Abstract
Very little is known about how cellular osmosensors monitor changes in osmolarity of the environment. Here, we report that in yeast, Sln1 osmosensor histidine kinase monitors changes in turgor pressures. Reductions in turgor caused by either hyperosmotic stress, nystatin, or removal of cell wall activate MAPK Hog1 specifically through the SLN1 branch, but not through the SHO1 branch of the high osmolarity glycerol pathway. The integrity of the periplasmic region of Sln1 was essential for its sensor function. We found that activity of the plant histidine kinase cytokinin response 1 (Cre1) is also regulated by changes in turgor pressure, in a manner identical to that of Sln1, in the presence of cytokinin. We propose that Sln1 and Cre1 are turgor sensors, and that similar turgor-sensing mechanisms might regulate hyperosmotic stress responses both in yeast and plants.
Keywords: signal transduction; high osmolarity stress; histidine kinase; two-component system; HOG MAPK pathway
Introduction
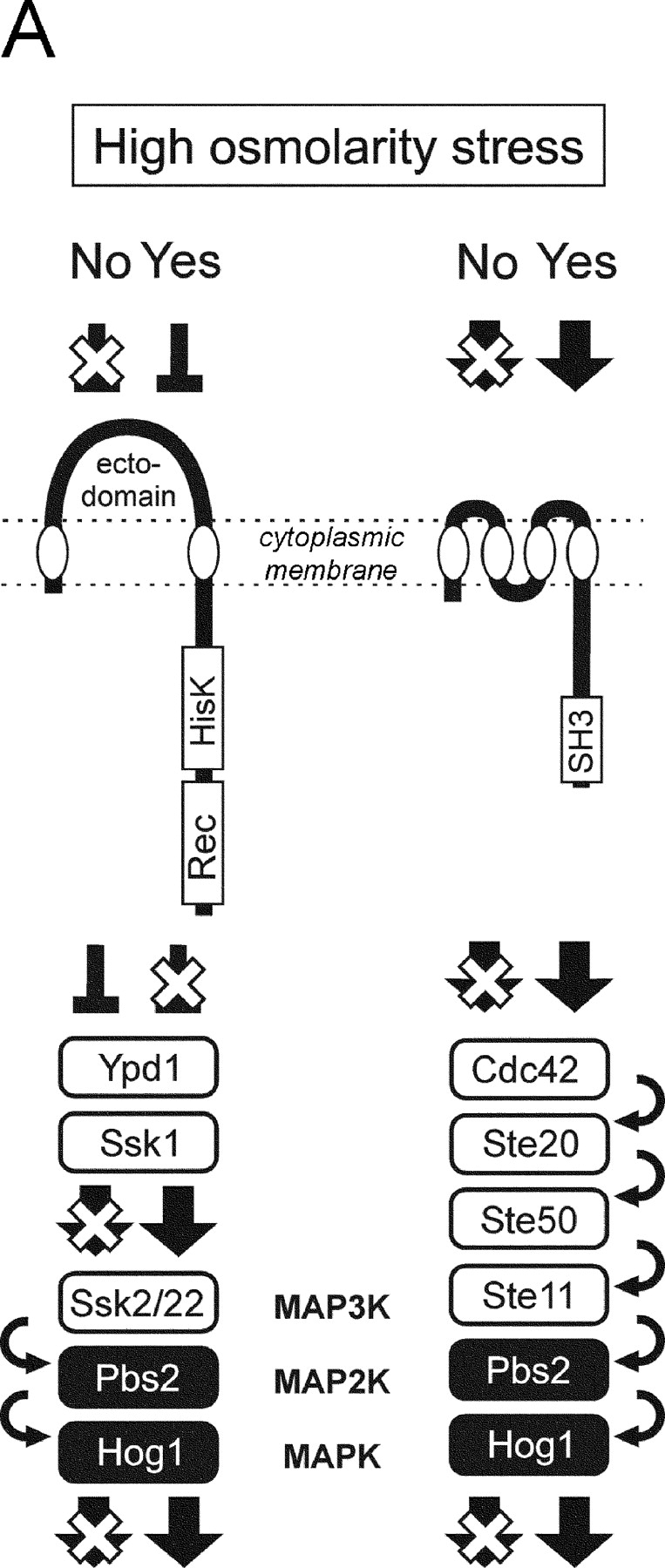
Cells have developed elaborate and sensitive protection systems that enable them to rapidly signal, respond, and properly adapt to osmotic changes. The high osmolarity glycerol (HOG)* MAPK signaling pathway in yeast and homologous p38 pathways in more complex eukaryotes play a central role in such osmoprotection systems. In budding yeast (Saccharomyces cerevisiae), exposure to a high osmolarity environment leads to rapid phosphorylation and activation of MAPK kinase Hog1 (Brewster et al., 1993; O'Rourke et al., 2002; Fig. 1 A). Hog1 is activated through phosphorylation by MAPK Pbs2, which is itself activated by upstream kinases, either through the SLN1 or SHO1 branch of the HOG pathway (O'Rourke et al., 2002; Fig. 1 A). Activated Hog1 rapidly (but transiently) accumulates in the nuclear compartment, where it participates in a modification of the transcriptional program in response to stress.
Figure 1.
