximity so they can be ligated together. A model of events that likely occurred to generate the trans-recombination events is shown in Fig. 7. Initially, a VDJ gene rearrangement occurred in cis on the C allele whereas no JH rearrangement occurred on the E allele. The lack of DJ or VDJ gene rearrangement on the E allele is not unexpected because Tunyaplin and Knight (31) showed that, in ≈40% of rabbit B cells, JH remains in germline configuration on the unexpressed allele. The B cell with this cis VDJ gene rearrangement would have synthesized VHa1 μ-chain, and then, after antigenic stimulation, the B-cell would have isotype switched to IgA. In this case, the DNA, presumably on both chromosomes, cleaved in or near the Sμ and near Sα5 of the E haplotype and Sα6 of the C haplotype. We suggest that the free ends of DNA from VDJ-Sμ of the C haplotype then ligated to the Sα5 region of the E haplotype, and simultaneously, the JH-Sμ of the E haplotype ligated to Sα6 of the C haplotype. We do not know the relative positions of Cα5 of the E haplotype and Cα6 of the C haplotype because the organization of genes in both haplotypes is not complete. However, it appears that the trans-recombination may not be necessarily between allelic homologues, e.g., Cα5 and Cα6.
Figure 7.
Model for trans-chromosomal DNA recombination during isotype switching in hybridoma M19.
High Frequency Trans-Chromosomal Recombination in Normal B Cells.
Although trans-chromosomal DNA recombination of Ig genes has been observed in in vitro studies and in transgenic animals (9–11), our study shows that such trans-chromosomal DNA recombination occurs in vivo and in normal B cells and at a high frequency. We found that 7 (7%) of the 106 IgA hybridomas that reacted with anti-IgA allotype antibodies synthesized trans-associated IgA heavy chains. This number is consistent with data obtained by two-color immunofluorescence studies of IgA-secreting plasma cells in mucosal tissues and in quantitative immunoprecipitation studies with purified secretory IgA that found that 3 to 8% of both purified IgA molecules and IgA-secreting plasma cells had trans-associated VH and Cα heavy chains (8, 32). In general, trans-chromosomal recombination in somatic cells is rare. The high frequency of trans-chromosomal recombination in the IgH locus might reflect the high number of double-strand DNA breaks on both alleles during isotype switch. Although double-strand DNA breaks also may occur on both alleles during recombination of D and J gene segments, trans-chromosomal recombination seems to occur only infrequently at this stage. Presumably, this reflects differences in the mechanism by which the V(D)J and switch recombinations occur (33, 34).
Trans-Association of VH and Cα Allotypes Through Gene Conversion.
In hybridoma F41, we found that the VDJ and Cα genes originated from the same IgH haplotype (C), even though the reaction of anti-allotype antibodies with the secreted IgA indicated that they originated from different haplotypes. The trans-associated VHa2 allotype appeared to result from a trans-chromosomal gene conversion event with an a2-encoding gene like VH7 or VH9 from the E haplotype as donor. Both VH7 and VH9 have a2-encoding nucleotides identical to those found in FR3 of hybridoma F41, and both have been identified as potential gene conversion donors of a2 allotype-encoding nucleotides in VDJ genes (35, 36). Neither VH7 nor VH9 has been found to be used in VDJ gene rearrangements, and, consequently, we assume that the VH7 or VH9 donor of a2-encoding nucleotides was in germline configuration. Although we conclude that the a2-encoding nucleotides originated from an interchromosomal gene conversion, we cannot rule out the possibility that they originated from an intrachromosomal gene conversion using as donor a VH gene in cis that encodes a2-associated amino acids in FR3. However, in a1 rabbits, no VH gene that encodes a2-allotype associated amino acids has been found.
Surprisingly, the IgA of hybridoma F41 did not react with anti-a1 allotype antibody even though all four a1 allotype-associated amino acids in FR1 and FR3 were present. By three-dimensional modeling of the VH region of rabbit Ig (37), the VHa allotype-associated amino acids in FR1 and FR3 appear to be in close proximity, and together, they may form one epitope. We suggest that the three-dimensional folding of the F41 a1 molecule with the a2 allotype-associated amino acids results in the disruption of the a1 epitope so that the molecule no longer reacts with anti-a1 antibody.
Trans-Chromosomal Recombination and B-Cell Tumors.
The high frequency of recombination in trans during isotype switching may explain the high incidence of B-lineage tumors that arise from various chromosomal translocations into the switch region of the IgH locus. Translocations of c-myc, bcl-2, bcl-3, and bcl-6 frequently are associated with tumors such as murine plasmacytoma (38, 39), Burkitt’s lymphoma (39), chronic lymphocytic leukemia (40–42), follicular lymphoma (43), and diffuse large-cell lymphoma (44). Further, Bergsagel et al. (45) reported that 15 of 21 multiple myeloma cell lines had undergone IgH switch region translocations involving genes from at least six different chromosomal loci.
