Abstract
The protein PCM-1 localizes to cytoplasmic granules known as “centriolar satellites” that are partly enriched around the centrosome. We inhibited PCM-1 function using a variety of approaches: microinjection of antibodies into cultured cells, overexpression of a PCM-1 deletion mutant, and specific depletion of PCM-1 by siRNA. All approaches led to reduced targeting of centrin, pericentrin, and ninein to the centrosome. Similar effects were seen upon inhibition of dynactin by dynamitin, and after prolonged treatment of cells with the microtubule inhibitor nocodazole. Inhibition or depletion of PCM-1 function further disrupted the radial organization of microtubules without affecting microtubule nucleation. Loss of microtubule organization was also observed after centrin or ninein depletion. Our data suggest that PCM-1–containing centriolar satellites are involved in the microtubule- and dynactin-dependent recruitment of proteins to the centrosome, of which centrin and ninein are required for interphase microtubule organization.
Keywords: centrosome; microtubules; pericentriolar material; RNAi; dynein
Introduction
Microtubule organization is essential for directional intracellular transport, for the modulation of cell morphology and locomotion, and for the formation of the spindle apparatus during cell division. With the exception of plants, most cells organize their microtubule network using specialized structures, such as the spindle pole body in yeast cells, the basal body of cilia and flagella in protozoan organisms, and the centrosome in animal cells. The centrosome consists of two centriolar cylinders surrounded by electron-dense pericentriolar material. The centriolar cylinders have diameters of ∼0.2 μm and are each composed of nine triplets of short microtubules, arranged to form the wall of the cylinder. In addition to various tubulin isoforms (McKean et al., 2001), centrin, a member of a larger calcium-binding protein superfamily, has been found associated with the centrioles (Paoletti et al., 1996). However, 95% of centrin in human cells is not bound to the centrioles, but fractionates with the cytoplasm or with nuclei in biochemical experiments (Baron et al., 1994; Paoletti et al., 1996). A large variety of proteins is attached to the periphery of the centrioles as part of the pericentriolar material (Kalt and Schliwa, 1993). Some of the proteins in this matrix have been characterized in recent years (for review see Doxsey, 2001). The primary function of the pericentriolar material is to nucleate microtubules, which are radially arranged from the centrosomal surface or subsequently released and anchored in other places of the cell (Mogensen, 1999). The initial step of microtubule nucleation is dependent on the function of 25S ring complexes of the protein γ-tubulin and associated proteins (Zheng et al., 1995). However, for stable anchoring of microtubules, another protein (ninein) is required (Bouckson-Castaing et al., 1996; Mogensen et al., 2000; Piel et al., 2000; Ou et al., 2002). Ninein is also found at noncentrosomal sites in specialized cell types such as polarized epithelial cells that undergo a change from a radial microtubule organization into an arrangement of fibers from the apical to the basal pole (Bacallao et al., 1989; Mogensen et al., 1989; Mogensen, 1999). To facilitate microtubule nucleation, it has been proposed that γ-tubulin complexes are embedded in the pericentriolar material, in a lattice formed by pericentrin (Dictenberg et al., 1998). Pericentrin is a large protein of which two isoforms have been described: a 220-kD form (pericentrin A; Doxsey et al., 1994), as well as a newly identified 350-kD form (pericentrin B; Li et al., 2001). It is transported to the centrosome by the microtubule-dependent motor dynein, a process apparently mediated through direct binding of pericentrin to the dynein light intermediate chain (Purohit et al., 1999; Tynan et al., 2000). Pericentrin has recently been found to bind to PCM-1, a 228-kD protein that localizes to small 70–100-nm granules in the cytoplasm of interphase cells (Balczon et al., 1994; Kubo et al., 1999; Li et al., 2001). These granules can move along microtubules in a dynein-dependent way and often concentrate near the microtubule organizing center (Balczon et al., 1999; Kubo et al., 1999). Detailed morphological analysis revealed that these PCM-1 containing granules are identical to structures previously described as “centriolar satellites” (Kubo et al., 1999). Although centriolar satellites have been extensively studied by electron microscopy (Rattner, 1992), their function is so far unknown. In this paper, we test the potential role of their component protein PCM-1 in centrosome assembly and the organization of microtubule networks.
Results
Microinjection of antibodies against PCM-1 causes accumulation of centrin and pericentrin
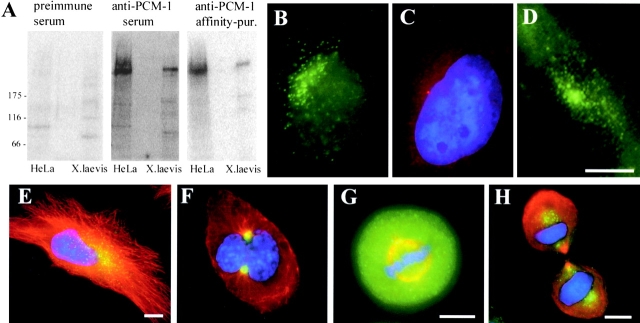
To examine the cellular function of the protein PCM-1, we raised antibodies in rabbits against a hexahistidine-tagged fusion protein containing amino acids 1665–2024 of human PCM-1. As shown in Fig. 1 A, these antibodies specifically recognized a 230-kD band in HeLa cell extract, characteristic of full-length PCM-1. Because the region of the PCM-1 fusion protein used for immunization is highly conserved among different vertebrate species (Kubo et al., 1999), our antibodies also cross reacted with PCM-1 from other animals, including Xenopus laevis. PCM-1 staining in HeLa cells displays a pattern of cytoplasmic granules that are partly enriched near the centrosome, but clearly distinct from the strong centrosomal staining of γ-tubulin (Fig. 1, B and C). Although this is in agreement with data from Kubo et al. (1999), it contrasts with immunofluorescence data initially provided by Balczon et al. (1994), showing a concentrated staining of PCM-1 at the centrosome of CHO cells. Testing various cell lines, we found that the amount and localization of cytoplasmic granules varied between different cell types (unpublished data).
Figure 1.
PCM-1 localizes to cytoplasmic granules that show a dynamic distribution during the cell cycle. (A) immunoblots of HeLa cell extracts and Xenopus egg extracts probed with rabbit preimmune serum, immune serum against PCM-1, and the same immune serum after affinity purification. Positions of molecular mass markers are indicated on the left. (B and C) HeLa cell stained with antibodies against (B) PCM-1, and against (C) γ-tubulin. (D) Live image of a HeLa cell transfected with full-length PCM-1, tagged with GFP at the carboxy terminus. (E–H) Double immunofluorescence of PCM-1 (green) and tubulin (red), and staining of the DNA (blue) of HeLa cells (E) in interphase, (F) prophase, (G) metaphase, and (H) telophase. PCM-1 signal in E–H was photographed at identical exposure levels. Bars: (D, E, G, and H) 10 μm; same magnifications in B–D and F and G, respectively.