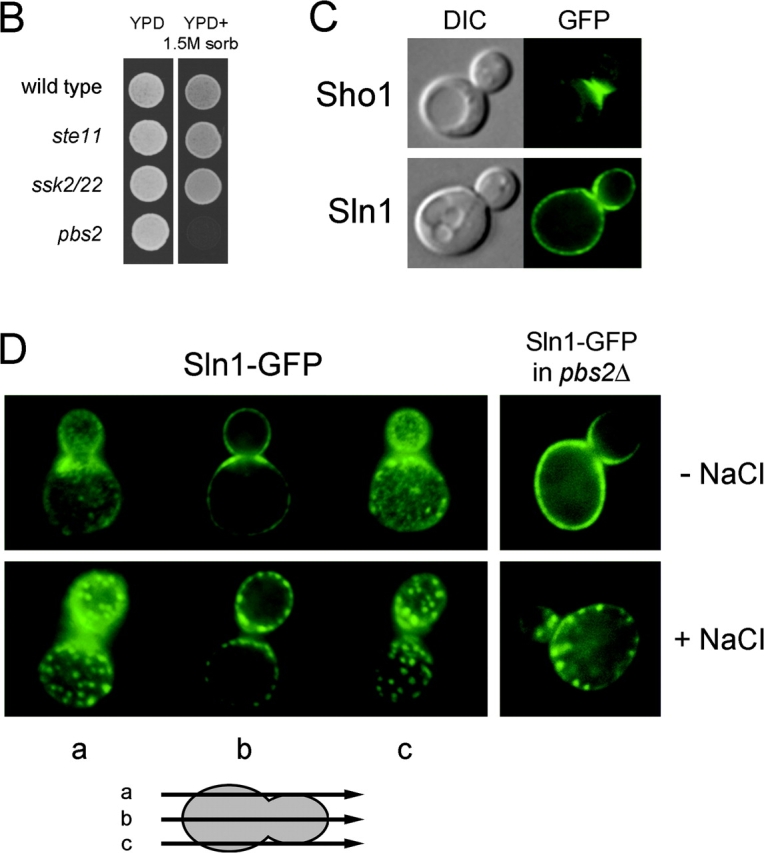
Sln1 and Sho1 have distinct cellular distributions. (A) Architecture of the SLN1 and SHO1 branches of the HOG pathway. (B) Either the SLN1 or SHO1 branch is sufficient to survive on high osmolarity. The wild-type, ste11Δ, ssk2Δ ssk22Δ, or pbs2Δ mutant cells were spotted on low (YPD) and high (YPD + 1.5 M sorbitol) osmolarity media plates, and were grown for 2 d at 30°C. (C) Sln1 has a nearly uniform cytoplasmic membrane distribution. The localization of the Sln1–GFP and the Sho1–GFP fusion proteins was analyzed by fluorescent microscopy in unstressed cells. GFP, fluorescence images; DIC, differential interference contrast images. (D) Hyperosmotic stress induces a clustering of Sln1. Wild-type or pbs2Δ mutant cells expressing Sln1–GFP were observed by fluorescent microscopy before and after (5 min) addition of 0.4 M NaCl.
Osmosensors are proteins whose primary role is to monitor fluctuations in external osmolarity and initiate an activation of signaling pathways for osmo-adaptation. Although a wealth of information is available on regulation of signaling pathways controlled by osmosensors in both prokaryotes and eukaryotes (Pratt et al., 1996; Gustin et al., 1998; Karin, 1998; Hohmann, 2002), our understanding of how osmosensors actually respond to osmotic changes is limited. Here, we studied the principles of a mechanism that monitors external osmolarity in yeast.
Results and discussion
The two upstream branches (henceforth called the SLN1 and SHO1 branches) in the HOG pathway respond independently to osmotic status of the environment and are apparently redundant (Fig. 1 B; Maeda et al., 1995). However, the unique compositions of the SLN1 and SHO1 branches suggest they have distinct cellular functions. In the SLN1 branch, a transmembrane (TM) histidine kinase Sln1 serves as an osmosensor, and transmits the signal through the Sln1–Ypd1–Ssk1 multistep phosphorelay to the redundant pair of kinases Ssk2 and Ssk22 (Maeda et al., 1994; Posas et al., 1996). In contrast, another TM protein (Sho1) serves as a facilitator of signaling module assembly that includes Pbs2, Ste11, Ste20, and Cdc42 (Raitt et al., 2000; Reiser et al., 2000).
The subcellular distributions of Sln1 and Sho1 are consistent with such functional specialization. Sho1 is predominantly associated with the cytoplasmic membrane at the places of polarized growth, and does not change its localization on osmotic shock (Fig. 1 C). In contrast, Sln1 is distributed uniformly throughout the cytoplasmic membrane, except perhaps the regions where Sho1 is localized (Fig. 1, C and D). Furthermore, the localization of Sln1 changes in response to hyperosmotic stress by rapidly clustering into dotlike structures (Fig. 1 D). This relocalization of Sln1 is transient and is independent of Hog1 kinase activation (Fig. 1 D, right panels).