In summary, we demonstrated that trans-chromosomal recombination between homologous chromosome pairs occurs in B cells of normal animals. In at least one case, we showed that this trans-chromosomal recombination occurred in the Ig switch regions, presumably during isotype switching. We also found one example of trans-association of VH and CH regions that can be explained by a trans-chromosomal gene conversion event. We conclude that trans-chromosomal recombination within the IgH locus occurs at a high frequency in normal animals.
Acknowledgments
This work was supported by National Institutes of Health Grants AI11234 and AI16611.
ABBREVIATION
- RFLP
restriction fragment length polymorphism
- kb
kilobase
Footnotes
References
- 1. Hardy R R, Carmack C E, Shinton S A, Kemp J D, Hayaka K. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis M M, Kim S K, Hood L E. Science. 1980;209:1360–1365. doi: 10.1126/science.6774415. [DOI] [PubMed] [Google Scholar]
- 3.Davis M M, Calame K, Early P W, Livant D L, Joho R, Weissman I L, Hood L. Nature (London) 1980;283:773–779. doi: 10.1038/283733a0. [DOI] [PubMed] [Google Scholar]
- 4.Sakano H, Maki R, Kurosawa Y, Roeder W, Tonegawa S. Nature (London) 1980;286:676–683. doi: 10.1038/286676a0. [DOI] [PubMed] [Google Scholar]
- 5.Nikaido T, Yamawaki-Kataoka Y, Hongo T. J Biol Chem. 1982;257:7322–7329. [PubMed] [Google Scholar]
- 6.Landucci-Tosi S, Mage R G, Dubiski S. J Immunol. 1970;104:641–647. [PubMed] [Google Scholar]
- 7.Pernis B, Forni L, Dubiski S, Kelus A S, Mandy W J, Todd C W. Immunochemistry. 1973;10:281–285. doi: 10.1016/0019-2791(73)90023-2. [DOI] [PubMed] [Google Scholar]
- 8.Knight K L, Malek T, Hanly W C. Proc Natl Acad Sci USA. 1974;71:1169–1173. doi: 10.1073/pnas.71.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kipps T J, Herzenberg L A. EMBO J. 1986;5:263–268. doi: 10.1002/j.1460-2075.1986.tb04208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerstein R M, Frankel W N, Hsieh C L, Durdik J M, Rath S, Coffin J M, Nisonoff A, Selsing E. Cell. 1990;63:537–548. doi: 10.1016/0092-8674(90)90450-s. [DOI] [PubMed] [Google Scholar]
- 11.Umar A, Gearhart P J. Eur J Immunol. 1995;25:2392–2400. doi: 10.1002/eji.1830250840. [DOI] [PubMed] [Google Scholar]
- 12.Giusti A M, Coffee R, Manser T. Proc Natl Acad Sci USA. 1992;89:10321–10325. doi: 10.1073/pnas.89.21.10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giusti A M, Manser T. J Exp Med. 1994;179:235–248. doi: 10.1084/jem.179.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y-W, Word C, Dev V, Uhr J W, Vitetta E S, Tucker P W. J Exp Med. 1986;164:562. doi: 10.1084/jem.164.2.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimizu A, Nussenzweig M C, Han H, Sanchez M, Honjo T. J Exp Med. 1991;173:1385–1393. doi: 10.1084/jem.173.6.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shimizu A, Honjo T. FASEB J. 1993;7:149–154. doi: 10.1096/fasebj.7.1.7916698. [DOI] [PubMed] [Google Scholar]
- 17.Fujieda S, Lin Y Q, Saxon A, Zhang K. J Immunol. 1996;157:3450–3459. [PubMed] [Google Scholar]
- 18.Knight K L, Hanly W C. Contemp Top Mol Immunol. 1975;4:55–88. doi: 10.1007/978-1-4615-8930-3_3. [DOI] [PubMed] [Google Scholar]
- 19.Mage R G, Bernstein K E, McCartney-Francis N, Alexander C B, Young-Cooper G O, Padlan E A, Cohen G H. Mol Immunol. 1984;21:1067–1081. doi: 10.1016/0161-5890(84)90117-2. [DOI] [PubMed] [Google Scholar]
- 20.Knight K L, Burnett R C, McNicholas J M. J Immunol. 1985;134:1245–1250. [PubMed] [Google Scholar]
- 21.Burnett R C, Hanly W C, Zhai S K, Knight K L. EMBO J. 1989;8:4041–4047. doi: 10.1002/j.1460-2075.1989.tb08587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knight K L, Kingzette M, Crane M A, Zhai S-K. J Immunol. 1995;155:684–691. [PubMed] [Google Scholar]