Expression of a highly conserved homologue of full-length PCM-1 from chicken (66% identity, 75% homology to human PCM-1 over the entire length of the protein) that was tagged with GFP at the carboxy-terminal end gave the same pattern of cytoplasmic granules as seen in our immunofluorescence, therefore excluding a staining artifact of our antibodies (Fig. 1 D). After centrosome duplication, we could see PCM-1 concentrating in two large foci, with the highest concentration as cells entered mitosis (compare Fig. 1 E with Fig. 1 F). During metaphase, a fraction of PCM-1 granules concentrated at the spindle poles, but the majority of the protein was found dispersed in the cytoplasm (Fig. 1 G). In telophase, PCM-1 could be seen enriched in two areas of each daughter cell: (1) distal from the cleavage site, in the area of the centrosomal microtubule organizing center, as well as (2) proximal to the cleavage site, in an area where the minus-ends of midbody microtubules terminate (Fig. 1 H).
We used affinity-purified rabbit antibodies for microinjection into the cytoplasm of cultured Xenopus A6 cells. 24–48 h after microinjection, we found that PCM-1 granules were no longer detectable in 89% of the cells (n = 88), using a mouse antibody against PCM-1 for immunofluorescence (Fig. 2 B). Instead, only a weak staining in the centrosomal area remained. This could mean that the PCM-1 epitopes were masked by the microinjected antibody, and therefore, no longer detectable by immunofluorescence, or that the PCM-1 granules were dispersed upon microinjection. No apparent morphological defect was seen in injected cells, but when examining the distribution of other proteins, we observed large cytoplasmic aggregates of the centrosomal protein centrin, in addition to centrosome staining, in 67% of the cells (n = 284; Fig. 2, D–F). In 10% of the injected cells, these aggregates had acquired a filamentous or ribbonlike structure (Fig. 2, E and F). Further, there was a weak effect on pericentrin, with 17% of cells (n = 283) exhibiting small pericentriolar aggregates in addition to centrosome staining (Fig. 2 H). By contrast, the localization of γ-tubulin was not significantly affected by microinjection of PCM-1 antibodies (Fig. 2 J). Microinjection of control antibodies had no significant effect on the localization of centrosomal proteins or PCM-1 (Fig. 2, A, C, G, and I).
Figure 2.
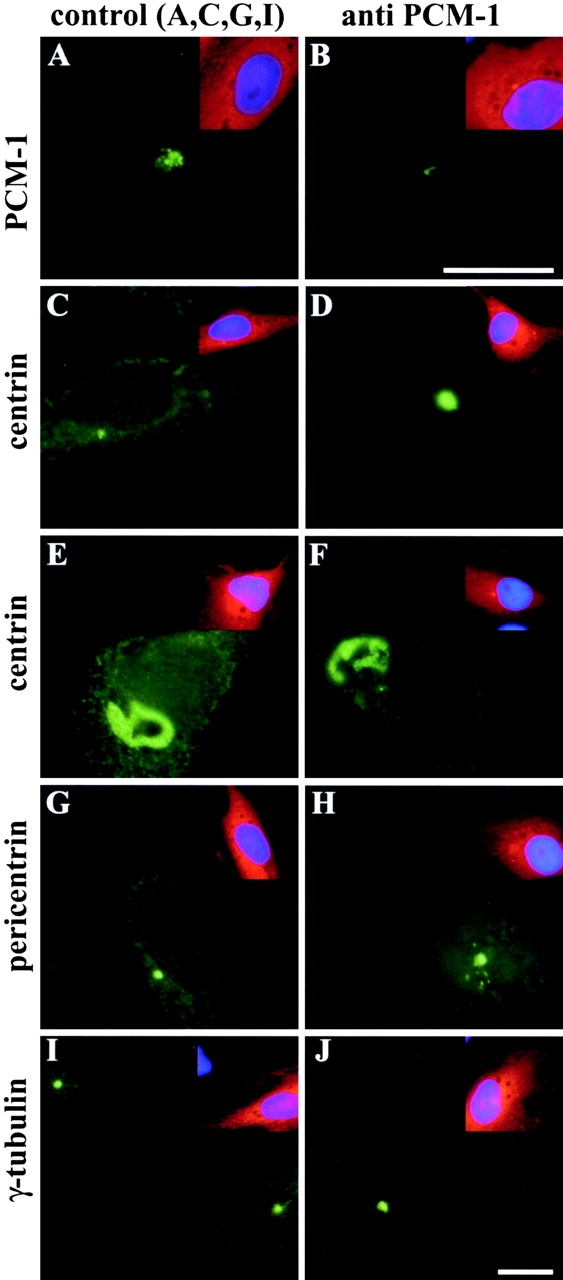
Microinjection of antibodies against PCM-1 causes aggregation of centrin and pericentrin. Xenopus A6 cells were microinjected with affinity-purified antibodies against PCM-1 (B, D–F, H, and J), or with control antibodies (A, C, G, and I). Cells were stained for immunofluorescence of (A and B) PCM-1, (C–F) centrin, (G and H) pericentrin, or (I and J) γ-tubulin. Insets show the same cells stained with Texas red–labeled anti–rabbit antibody to identify microinjected cells, and stained with DAPI to detect DNA (blue). Bars: (B and J) 10 μm; same magnifications in A and B and C–J, respectively.
Overexpression of a PCM-1 deletion mutant causes aggregation of a subset of centrosomal proteins
Our microinjection data suggest that antibodies against PCM-1 can affect the intracellular distribution of centrosome components such as centrin and pericentrin. To examine the role of PCM-1 in centrosomal protein targeting using a different approach, we generated a set of PCM-1 deletion mutants lacking various parts of their carboxy-terminal end. Whereas full-length PCM-1 (aa 1–1904) localized to small cytoplasmic granules characteristic of endogenous PCM-1 (Fig. 1 D), mutants comprising amino acids 1–1468 or 1–1148 formed large cytoplasmic protein aggregates of various sizes, up to ten times the size of normal PCM-1 granules. When overexpressing these mutants, we found that all endogenous PCM-1 was segregated to these protein aggregates (Fig. 3, C and D). We were able to distinguish between the mutant and the endogenous form of PCM-1 using an antibody raised against the carboxy terminus, present only in the endogenous full-length PCM-1 (Fig. 3 D), and a species-specific antibody raised against amino acids 1–114 of the chicken homologue of PCM-1, from which the mutant expression construct was derived (Fig. 3 C). Overexpression of mutant PCM-1 also affected the correct localization of centrosomal proteins; the majority of centrin accumulated at the same large cytoplasmic aggregates that contained PCM-1 and the deletion mutant (Fig. 3, E and F; detected in 96% of the transfected cells [n = 200]).
Figure 3.