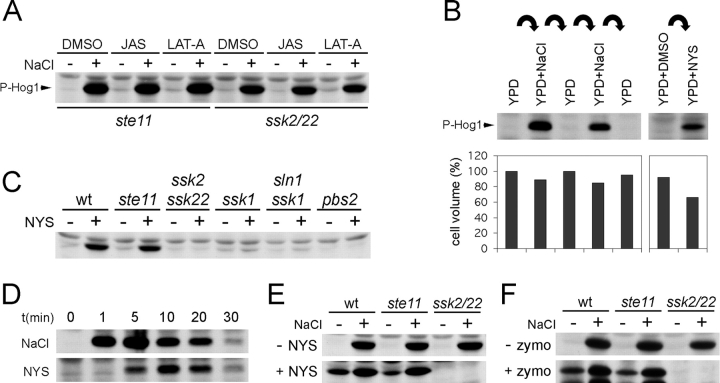
Activation of the HOG pathway in response to high osmolarity stress is accompanied by a dynamic reorganization of the actin cytoskeleton (Brewster and Gustin, 1994). Our data (Fig. 2 A) show that the levels of Hog1 activity are affected neither by actin-destabilizing drug latrunculin A (Ayscough et al., 1997), nor by actin-stabilizing drug jasplakinolide (Ayscough, 2000). Thus, the dynamic actin structures are not involved in regulation of the HOG pathway.
Figure 2.
The SLN1 branch of the HOG pathway is stimulated by turgor reduction. (A) The actin cytoskeleton dynamics does not affect the HOG pathway regulation. The ste11Δ or ssk2Δ ssk22Δ mutant cells were preincubated for 30 min in the presence of latrunculin-A (LAT-A, 100 μM), jasplakinolide (JAS, 10 μM), or the vehicle DMSO, and the Hog1 phosphorylation was assayed before and after (5 min) addition of 0.4 M NaCl. In each strain, the multi-drug resistance gene PDR5 was deleted to increase drug uptake. (B) The activation of Hog1 correlates with turgor-dependent cell volume shrinkage. The wild-type cells were exposed to cycles of high (YPD + 0.4 M NaCl) and low (YPD) osmolarity media in 2-min intervals. For each cycle, samples were withdrawn to determine the Hog1 phosphorylation and the relative cell volume. The wild-type cells treated with nystatin (10 μM, 5 min) were analyzed similarly. (C) Nystatin activates the SLN1 branch of the HOG pathway. The phosphorylation of Hog1 was determined in the mutant strains after treatment with nystatin (NYS; 10 μM, 5 min). (D) Time course of Hog1 activation by nystatin. Wild-type cells were treated with 0.4 M NaCl or 10 μM nystatin (NYS), and the Hog1 phosphorylation was analyzed. (E and F) Treatment by nystatin or removal of cell wall stimulates the SLN1 branch of the HOG pathway. The wild-type, ste11Δ, or ssk2Δ ssk22Δ cells were treated with nystatin (E; +NYS, 10 μM) or zymolyase (F; +zymo). The control samples (−NYS and −zymo) were prepared identically, except the nystatin or zymolyase addition. The samples were analyzed for the Hog1 phosphorylation directly or after (0.4 M NaCl, 5 min) osmotic stress.
High osmolarity stress also causes a rapid reduction of turgor pressure with associated reduction in cell volume (Fig. 2 B; Albertyn et al., 1994; Gervais and Beney, 2001). Thus, we tested if a turgor pressure manipulation, without any application of osmotic stress, could activate the HOG pathway. For this purpose, we used a membrane-permeabilizing antifungal drug, nystatin (Bolard, 1986). Leakage of low mol wt cytosolic components imitates a reduction in turgor caused by water efflux during high osmolarity stress. Nystatin was indeed an effective inducer of cell volume shrinkage, an indication of reduced turgor, in yeast (Fig. 2 B). More important, a treatment of the cells with nystatin strongly activated the HOG pathway (Fig. 2 B). We also observed that nystatin activates Hog1 in both wild-type and ste11 mutant cells, but not in the ssk2 ssk22 double mutant or the ssk1 mutant (Fig. 2, C and E), suggesting that nystatin selectively stimulates the SLN1 branch (Fig. 1 A). The nystatin effect is specific, because (1) Hog1 activation occurred at nystatin concentrations that are too low to evoke a genuine osmo-stress response; (2) nystatin activates only the SLN1 branch; and (3) activation of the SLN1 branch occurred before nystatin halted the cell growth (Fig. 2 D; unpublished data). The Hog1 activation by nystatin is not caused by alternations in membrane ergosterols because levels of both uninduced and induced Hog1 phosphorylation in ergosterol biosynthesis mutants (erg6; Gaber et al., 1989) are similar to those found in the wild-type strain (Fig. S1 A, available at http://www.jcb.org/cgi/content/full/jcb.200301099/DC1). The nystatin finding is consistent with a concept of turgor-dependent regulation of the Sln1 osmosensor. It is also consistent with an observation that water stress is not, per se, an inducer of the yeast osmoresponse (Tamas et al., 2000).
Reduction in turgor, induced by high osmolarity, leads concomitantly to shrinkage of cytosolic volume and an increase in the distance between plasma membrane and cell wall. Thus, monitoring of turgor pressure by the Sln1 osmosensor might be affected through the contact between plasma membrane and cell wall. To simulate the conditions of membrane detached from cell wall, we enzymatically removed the yeast cell wall using zymolyase. To keep the spheroplasts from rupturing, they are formed in media containing 1 M sorbitol. Although this concentration of sorbitol induces a transient activation of Hog1, the Hog1 activity returns to its basal level during the 2-h preincubation in sorbitol media before addition of zymolyase (in such adapted cells, Hog1 can be reactivated with additional osmotic stress). Removal of the cell wall gave rise to Hog1 activation in spheroplasts (Fig. 2 F), and this activation was through the SLN1 pathway because it is abrogated in ssk2 ssk22 double mutant, but not in ste11 mutant. Our preparation of spheroplasts could respond to high osmolarity stress by further activation of Hog1 (Fig. 2 F), likely due to remaining patches of intact cell wall in the spheroplasts. The activation of the SLN1 branch is not indirectly induced by degradation of the membrane Sln1 protein by proteolytic activity in zymolyase preparations (Fig. S1, B and C). Together, these observations indicate that there is a causal link between the pressure of the plasma membrane against the cell wall and the stimulation of Hog1 through the SLN1 branch. Perhaps consistent with this model, nystatin treatment did not activate the mammalian osmo-stress responsive p38 MAPK, an orthologue of Hog1, in wall-less HeLa cells (Fig. 3 C).