- 23.Spieker-Polet H, Setupathi P, Yam P-C. Proc Natl Acad Sci USA. 1995;92:9348–9352. doi: 10.1073/pnas.92.20.9348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schneiderman R D, Hanly W C, Knight K L. Proc Natl Acad Sci USA. 1989;86:7561–7565. doi: 10.1073/pnas.86.19.7561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim U J, Shizuya H, de Jong P J, Birren B, Simon M I. Nucleic Acids Res. 1992;20:1083–1085. doi: 10.1093/nar/20.5.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knight K L, Martens C L, Stoklosa C M, Schneiderman R D. Nucleic Acids Res. 1984;12:1657–1670. doi: 10.1093/nar/12.3.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knight K L, Spieker-Polet H, Kazdin D S, Oi V T. Proc Natl Acad Sci USA. 1988;85:3130–3134. doi: 10.1073/pnas.85.9.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Aquila R T, Bechtel L J, Videler J A, Eron J J, Gorczyca P, Kaplan J C. Nucleic Acids Res. 1991;19:3749–3750. doi: 10.1093/nar/19.13.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lang R B, Stanton L W, Marcu K B. Nucleic Acids Res. 1982;10:611–630. doi: 10.1093/nar/10.2.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Radbruch A, Muller W, Rajewsky K. Proc Natl Acad Sci USA. 1986;83:3954–3957. doi: 10.1073/pnas.83.11.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tunyaplin C, Knight K L. J Immunol. 1997;158:4805–4811. [PubMed] [Google Scholar]
- 32.Martens C L, Gilman-Sachs A, Knight K L. In: The Immune System 1: Past and Future. Steinberg C M, Lefkovits I, editors. Vol. 1. Basel: Karger; 1981. pp. 291–298. [Google Scholar]
- 33.Ramsden D A, van Gent D C, Gellert M. Curr Opin Immunol. 1997;9:114–120. doi: 10.1016/s0952-7915(97)80167-7. [DOI] [PubMed] [Google Scholar]
- 34.Casellas R, Nussenzweig A, Wuerffel R, Pelanda R, Reichlin A, Suh H, Qin X F, Besmer E, Kenter A, Rajewsky K, et al. EMBO J. 1998;17:2404–2411. doi: 10.1093/emboj/17.8.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crane M A, Kingzette M, Knight K L. J Exp Med. 1996;183:2119–22127. doi: 10.1084/jem.183.5.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sehgal D, Mage R G, Schiaffella E. J Immunol. 1998;160:1246–1255. [PubMed] [Google Scholar]
- 37.Jaton J-C, Schweizer M, Knight K L. Eur J Immunol. 1976;6:878–882. [Google Scholar]
- 38.Shen-Ong G L, Keath E J, Piccoli S P, Cole M D. Cell. 1982;31:443–452. doi: 10.1016/0092-8674(82)90137-4. [DOI] [PubMed] [Google Scholar]
- 39.Taub R, Kirsch I, Morton C, Lenoir G, Swan D, Tronick S, Aaronson S, Leder P. Proc Natl Acad Sci USA. 1982;79:7837–7841. doi: 10.1073/pnas.79.24.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsujimoto Y, Finger L R, Yunis J, Nowell P C, Croce C M. Science. 1984;226:1097–1099. doi: 10.1126/science.6093263. [DOI] [PubMed] [Google Scholar]
- 41.Ohno H, Doi S, Yabumoto K, Fukuhara S, McKeithan T W. Leukemia. 1993;7:2057–2063. [PubMed] [Google Scholar]
- 42.Crossen P E, Kennedy M A, Heaton D C, Morrison M J. Genes Chromosomes Cancer. 1993;8:60–62. doi: 10.1002/gcc.2870080110. [DOI] [PubMed] [Google Scholar]
- 43.Kadowaki N, Hayashi T, Amakawa R, Akasaka T, Yabumoto K, Ohno H, Fukuhara S, Okuma M. Int J Hematol. 1995;61:69–75. doi: 10.1016/0925-5710(94)00347-h. [DOI] [PubMed] [Google Scholar]
- 44.Ye B H, Chaganti S, Chang C C, Niu H, Corradini P, Chaganti R S, Dalla-Favera R. EMBO J. 1995;14:6209–6217. doi: 10.1002/j.1460-2075.1995.tb00311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bergsagel P L, Chesi M, Nardini E, Brents L A, Kirby S L, Kuehl W M. Proc Natl Acad Sci USA. 1996;93:13931–13936. doi: 10.1073/pnas.93.24.13931. [DOI] [PMC free article] [PubMed] [Google Scholar]