Overexpression of the PCM-1 deletion mutant 1–1468 delocalizes centrin, pericentrin, and endogenous PCM-1 to large cytoplasmic aggregates. (A and B) Control HeLa cells overexpressing β-galactosidase, stained in red; DNA stained in blue. (B) PCM-1 is stained in green. (C, E, G, and M) HeLa cells overexpressing PCM-1 deletion mutant 1–1468, stained in red; DNA stained in blue. The same cells were stained for (D) PCM-1, (F) centrin, (H) pericentrin, or (N) acetylated tubulin. The arrowhead in N indicates the position of the centriole pair. (I–L) CHO cells overexpressing PCM-1 mutant 1–1468 (I and K; red), stained for (J) γ-tubulin, or (L) ninein. Bars: (H and N) 10 μm; same magnifications in A, B, E–H and C, D, I–N, respectively.
These large protein aggregates also segregated significant amounts of pericentrin (Fig. 3, G and H; 93% of overexpressing cells [n = 401]). In addition to centrin and pericentrin, the localization of the centrosomal protein ninein was also affected; whereas ninein localizes to the pericentriolar material in control cells, overexpression of mutant PCM-1 led to dispersion into multiple small foci of ninein in the cytoplasm in 90% of cells (n = 249; Fig. 3, K and L), which partly colocalized with the large PCM-1 protein aggregates. The localization of γ-tubulin, on the other hand, was not significantly altered by mutant PCM-1 (Fig. 3, I and J). Next, we tested whether any of the protein aggregates induced by the PCM-1 deletion mutant 1–1468 represented newly replicated centrosomes. Therefore, we stained cells with an antibody against acetylated tubulin, a marker that labels stable microtubules, including those that form the 9 × 3 filaments of the centriolar cylinders. We found that cells expressing mutant PCM-1 stained for acetylated tubulin indistinguishably from controls; generally, acetylated tubulin was enriched at a double-dot representing one pair of centrioles, and at cytoplasmic fibers representing stable microtubules (Fig. 3, M and N). However, the large protein aggregates of mutant PCM-1 did not show any enriched staining of acetylated tubulin, indicating that no new centrioles, and therefore no additional centrosomes, had been formed in these cells. Overexpression of a control protein, β-galactosidase, had no effect on PCM-1 or the centrosome (Fig. 3, A and B).
RNA silencing of PCM-1 leads to reduced assembly of centrin, pericentrin, and ninein at the centrosome
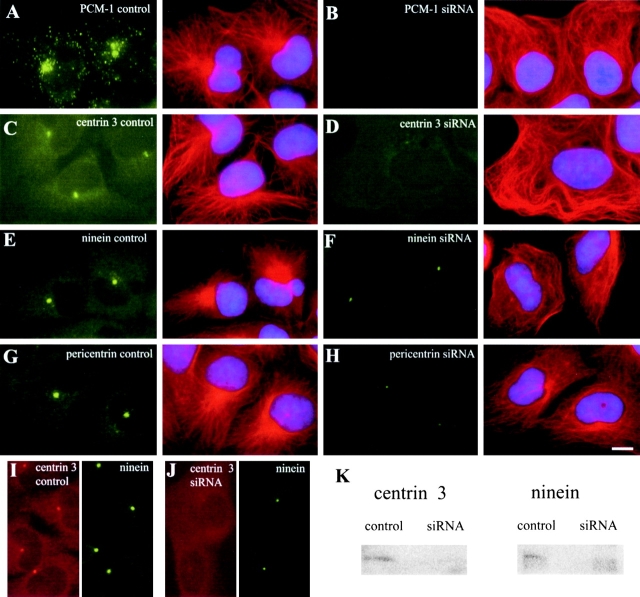
Because antibody microinjections and overexpression of PCM-1 mutants could have dominant secondary effects on other proteins in the cell by steric hindrance or by segregation of interacting components, we tested the role of PCM-1 in an approach based on depletion rather than inhibition of this protein. A recently published technique using transfection of double-stranded RNA oligomers of 21 base pairs has demonstrated that depletion of specific mRNAs is possible (Elbashir et al., 2001). Using oligomer pairs from two different regions of human PCM-1, as well as control oligomers (see Materials and methods), we reached transfection levels of 95% (n = 522; as judged using labeled control oligomers) and were able to remove 34% (siRNA PCM-1.1) or 82% (siRNA PCM-1.2) of the original amounts of PCM-1 in cultures of HeLa cells, U-2 OS human osteosarcoma cells, and C2C4 mouse myoblasts.
We proceeded with the RNA oligomer pair PCM-1.2 that had the strongest effect on PCM-1 depletion, corresponding to nucleotides 1464–1484 in human PCM-1 cDNA. Efficient depletion of PCM-1 was observed at time points longer than 90 h (Fig. 4 K), which required prolonged culturing and retransfection with siRNA at 48 h. This may reflect a slow turnover rate of PCM-1 in the cells. When analyzing individual cells, we found that PCM-1 depletion removed centriolar satellite staining almost completely, with a few PCM-1 granules occasionally remaining near the centrosome or in the cytoplasm (Fig. 4, B, D, F, H, and J). Photometric analysis of PCM-1 fluorescence revealed that the depletion levels in individual cells ranged from 69 to 99%, with an average depletion of 89% of PCM-1 protein. As a consequence of PCM-1 depletion, we again saw an effect on the assembly of centrin, pericentrin, and ninein, but no significant effect on γ-tubulin (Fig. 4, A–H). In all cell types examined, we found that the amounts of centrin, pericentrin, and ninein at the centrosome were significantly reduced after PCM-1 depletion. We quantified the fluorescence intensity of these proteins in the centrosomal region of U-2 OS cells (see Materials and methods), and determined that only 39% of centrin, 36% of pericentrin, and 38% of ninein remained localized at the centrosome as compared with control cells. In contrast, the levels of γ-tubulin remained largely constant (99%). Because previous work has indicated an interaction between PCM-1 and dynactin (Balczon et al., 1999), we also measured the centrosomal levels of the dynactin component p150/glued, which remained largely unaffected after PCM-1 depletion (82%; Fig. 4, I and J). Culturing of cells in the presence of PCM-1 siRNA for periods longer than 120 h led to extensive cell death.
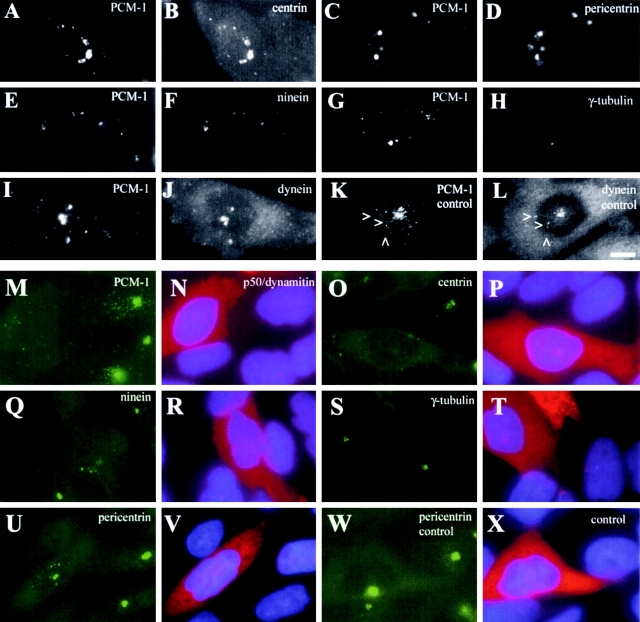
Figure 4.