Figure 3.
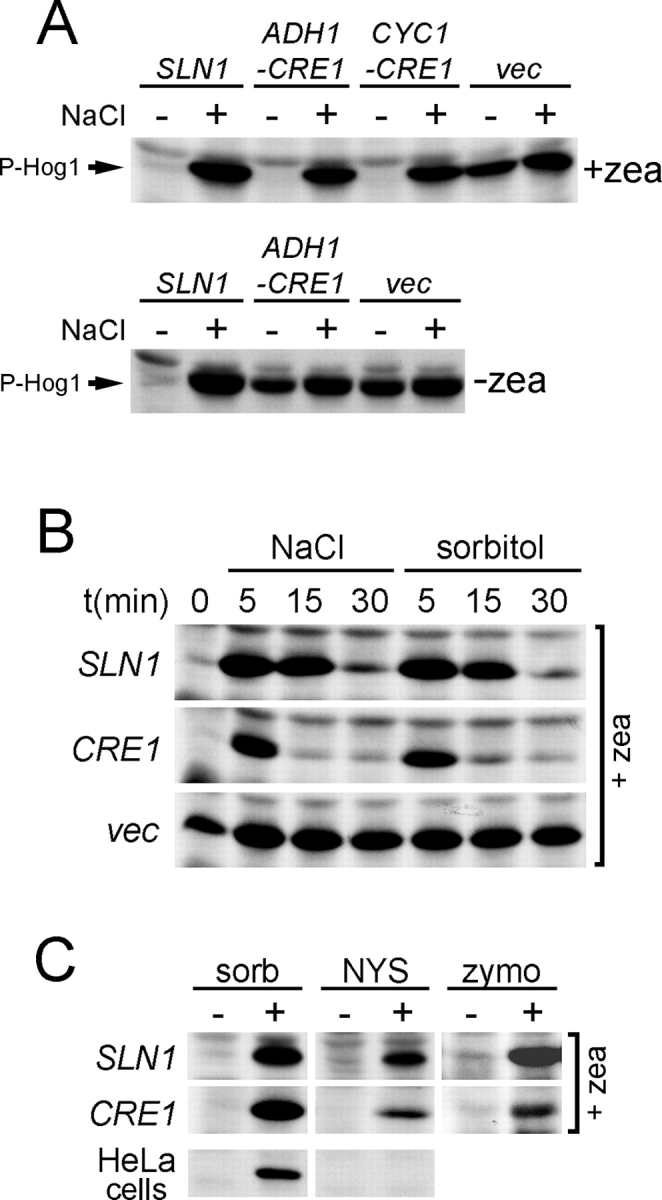
Plant histidine kinase Cre1 responds to changes in turgor pressure. (A) The active (zeatin-bound) Cre1 histidine kinase is inhibited by high osmolarity stress. The double mutant sln1Δ ste11Δ GAL1-PTP2 (strain BVRY179) expressing plasmid encoding for SLN1 (sln1Δ SLN1), CRE1 (sln1Δ ADH1-CRE1 or sln1Δ CYC1-CRE1), or empty vector (sln1Δ vec), was grown in glucose media (see Materials and methods for the experimental details) in the presence (+zea) or absence (−zea) of zeatin. The activity of Hog1 was then determined in these strains before and after (0.4 M NaCl, 5 min) high osmolarity stress. (B) The time course of Hog1 activation by NaCl or sorbitol in cells expressing Cre1. The Hog1 activity was analyzed in the sln1Δ ste11Δ GAL1-PTP2 strain (BVRY179) carrying a plasmid encoding SLN1 or CRE1 or vector alone (vec). These strains were grown in glucose media (as in A) supplemented with 10 μM zeatin and stressed by 0.4 M NaCl or by 1.0 M sorbitol for indicated times. (C) Cre1 activity responds to reduction in turgor pressure. The sln1Δ ste11Δ GAL1-PTP2 strain (BVRY179) expressing SLN1 or CRE1 was incubated with sorbitol (sorb; 1.0 M, 5 min), nystatin (NYS; 10 μM, 5 min), or zymolyase (zymo; as in Fig. 2 F) before the samples were assayed for Hog1 phosphorylation. HeLa cells were treated with 1 M sorbitol for 15 min before cell extracts were prepared to determine p38 MAPK activation by immunoblot analysis using an anti-phospho-p38 antibody.
Cytokinin response 1 (Cre1) is a plant (Arabidopsis thaliana) hybrid histidine kinase, which has been identified as a receptor for the plant hormone cytokinin (Inoue et al., 2001; Suzuki et al., 2001; Ueguchi et al., 2001; Yamada et al., 2001), and has been shown to be involved in plant vascular morphogenesis (Mähönen et al., 2000). Cre1 and Sln1 have similar domain organization; however, high level sequence similarity is limited only to the cytoplasmic histidine kinase and receiver domains (Saito, 2001). When expressed in yeast in the presence of a cytokinin (e.g., zeatin), Cre1 functionally interacts with the yeast Ypd1–Ssk1 phosphorelay system and suppresses the Hog1 hyperactivation in the sln1 deletion mutant (Fig. 3 A and Fig. S3 A; Inoue et al., 2001). To find functional difference between Sln1 and Cre1, we measured the activity of Hog1 in response to osmotic stress in sln1Δ mutant expressing CRE1. Surprisingly, high osmolarity stress rapidly activated the HOG pathway in CRE1-expressing cells in the presence of zeatin, indicating that the active form of Cre1 (i.e., bound to zeatin) has transformed to inactive in response to sudden increase of external osmolarity (Fig. 3 A, top). This was neither an indirect consequence of zeatin dissociating from its receptor at higher ionic concentrations (because nonionic sorbitol had the same effect; Fig. 3 B), nor due to interference of zeatin with yeast osmosensing system (Fig. 3 A, bottom). Cre1 activity could also be modulated by turgor pressure. The sln1Δ mutants expressing CRE1, or SLN1 as a control, were treated either with nystatin or with zymolyase. The both treatments induced Hog1 activation in sln1Δ CRE1 cells (Fig. 3 C). Together, these results indicate that cytokinins and hyperosmotic stress (or reduction of turgor) regulate antagonistically the activity of Cre1 in yeast. The remarkable similarities between Sln1 and Cre1, and the fact that plant cells are also surrounded by the cell wall, suggest that Cre1 could have a dual sensor function as a cytokinin receptor and an osmosensor in plants (Fig. S3, A and B). However, this possibility needs to be tested in the plant system. It has been proposed that another plant histidine kinase, ATHK1, might be involved in osmosensing in Arabidopsis (Urao et al., 1999).
How do Sln1 and Cre1 histidine kinases monitor changes in turgor pressure? It is possible that Sln1 and Cre1 mediate a physical contact between cell wall and plasma membrane, perhaps through a specific binding site in their periplasmic ectodomains. However, the results of systematic deletion analysis in the Sln1 ectodomain argues, rather, against this possibility. We identified only a sequence of 13 amino acids (aa 138–150; Sln1Δ9) in the Sln1 ectodomain that was essential for its function (Fig. 4 A and Fig. S4 A). However, because Sln1Δ9 had altered cellular distribution as well as the total protein level (Fig. 4, C and D), we believe that these defects are responsible for the loss-of-function phenotype. The fact that the phenotype of sln1Δ9 could be suppressed by overexpression of Vps10, a protein involved in protein sorting, supports this explanation (Fig. 4, D and E). Although contact of Sln1 with the cell wall could be, in principle, mediated by N-linked oligosaccharides, this is also unlikely because the mutations of the two putative N-glycosylation sites in the essential part of ectodomain did not affect the function of Sln1 (Fig. 4 B and Fig. S4 B).
Figure 4.
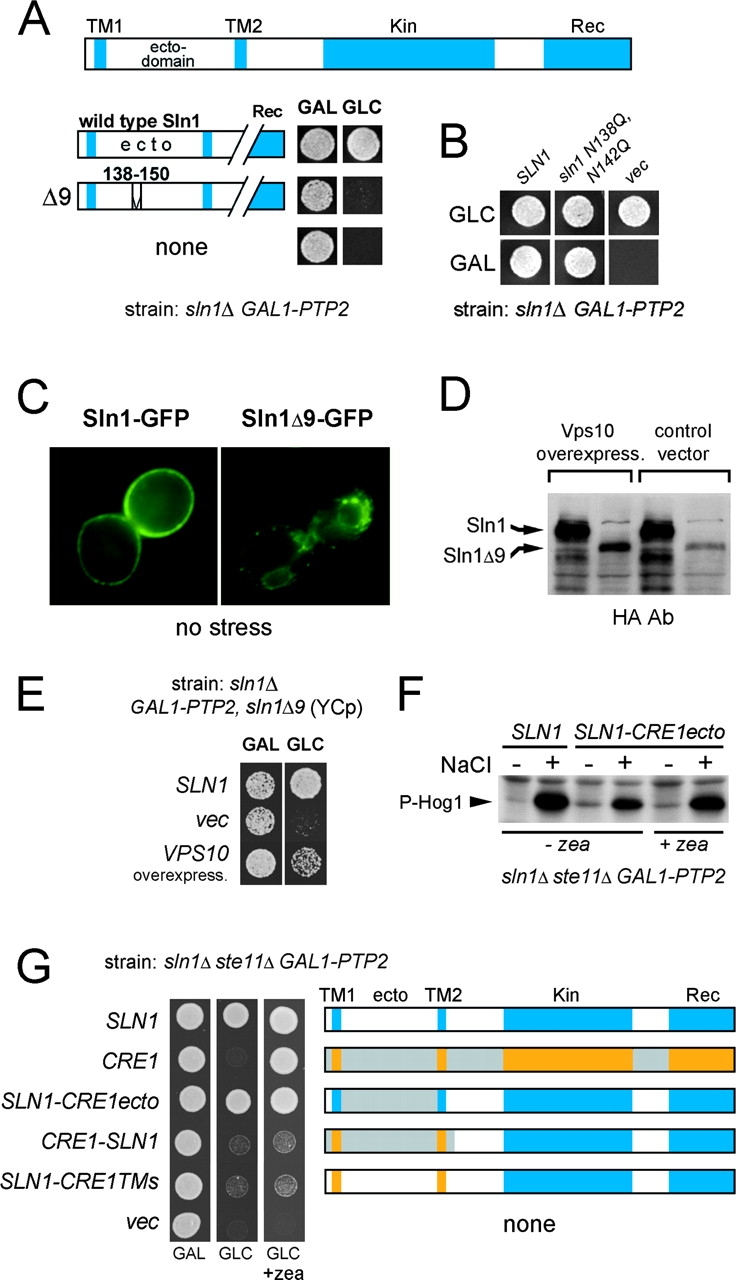