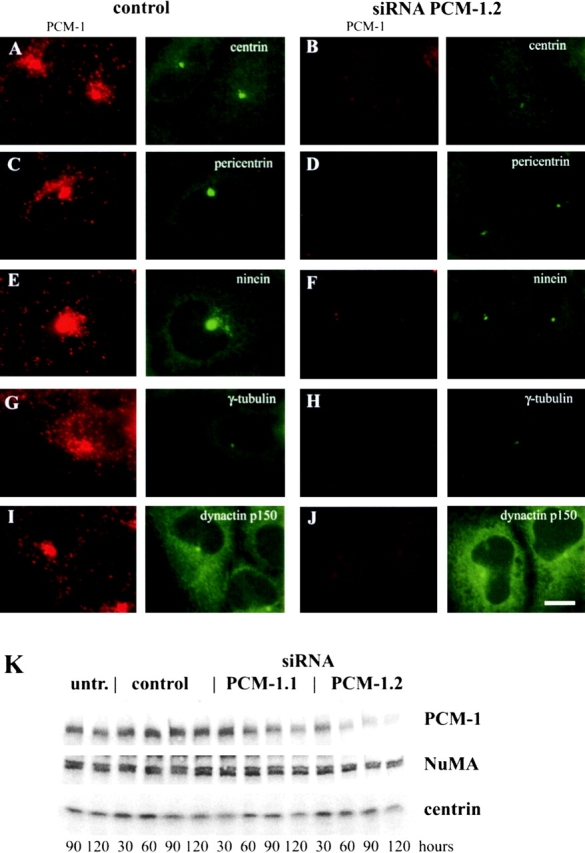
Depletion of PCM-1 by RNA silencing reduces centrosomal localization of centrin, pericentrin, and ninein, but not γ-tubulin or dynactin. A–J show U-2 OS cells treated with control or PCM-1 siRNA oligonucleotides, as indicated. Image pairs show cells double stained for (A and B) PCM-1 and centrin, (C and D) PCM-1 and pericentrin, (E and F) PCM-1 and ninein, (G and H) PCM-1 and γ-tubulin, and (I and J) PCM-1 and dynactin p150/glued. The amount of centrosomal protein localization after PCM-1 depletion was determined by photometric analysis to be 39% of centrin (± 17), 36% of pericentrin (± 21), 38% of ninein (± 20), 99% of γ-tubulin (± 58), and 82% of dynactin (± 37), as compared with control cells (n = 34 cells/each). (K) Immunoblots of extracts from untreated cells (untr.), and cells treated with control RNA oligomers siRNA PCM-1.1 or siRNA PCM-1.2, for different lengths of time as indicated. Blots were probed with antibodies against PCM-1, NuMA ,and centrin-3. Bar (J), 10 μm.
PCM-1 and centrosomal proteins colocalize in a subset of centriolar satellites
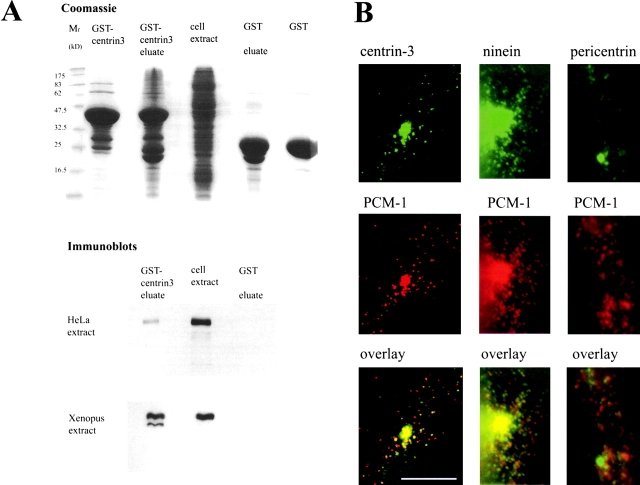
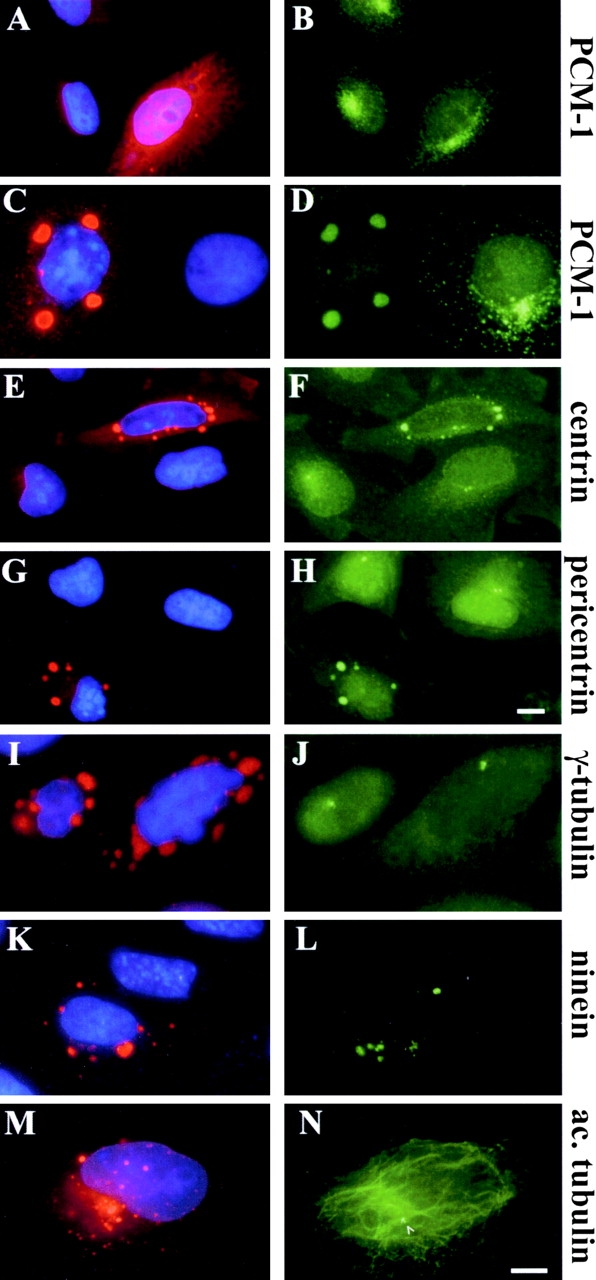
Because antibody microinjection experiments, mutant overexpression experiments, and RNA silencing experiments consistently showed a dependence of centrin assembly on PCM-1, we wanted to test directly for an interaction between PCM-1 and centrin using biochemical methods. Therefore, we used glutathione-Sepharose beads on HeLa cell extract or Xenopus egg extract, preincubated with GST-tagged centrin isoform 3. As shown in Fig. 5 A, centrin-3 loaded beads, but not control beads, were able to copellet PCM-1 from both extracts, indicating that PCM-1 and centrin can bind to each other. Because previous reports have shown centrin localizing to electron-dense cytoplasmic granules during ciliogenesis and to dynamic pericentriolar spots in PtK2 cells (Baron et al., 1994), we examined whether PCM-1 colocalized with centrin in these cells. As shown in Fig. 5 B, ∼79% of cytoplasmic granules of centrin-3 colocalized with PCM-1 (n = 518). We noticed that in specialized cell types, such as mouse myoblasts, the centrosomal proteins ninein and pericentrin could also be seen in small satellites surrounding the centrosome. As with centrin, these also partly colocalized with PCM-1 (Fig. 5 B).
Figure 5.
PCM-1 binds to centrin, and partly colocalizes with centriolar satellites of centrin, ninein, and pericentrin. (A) Binding assay using glutathione beads on cell extracts preincubated with GST-tagged centrin-3 or with GST alone. Lanes on a Coomassie-stained gel show relative molecular mass markers (Mr), purified GST-centrin-3, the eluate from glutathione beads incubated with Xenopus egg extract and GST-centrin-3, egg extract alone, the eluate from glutathione beads incubated with egg extract and GST, and purified GST alone. Positions of molecular weight markers are indicated on the left. Shown below are immunoblots of corresponding lanes, probed with antibodies against PCM-1. (B) Immunofluorescence staining of centriolar satellites. Left column, confocal section of a PtK2 cell stained for centrin-3 (green) and PCM-1 (red). Middle and right columns, conventional immunofluorescence of mouse myoblast cells stained for (middle) ninein and PCM-1, or (right) pericentrin and PCM-1. Bar (B), 10 μm.
Centrin, pericentrin, and ninein require PCM-1, dynactin, and microtubules for centrosomal localization
Together, our findings suggest that assembly of specific pericentriolar components depends on PCM-1. Because Kubo et al. (1999) have reported shuttling of PCM-1 granules in and out of the centrosome in a dynein-dependent manner, and because other reports provided evidence for pericentrin and PCM-1 transport dependent on microtubules and dynein–dynactin (Balczon et al., 1999; Purohit et al., 1999; Tynan et al., 2000), we wanted to examine whether prolonged treatment of cells with microtubule-destabilizing drugs would have similar effects on the localization of centrosomal proteins as PCM-1 inhibition. When CHO cells were treated with 17 μM nocodazole for 2 h, leading to complete depolymerization of microtubules, we observed large protein aggregates in the cytoplasm that contained PCM-1 together with centrin, pericentrin, ninein, and also the motor protein dynein (Fig. 6, A–F, I, and J), but not γ-tubulin (Fig. 6, G and H). Interestingly, a fraction of dynein also colocalized with PCM-1 granules in untreated cells (Fig. 6, K and L, arrows). We then addressed the question whether assembly of these centrosomal proteins could be directly inhibited by destabilizing the dynein activating complex of dynactin. For this purpose, we microinjected CHO cells with purified p50/dynamitin, a dynactin subunit that sequesters other dynactin components and leads to dynactin disassembly when added in excess (Echeverri et al., 1996; Quintyne et al., 1999). We noticed that dynamitin led to the dispersion of most PCM-1 in injected cells (Fig. 6, M and N), and caused similar defects in centrosomal protein assembly as observed with PCM-1 inhibition or depletion; centrin, pericentrin, and ninein were dispersed or formed small cytoplasmic aggregates, whereas γ-tubulin localization was unaffected (Fig. 6, O–X).
Figure 6.
Microtubules and dynactin are essential for the centrosomal accumulation of PCM-1, centrin, pericentrin, and ninein, but not γ-tubulin. (A–J) Image pairs of CHO cells after nocodazole treatment are shown, stained for (A and B) PCM-1 and centrin, (C and D) PCM-1 and pericentrin, (E and F) PCM-1 and ninein, (G and H) PCM-1 and γ-tubulin, and (I and J) PCM-1 and dynein intermediate chain. K and L show an untreated cell stained for PCM-1 and dynein intermediate chain. Arrowheads indicate pericentriolar dynein spots colocalizing with PCM-1 granules. (M–V) Image pairs of CHO cells microinjected with p50/dynamitin are shown. (M, O, Q, S, and U) Cells were stained for PCM-1, centrin-3, ninein, γ-tubulin, and pericentrin, respectively. (N, P, R, T, and V) Corresponding images showing dynamitin-injected cells (red) and DNA staining (blue). Dynamitin-dependent inhibition of centrosomal localization varied for different proteins; PCM-1 dispersed in 77% of injected cells (n = 106, controls 2%, n = 182), centrin was affected in 60% (n = 50, controls 2%, n = 191), ninein in 45% (n = 110, controls 3%, n = 169), pericentrin in 33% (n = 470, controls 5%, n = 73),and γ-tubulin in 3% (n = 76, controls 1%, n = 135). (W and X) Image pair of a control cell injected with labeled goat anti–rabbit antibody and stained for pericentrin. Bar (L), 10 μm.
Organization of a radial microtubule network depends on PCM-1
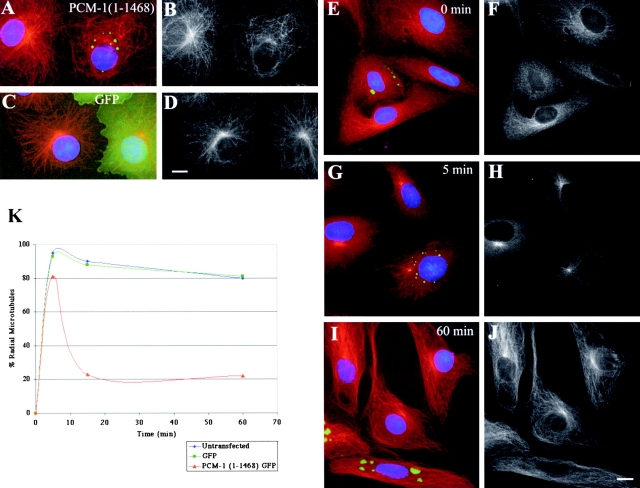
In a final set of experiments, we wanted to test whether the inhibition of PCM-1 had an effect on centrosome function, such as microtubule nucleation or organization of a radial microtubule network. In these experiments, we used Cos-7, U-2 OS, and PtK2 cells, all of which contain a well focused microtubule network, radiating from a single microtubule-organizing center at the centrosome (Clark and Meyer, 1999; Quintyne et al., 1999). Transfection of PCM-1 deletion construct 1–1468 disrupted this microtubule organization, with most microtubules now randomly distributed throughout the cytoplasm (Fig. 7, A and B). To test whether this is due to a lack of microtubule nucleation at the centrosome, we performed a microtubule regrowth assay. Transfected cells were treated for 40 min with 25 μM nocodazole on ice, to depolymerize all microtubules (Fig. 7, E and F), after which time the drug was washed out to allow regrowth of microtubules at 37°C. As shown in Fig. 7 (G and H), both untreated control cells as well as cells overexpressing mutant PCM-1 showed initial growth of small centrosomal microtubule asters. Within 15 min, however, the radial microtubule organization in mutant expressing cells was lost, and the microtubules became randomly distributed in the cytoplasm, as compared with control cells (Fig. 7, I–K). This indicates that although centrosomal microtubule nucleation was not affected by inhibition of PCM-1, the ability of the centrosomes to anchor microtubules was disturbed. No microtubules were seen nucleated or anchored at the protein aggregates formed by mutant PCM-1, suggesting that proteins segregated to these structures were not competent to nucleate or anchor microtubules by themselves.