Integrity of extracellular domain is essential for Sln1 function. (A) Deletion of aa 138–150 in the periplasmic domain impairs the Sln1 function. The culture of the sln1Δ GAL1-PTP2 strain (TM182) grown in galactose media and carrying centromeric plasmid encoding for wild-type SLN1 or the mutant SLN1 with a deletion of aa 138–150 (sln1Δ9) was dropped onto media containing galactose (GAL) or glucose (GLC). (B) Absence of N-linked glycosylation is not responsible for the phenotype of the sln1Δ9 mutant. The sln1N138, 142Q mutant was tested for the complementation of the sln1Δ strain (TM182) as described in A. (C) The Sln1Δ9–GFP fusion protein has abnormal cellular distribution. The localization of Sln1–GFP and Sln1Δ9–GFP expressed from centromeric plasmid was determined in the sln1Δ strain (TM182) by fluorescent microscopy. (D and E) The phenotype of sln1Δ9 is suppressed by VPS10 overexpression. VPS10 was identified as a multicopy suppressor gene of the sln1Δ9 mutant (tested as in A). (E) The protein levels of HA-tagged Sln1 or Sln1Δ9 were compared by immunoblot analysis in the sln1Δ strain (TM182) transformed with a VPS10 multicopy plasmid or the empty vector (D). (F) The periplasmic domains of Sln1 and Cre1 are interchangeable. The culture of the sln1Δ ste11Δ GAL1-PTP2 strain (BVRY179) transformed with the SLN1 variant containing the periplasmic domain from Cre1 (SLN1-CRE1ecto) was analyzed for Hog1 phosphorylation before and after osmotic stress (0.4 M NaCl) in the absence or presence of zeatin in glucose media as in Fig. 3 A. (G) Domain swapping analysis between Sln1 and Cre1. The centromeric plasmids encoding Sln1–Cre1 hybrids were tested for the complementation of the sln1Δ ste11Δ GAL1-PTP2 strain (BVRY179) as described in A.
To further dissect a relative importance of the Sln1 periplasmic and TM segments, we constructed and examined the capacity of Sln1–Cre1 hybrid constructs to complement sln1Δ phenotype. Interestingly, despite the significant differences in primary structures in the ectodomain between Sln1 and Cre1, the Sln1–Cre1 hybrid with the periplasmic domain derived from Cre1 (SLN1-CRE1ecto) behaved indistinguishably from wild-type Sln1 under both unstressed and stressed growth conditions (Fig. 4 F). The SLN1-CRE1ecto was completely independent of cytokinin (Fig. 4, F and G). On the other hand, the hybrids with swapped TM segments only (SLN1-CRE1TMs), or TM and periplasmic regions (CRE1-SLN1), failed to complement the sln1Δ mutant (Fig. 4 G). Thus, a particular TM–cytoplasmic domain combination is unique for each histidine kinase and cannot be separated, whereas periplasmic domains are compatible with heterologous TM–cytoplasmic domain combination. Our results also suggest that an integrity of the periplasmic domain as a whole, rather then a particular region or amino acid sequence, is essential for Sln1 function. This observation might point to common architectural characteristics among all histidine kinase–based osmosensors because a similar observation has been made for bacterial osmosensor EnvZ (Leonardo and Forst, 1996).
In summary, we have presented evidence that turgor pressure is a key factor that regulates the activity of yeast osmosensor Sln1. Loss of turgor inactivates the Sln1 histidine kinase activity, leading to subsequent phosphorylation and activation of the HOG MAPK pathway. Also, we have shown that the plant cytokinin receptor Cre1, when activated by cytokinin, can substitute the Sln1 osmosensing function, and that its kinase activity is similarly regulated by turgor pressure. The compatibility of Cre1 with yeast hyperosmotic stress signaling pathway suggests that a mechanistically analogous osmosensing system could exist in plants.
Materials and methods
General methods
Standard recombinant DNA techniques are described by Sambrook et al. (1989). Yeast media, growth conditions, and procedures were used as presented by Guthrie and Fink (1994). Amino acids were omitted as necessary to select for plasmids. SLN1 ectodomain deletion constructs and hybrids between SLN1 and CRE1 were prepared using an overlapping PCR technique (Innis, 1990). The exact sequences of oligonucleotides used is available on request. HA- and GFP-tagged Sln1 proteins were constructed using vectors based on pRS415 (Sikorski and Hieter, 1989). HA-Sln1 contains three tandem repeats of HA epitope (YPYDVPDYA). Sln1–GFP has the EGFP coding sequence inserted immediately before the SLN1 stop codon. STE11 gene, in the strain BVRY179 (α ura3 leu2 his3 sln1::hisG ste11::HIS3 GAL1::PTP2), was deleted by the microhomology PCR method (Manivasakam et al., 1995). CRE1 plasmid was a gift from T. Kakimoto (Osaka University, Toyonaka, Japan).
Microscopy
Microscopic analyses were performed as described previously (Reiser et al., 1999). Fluorescent and differential interference contrast images were acquired using a microscope (Eclipse TE300; Nikon) equipped with a GFP filter set and a differential interference contrast objective. Images were captured using the MetaMorph® imaging software (Universal Imaging Corp.).
Immunoblot analysis
Hog1 phosphorylation was determined by immunoblot analysis of cell lysates using an anti-phospho-p38 antibody (Cell Signaling). HA-tagged Sln1 variants were analyzed by an anti-HA antibody conjugated to HRP (Roche).
Zymolyase treatment and spheroplast preparation
Logarithmic cell cultures were spun down and resuspended in YPD media containing 1.0 M sorbitol, buffered with 20 mM sodium phosphate (pH 7.5), and were shaken gently (60 rpm) for 2 h. Zymolyase 100T (Seikagaku Co.) at final concentration 50 mU μl−1 and 10 mM 2-mercaptoethanol was added to cell suspension and incubated for 30 min. Formation of spheroplasts was scored by light microscopy as a relative number of cells sensitive to lysis by 2% SDS. Hog1 phosphorylation was analyzed in samples with >90% of cells converted to spheroplasts.
Cell volume measurement
Cells were fixed directly in media by addition of 3.7% formaldehyde for 15 min, washed several times with distilled water, sonicated to remove cell aggregates, and cell volume was measured using a particle counter (Coulter Counter® Z1 series; Coulter International Corporation). Each measurement was repeated three times.
Assay of Hog1 phosphorylation and cell growth in the sln1Δ strains
The sln1Δ mutant is nonviable due to a hyperactivation of Hog1 kinase (Fig. S2; Maeda et al., 1994). The lethality of the sln1Δ deletion is rescued in the strain TM182 (sln1Δ GAL1-PTP2), or its derivative BVRY179 (sln1Δ ste11Δ GAL1-PTP2), by overexpression of the Ptp2 protein tyrosine phosphatase, induced by galactose, but repressed by glucose (Fig. S2). For determination of Hog1 activity in the sln1Δ strains carrying various SLN1, CRE1, or SLN1-CRE1 hybrid DNA constructs, cells were grown overnight in galactose media (PTP2 expression on) and then shifted for 6 h to glucose media supplemented (or not in the control experiments) with 10 μM zeatin. At this time point, Ptp2 is diluted sufficiently to observe activation of Hog1, but cells are still viable (Fig. S2). For complementation analysis of the sln1Δ mutant growth phenotype on solid media, cell cultures were grown overnight in galactose media and then dropped directly onto tested plates containing galactose or glucose and grown for 3 d at 30°C.
Online supplemental material
Online supplemental material demonstrates that Hog1 activation is not induced by ergosterol depletion or proteolytic degradation (Fig. S1). The growth curve of the sln1Δ mutant is shown in Fig. S2. Fig. S3 illustrates regulation of Sln1 and Cre1 by high omolarity stress. Fig. S4 shows the details of Sln1 ectodomain deletion analysis. Online supplemental material available at http://www.jcb.org/cgi/content/full/jcb.200301099/DC1.
Supplemental Material
Acknowledgments
We thank T. Kakimoto for plasmids, P. Silver for advice, and E.A. Witten for excellent technical assistance.
This work was supported in part by grants from the National Institutes of Health (GM56699); the Ministry of Education, Culture, Sports, Science, and Technology of Japan; and the Asahi Glass Foundation (to H. Saito). V. Reiser is a recipient of Human Frontiers Scientific Program Long-term Fellowship (LT0312/2000 M).
The online version of this article includes supplemental material.
Footnotes
Abbreviations used in this paper: Cre1, cytokinin response 1; HOG, high osmolarity glycerol; TM, transmembrane.
References
- Albertyn, J., S. Hohmann, and B.A. Prior. 1994. Characterization of the osmotic-stress response in Saccharomyces cerevisiae: osmotic stress and glucose repression regulate glycerol-3-phosphate dehydrogenase independently. Curr. Genet. 25:12–18. [DOI] [PubMed] [Google Scholar]
- Ayscough, K.R. 2000. Endocytosis and the development of cell polarity in yeast require a dynamic F-actin cytoskeleton. Curr. Biol. 10:1587–1590. [DOI] [PubMed] [Google Scholar]
- Ayscough, K.R., J. Stryker, N. Pokala, M. Sanders, P. Crews, and D.G. Drubin. 1997. High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 137:399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolard, J. 1986. How do the polyene macrolide antibiotics affect the cellular membrane properties? Biochim. Biophys. Acta. 864:257–304. [DOI] [PubMed] [Google Scholar]
- Brewster, J.L., and M.C. Gustin. 1994. Positioning of cell growth and division after osmotic stress requires a MAP kinase pathway. Yeast. 10:425–439. [DOI] [PubMed] [Google Scholar]
- Brewster, J.L., T. de Valoir, N.D. Dwyer, E. Winter, and M.C. Gustin. 1993. An osmosensing signal transduction pathway in yeast. Science. 259:1760–1763. [DOI] [PubMed] [Google Scholar]
- Gaber, R.F., D.M. Copple, B.K. Kennedy, M. Vidal, and M. Bard. 1989. The yeast gene ERG6 is required for normal membrane function but is not essential for biosynthesis of the cell-cycle-sparking sterol. Mol. Cell. Biol. 8:3447–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervais, P., and L. Beney. 2001. Osmotic mass transfer in the yeast Saccharomyces cerevisiae. Cell. Mol. Biol. (Noisy-le-grand). 47:831–839. [PubMed] [Google Scholar]
- Gustin, M.C., J. Albertyn, M. Alexander, and K. Davenport. 1998. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1264–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and G.R. Fink. 1994. Guide to yeast genetics and molecular and cell biology. Academic Press, San Diego. 933 pp.