Figure 7.
Microtubule anchoring to the centrosome depends on PCM-1. (A and B) Cos-7 cells expressing GFP-tagged PCM-1 deletion mutant 1–1468 (A, green). (A) Microtubules are stained in red, B shows microtubules only. (C and D) Control cells overexpressing GFP only (C, green). (C) Microtubules in red, (D) microtubules only. (E–J) Microtubule regrowth after nocodazole treatment of PtK2 cells. (E, G, and I) Staining of microtubules in red, GFP-tagged PCM-1 mutant 1–1468 in green; (F, H, and J) microtubules only. Time points at (E and F) 0 min, (G and H) 5 min, and (I and J) 60 min after removal of the drug. (K) Graph showing percentage of cells with radial microtubule organization at different time points after removal of nocodazole in untransfected cells (blue), cells overexpressing GFP (green), and cells overexpressing GFP-tagged PCM-1 mutant 1–1468 (red). Between 400 and 600 cells were counted for each time point. Bars: 10 μm (D and J).
Depletion of PCM-1, centrin, or ninein inhibits anchorage of microtubules to the centrosome
Then, we tested whether removal of PCM-1 had the same effect on radial microtubule organization as overexpression of mutant PCM-1. As shown in Fig. 8 (A and B), PCM-1 depletion from U-2 OS cells by siRNA induced loss of centrosomal microtubule anchorage, leaving only 34% of the cells with a radial microtubule network (n =1,001, controls 74%, n =1,001). Similar results were obtained in mouse C2C4 myoblasts (unpublished data). Because we described earlier in this paper that removal of PCM-1 affected the localization of centrin, ninein, and pericentrin to the centrosome, we examined whether loss of microtubule anchorage was mediated by one or more of these proteins. For this purpose, we depleted centrin, ninein, or pericentrin individually, using specific siRNA oligomers. Protein levels were reduced to 23 (centrin), 29 (ninein), or 20% (pericentrin) of control levels, as measured by quantitative immunoblotting (Fig. 8 K) or photometric analysis (available antibodies against pericentrin were unable to recognize the denatured protein by immunoblotting). Reduction of centrin-3 and ninein levels similarly affected microtubule organization (Fig. 8, C–F), with only 18 (n = 1,001) and 31% (n = 934) of cells, respectively, exhibiting a radial network. In contrast, pericentrin depletion had no significant effect on microtubule organization (Fig. 8, G and H). Radial microtubules were seen in 77% of the cells (n = 833), approximately at the same level as in control cells (74%). However, we noticed that removal of pericentrin resulted in a reduced density of microtubules emanating from the centrosome (Fig. 8 H).
Figure 8.
Depletion of PCM-1, centrin-3, or ninein results in loss of microtubule anchoring at the centrosome. (A C, E, and G) U-2 OS cells were transfected with control dsRNA oligomers and stained in green for (A) PCM-1, (C) centrin-3, (E) ninein, and (G) pericentrin. Microtubules were stained in red, DNA in blue. (B, D, F, and H) Corresponding image pairs showing cells after treatment with siRNA against (B) PCM-1, (D) centrin-3, (F) ninein, (H) pericentrin. I, J, U-2 OS cells treated with (I) control oligomers or (J) centrin-3 siRNA. Red, centrin-3 immunofluorescence; green, ninein immunofluorescence. K, immunoblots of cells treated with control RNA or siRNA against (left) centrin-3, or (right) ninein, stained for centrin-3 or ninein, respectively. Bar in H, 10 μm.
Because centrin-3 is not known to bind directly to microtubules, we tested whether loss of microtubule anchorage at the centrosome after centrin depletion could be due to an indirect effect mediated by ninein. As shown in Fig. 8 (I and J), removal of centrin-3 from U-2 OS cells also resulted in loss of ninein localization to the centrosome.
Discussion
Microtubule nucleation and anchoring of the microtubule filament network are two functions associated with the centrosome. Our present work has directly shown that these two functions can be separated, and that microtubule anchoring is dependent on the protein PCM-1. Most interestingly, PCM-1 itself is not a classical component of the centrosome, but instead localizes to electron-dense protein granules in the cytoplasm. Because these were most easily recognized in electron micrographs near the centrosome, they were termed centriolar satellites (Rattner, 1992; Kubo et al., 1999). This raises the question of how a satellite component such as PCM-1 functions in anchoring microtubules to the centrosomal surface. Our data suggest that the role of PCM-1 in microtubule anchoring is an indirect one, most likely mediated through other proteins that assemble at the centrosome in a PCM-1–dependent manner. As shown in this paper, the correct assembly of a subset of centrosomal proteins, including centrin, pericentrin, and ninein, depends on PCM-1 function. Beyond this, we have demonstrated that PCM-1 binds to an isoform of the protein centrin. Supporting evidence was also provided by Li et al. (2001), who reported that PCM-1 binds to pericentrin-B, and by Balczon et al. (2002), who noted changes in centrosome morphology after PCM-1 antibody injection into mouse oocytes.
To understand the role of PCM-1, it is important to note that the distribution of centrin, pericentrin and PCM-1 is very dynamic (Baron et al., 1994; Kubo et al., 1999; Young et al., 2000) and dependent on the action of dynein–dynactin motor complexes (Balczon et al., 1999; Kubo et al., 1999; Purohit et al., 1999). Intriguingly, small granules of GFP-tagged PCM-1 have been directly followed by video microscopy, shuttling along microtubules between the cytoplasm and the centrosome (Kubo et al., 1999). Therefore, a possible function of PCM-1 could be to mediate the transport of centrosome components from the cytoplasm to the centrosome, along microtubules. PCM-1 may serve as a carrier that associates with centrin, pericentrin, or ninein, and docks onto dynein–dynactin. Consistent with this idea is our observation that depolymerization of microtubules, as well as dynactin inhibition, led to dispersion of centrosomal proteins and cytoplasmic protein aggregates that contain centrin, pericentrin and ninein, as well as dynein and PCM-1. Transport complexes would contain only a small proportion of cellular centrin, pericentrin, or ninein, explaining why most cell types do not show significant centriolar satellite staining of these proteins. Specific cell types such as PtK2 or mouse myoblasts do exhibit recognizable cytoplasmic granules of these proteins, and we show that they colocalize with PCM-1. Centrin granules are very dynamic structures that can rapidly fuse with the pericentriolar material (Baron et al., 1994), consistent with our transport model. A role of PCM-1 in facilitating transport of centrosomal proteins could be important for the duplication of centrosomes during the cell cycle, when new pericentriolar material is recruited to the centrosomal surface, and to increase the potential of centrosomes to organize microtubules into mitotic spindles. This would explain why PCM-1 staining before mitosis is particularly concentrated at the centrosomes, with fewer cytoplasmic granules visible than in interphase, and why the signal becomes again more dispersed in metaphase, after spindle poles have fully formed.
Another explanation of our data could be that PCM-1 granules in the cytoplasm represent sites at which centrosomal proteins associate temporarily to undergo proper folding, or to assemble into complexes with other proteins. The two interpretations on PCM-1 function are not mutually exclusive. Several centrosomal proteins are not simply confined to the centrosome, but are also present in a large cytoplasmic pool (Moudjou et al., 1996; Paoletti et al., 1996). Cytoplasmic factors that support folding and assembly as well as factors that aid transport would contribute to a dynamic equilibrium between centrosome-bound and free protein (Baron et al., 1994).
As shown in this paper, not all centrosomal proteins follow a PCM-1–dependent assembly pathway. In particular, we show that recruitment of γ-tubulin to the centrosome is independent of PCM-1, and apparently of dynein–dynactin-dependent transport. Our data are further consistent with previous reports by Khodjakov and Rieder (1999), Hannak et al. (2001), as well as earlier biochemical studies by Klotz et al. (1990), Felix et al. (1994), Moritz et al. (1995), and Schnackenberg et al. (1998), that centrosomal targeting of γ-tubulin and other potential microtubule nucleation factors is independent of microtubules. In contrast, work by Quintyne et al. (1999) and Young et al. (2000) clearly demonstrates a requirement for dynactin in γ-tubulin assembly. These seemingly contradictory findings may be reconciled by the existence of different pools of γ-tubulin at the centrosome with different rates of exchange with the cytoplasm, as shown by Khodjakov and Rieder (1999). If only the slowly exchanging pool of γ-tubulin required dynactin function, for example, as a microtubule anchor rather than as a transporter, then effects on γ-tubulin localization would only be observed after prolonged treatment of cells with dynactin inhibitors, as in the experiments of Quintyne et al. (1999) and Young et al. (2000), and not over the shorter time frame of a few hours in our experiments. There may also be differences in the dynamics of centrosomal components between different cell types and cell cycle stages, and these may explain the observation of dynactin-independent pericentrin assembly by Quintyne et al. (1999), in contrast to data from this study and Young et al. (2000).
Consistent with our finding of γ-tubulin assembly independent of PCM-1, microtubule nucleation at the centrosome is not affected when PCM-1 is inhibited. Our data highlight the notion that microtubule nucleation and the organization of the microtubule network are distinct events. Earlier studies by Keating et al. (1997) provided direct evidence for microtubule release from the centrosome. In addition, Mogensen et al. (2000) have shown that in polarized cell types, microtubules can be transferred after nucleation from the centrosome to apical regions of the cell. It has been suggested that a protein involved in microtubule anchorage at these sites is ninein, previously identified as a component of the pericentriolar material (Bouckson-Castaing et al., 1996). It has further been shown in a paper by Piel et al. (2000) that immature daughter centrioles, lacking ninein localization, are able to nucleate microtubules, but fail to anchor them. Here, we provide direct evidence for a role of ninein in microtubule anchorage to the centrosome by demonstrating that depletion of ninein causes loss of centrosomal microtubule organization. Our data further suggest that the effects of PCM-1 or centrin depletion on the microtubule network organization are mediated through ninein, because ninein levels at the centrosome decrease when PCM-1 or centrin are depleted.
Therefore, the potential of the centrosome to anchor microtubules may depend on the correct assembly of a subset of proteins; PCM-1 may be involved in targeting centrin to the centrosome, where it would be necessary for the assembly of ninein, and thereby regulate microtubule anchorage. As discussed above, this targeting of microtubule-anchoring factors appears to be mediated by dynein–dynactin-dependent transport, consistent with observations by Quintyne et al. (1999) and Clark and Meyer (1999), who showed that dynactin inhibition interferes with microtubule organization at the centrosome. Pericentrin was also found to assemble at the centrosome in a PCM-1–dependent manner, but in contrast to ninein or centrin, its absence did not interfere with radial microtubule organization. Instead, the density of microtubules in the cell decreased after pericentrin depletion, indicating that pericentrin either stabilizes microtubules or aids microtubule nucleation, as suggested by Dictenberg et al. (1998), due to its close association with γ-tubulin. Additional factors may be involved in the regulation of microtubule anchorage, such as the microtubule-severing protein katanin (Hartman et al., 1998), the dynactin component p150/glued (Quintyne et al., 1999), or Cep 135 and MIR1, two recently identified novel centrosomal proteins (Ohta et al., 2002; Stein et al., 2002). Further studies of microtubule-anchoring proteins should provide valuable insights in the remodeling of the cytoskeleton during cell differentiation and morphogenesis.
Materials and methods
Cell culture
HeLa, U-2 OS human osteosarcoma cells, C2C4 mouse myoblasts, PtK2, and COS-7 cells were cultured in DME, CHO cells in McCoy's 5A medium, and Xenopus A6 cells in 0.6x L15 medium, all supplemented with 10% FBS. Cells were grown at 37°C and at 5% carbon dioxide, except A6 cells, which were grown at RT under atmospheric conditions. Transient transfections were performed by calcium phosphate precipitation as described in Sambrook et al. (1989).
Cloning of chicken PCM-1 cDNA and construction of expression vectors
An EST clone containing the middle 3.5-kb fragment of the chicken homologue of PCM-1 was obtained from a Bursal EST collection then managed by Dr. Jean-Marie Buerstedde (University of Hamburg, Hamburg Germany; clone 4d19r1, GenBank/EMBL/DDBJ accession no. AJ398048 [now obtainable through RZPD]). Clones containing the 5′ and 3′ ends of PCM-1 were obtained by screening a DU249 λZAP cDNA library (provided by S. Kandels-Lewis, University of Edinburgh, Edinburgh UK) with hybridization probes derived from this EST, and the full-length cDNA (GenBank/EMBL/DDBJ accession no. AJ508717) assembled in the cloning vector pBluescript® in a series of cloning steps.
A full-length PCM-1–GFP expression construct was then generated by modifying the cDNA insert at its 3′ end by PCR to remove the stop codon, and cloning it in frame into the multiple cloning site of pEGFP-N (CLONTECH Laboratories, Inc.). The deletion construct 1–1468, containing nucleotides 1–4423 after the start codon, was generated by PCR, and the GFP tag removed by cutting the vector with SmaI and NotI, blunting, and religating. A control vector for the expression of β-galactosidase was obtained from Dr. Adrian Bird (University of Edinburgh, Edinburgh UK).