- Hohmann, S. 2002. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66:300–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innis, M.A. 1990. PCR protocols: a guide to methods and applications. Academic Press, Inc., San Diego, CA. 482 pp.
- Inoue, T., M. Higuchi, Y. Hashimoto, M. Seki, M. Kobayashi, T. Kato, S. Tabata, K. Shinozaki, and T. Kakimoto. 2001. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature. 409:1060–1063. [DOI] [PubMed] [Google Scholar]
- Karin, M. 1998. Mitogen-activated protein kinase cascades as regulators of stress responses. Ann. NY Acad. Sci. 851:139–146. [DOI] [PubMed] [Google Scholar]
- Leonardo, M.R., and S. Forst. 1996. Re-examination of the role of the periplasmic domain of EnvZ in sensing of osmolarity signals in Escherichia coli Mol. Microbiol. 3:405–413. [DOI] [PubMed] [Google Scholar]
- Maeda, T., S.M. Wurgler-Murphy, and H. Saito. 1994. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 369:242–245. [DOI] [PubMed] [Google Scholar]
- Maeda, T., M. Takekawa, and H. Saito. 1995. Activation of yeast PBS2 MAPKK by MAPKKKs or by binding of an SH3- containing osmosensor. Science. 269:554–558. [DOI] [PubMed] [Google Scholar]
- Mähönen, A.P., M. Bonke, L. Kauppinen, M. Riikonen, P.N. Benfey, and Y. Helariutta. 2000. A novel two-component hybrid molecule regulates vascular morphogenesis of the Arabidopsis root. Genes Dev. 14:2938–2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manivasakam, P., S.C. Weber, J. McElver, and R.H. Schiestl. 1995. Micro-homology mediated PCR targeting in Saccharomyces cerevisiae. Nucleic Acids Res. 23:2799–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Rourke, S.M., I. Herskowitz, and E.K. O'Shea. 2002. Yeast go the whole HOG for the hyperosmotic response. Trends Genet. 18:405–412. [DOI] [PubMed] [Google Scholar]
- Posas, F., S.M. Wurgler-Murphy, T. Maeda, E.A. Witten, T.C. Thai, and H. Saito. 1996. Yeast HOG1 MAP kinase cascade is regulated by a multistep phosphorelay mechanism in the SLN1-YPD1-SSK1 “two-component” osmosensor. Cell. 86:865–875. [DOI] [PubMed] [Google Scholar]
- Pratt, L.A., W. Hsing, K.E. Gibson, and T.J. Silhavy. 1996. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol. Microbiol. 20:911–917. [DOI] [PubMed] [Google Scholar]
- Raitt, D.C., F. Posas, and H. Saito. 2000. Yeast Cdc42 GTPase and Ste20 PAK-like kinase regulate Sho1-dependent activation of the Hog1 MAPK pathway. EMBO J. 19:4623–4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser, V., H. Ruis, and G. Ammerer. 1999. Kinase activity-dependent nuclear export opposes stress-induced nuclear accumulation and retention of Hog1 mitogen-activated protein kinase in the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 10:1147–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiser, V., S.M. Salah, and G. Ammerer. 2000. Polarized localization of yeast Pbs2 depends on osmostress, the membrane protein Sho1 and Cdc42. Nat. Cell Biol. 2:620–627. [DOI] [PubMed] [Google Scholar]
- Saito, H. 2001. Histidine phosphorylation and two-component signaling in eukaryotic cells. Chem. Rev. 101:2497–2509. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y. 1659 pp.
- Sikorski, R.S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, T., K. Miwa, K. Ishikawa, H. Yamada, H. Aiba, and T. Mizuno. 2001. The Arabidopsis sensor His-kinase, AHK4, can respond to cytokinins. Plant Cell Physiol. 42:107–113. [DOI] [PubMed] [Google Scholar]
- Tamas, M.J., M. Rep, J.M. Thevelein, and S. Hohmann. 2000. Stimulation of the yeast high osmolarity glycerol (HOG) pathway: evidence for a signal generated by a change in turgor rather than by water stress. FEBS Lett. 472:159–165. [DOI] [PubMed] [Google Scholar]
- Ueguchi, C., S. Sato, T. Kato, and S. Tabata. 2001. The AHK4 gene involved in the cytokinin-signaling pathway as a direct receptor molecule in Arabidopsis thaliana. Plant Cell Physiol. 42:751–755. [DOI] [PubMed] [Google Scholar]
- Urao, T., B. Yakubov, R. Satoh, K. Yamaguchi-Shinozaki, M. Seki, T. Hirayama, and K. Shinozaki. 1999. A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell. 9:1743–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, H., T. Suzuki, K. Terada, K. Takei, K. Ishikawa, K. Miwa, T. Yamashino, and T. Mizuno. 2001. The Arabidopsis AHK4 histidine kinase is a cytokinin-binding receptor that transduces cytokinin signals across the membrane. Plant Cell Physiol. 42:1017–1023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.