Antibodies, immunofluorescence, and immunoblotting
The COOH terminus of human PCM-1 comprising nucleotides 4993–6095 after the start codon was amplified by PCR from a HeLa cDNA library (provided by S. Kandels-Lewis, University of Edinburgh, UK) using primers CTGAAAGACTGTGGAGAAGATC and GATGTCTTCAGAGGCTCATC, and cloned into the vector pGEM-T (Promega). The insert was then excised using PstI and NcoI and cloned into the bacterial expression vector pRSET-C (Invitrogen). Bacterial fusion protein was isolated using 8 M urea and purified over Nickel Sepharose (Amersham Biosciences) and hydroxyapatite (Bio-Rad Laboratories) columns, concentrated, and dialyzed against PBS before injection into two rabbits.
Affinity-purified antibody was obtained by passing serum over a column of the same antigen coupled to a CNBr-activated Sepharose column (Amersham Biosciences). The same antigen was also used to raise pAbs in mice. Chicken-specific pAbs were further generated in mice against the amino terminus of the chicken PCM-1 protein. For this, nucleotides 1–342 after the start codon were amplified by PCR from the chicken cDNA using primers AAGGATCCATGGCAACAGGAGGCG and AGAATTCACTGATCCAGATCACTGAAGTT, and cloned directly into the bacterial expression vector pRSET-A. Bacterial fusion protein was then purified as described above and injected into mice.
Mouse antibodies were raised against nucleotides 4002–6263 after the start codon of human pericentrin-B, a region common to both pericentrin-A and -B. This fragment was obtained by excision with BglII and NcoI from a partial cDNA clone provided by Dr. Harish Joshi (Emory University, Atlanta, GA), cloned into pRSET-C (Invitrogen), and expressed as above. All pAbs were used at a dilution of 1:100. Affinity-purified PCM-1 antibody was used at a concentration of 1 μg/ml for both immunofluorescence and immunoblotting. mAbs against dynein intermediate chain, NuMA, and β-galactosidase were obtained from CHEMICON International, Calbiochem, and Promega, respectively. mAbs against γ-tubulin and acetylated tubulin were from Sigma-Aldrich. Other antibodies were used as described previously; pAbs against γ-tubulin were a gift from Dr. Rebecca Heald (University of California, Berkeley, CA; Heald et al., 1997).
mAb 20H5 against centrin (Sanders and Salisbury, 1994) was a gift from Dr. Jeffrey Salisbury (Mayo Clinic, Rochester, MN). Rabbit pAbs against centrin-3 (anti-HsCen3p; Laoukili et al., 2000) and against ninein (Mogensen et al., 2000) were gifts from Dr. Michel Bornens (Institut Curie, Paris, France). Rabbit antibody against pericentrin-B (Li et al., 2001) was a gift from Dr. Harish Joshi (Emory University, Atlanta, GA), mouse antibody against human ninein (Ou et al., 2002) was a gift from Dr. Gordon Chan (Alberta Cancer Board, Canada), and mAb against β-tubulin was a gift from Dr. Don Cleveland (Ludwig Institute for Cancer Research, San Diego, CA). For immunofluorescence, cells were fixed for 10 min in methanol at −20°C, and processed and imaged using conventional fluorescence microscopy as described previously (Merdes et al., 2000). Cos-7 and PtK2 cells were fixed with 3.7% formaldehyde in 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, 10 mM Pipes, pH 6.8, to preserve microtubule integrity. Gel electrophoresis and immunoblotting were performed according to standard protocols.
siRNA experiments
RNA oligomers containing 21 nucleotides were synthesized in sense and antisense directions corresponding to human PCM-1 (Balczon et al., 1994) at nucleotides 2190–2208 (GGGCUCUAAACGUGCCUCC; PCM-1.1) and 1465–1483 (UCAGCUUCGUGAUUCUCAG; PCM-1.2) with dTdT overhangs at each 3′ terminus, deprotected, and desalted (Xeragon). Oligomers against centrin-3 (UGAAGUUGUGACAGACUGG), pericentrin (recognizing both pericentrin-A and -B; GCAGCUGAGCUGAAGGAGA), and ninein (UAUGAGCAUUGAGGCAGAG) were prepared accordingly. All oligomers were identical to both human and mouse sequences, except for PCM-1.1, which was human-specific. For annealing of siRNAs, 20-μM single strands were incubated in annealing buffer (100 mM potassium acetate, 2 mM magnesium acetate, 30 mM Hepes-KOH, pH 7.4) for 1 min at 90°C, followed by 1 h at 37°C (Elbashir et al., 2001).
Transfections were performed using Oligofectamine™ (Invitrogen) with 3 μg siRNA on HeLa, U-2 OS, or C2C4 cells grown overnight on 6-well dishes at 3 × 104 cells/well. For time points beyond 60 h, cells were split 48 h after the first transfection and then immediately subjected to a second treatment with siRNA. 3′ Rhodamine-labeled and unlabeled control oligonucleotides (CGUACGCGGAAUACUUCGA plus 3′ dTdT overhangs; control) were used to optimize transfection efficiency and to control for nonspecific effects due to the presence of siRNAs in cells, respectively. The level of protein depletion due to RNA silencing was determined by quantitative immunoblotting of cell extracts using 125I-labeled secondary antibody (Amersham Biosciences). Equal amounts of protein extracts were separated by SDS-PAGE, and quantification was performed on immunoblots using a PhosphorImager. Photometric quantification of immunofluorescence signals was performed from digital image files taken with a 40×/0.75-NA lens that allowed a large depth of focus. Mean pixel values of 1–2-μm2 areas were calculated using Adobe Photoshop®. Control cells stained with nonimmune serum and cells treated with control RNA, stained with the respective centrosomal antibodies, were used to calculate background levels and average control protein levels.
Microinjection experiments
Affinity-purified PCM-1 antibody was injected into Xenopus A6 cells cultured on glass coverslips at 2 mg/ml in injection buffer (100 mM KCl, 10 mM potassium phosphate, pH 7.4). At 24 or 48 h after injection, coverslips were fixed with methanol at −20°C and processed for immunofluorescence as above. Control injections were performed using rabbit IgG (Sigma-Aldrich) at the same concentration in injection buffer. Purified dynamitin (Wittmann and Hyman, 1999) at a concentration of 9 mg/ml was injected into CHO cells. After 2–4 h of incubation, cells were fixed and processed for immunofluorescence. Control cells were injected with fluorescently labeled secondary antibody.
Centrin copurification experiments
Human centrin-3 was obtained by PCR from a HeLa cDNA library (provided by S. Kandels-Lewis, University of Edinburgh, UK) using primers ATGGATCCATGAGTTTAGCTCTGAGAAGTGAGC and TAGAATTCTTAAATGTCACCAGTCATAATAGCA and cloned into the bacterial expression vector pGEX4T2 (Amersham Biosciences) using BamH1 and EcoR1. Sequencing confirmed it to be identical to the previously published human centrin-3 sequence (Middendorp et al., 1997). Bacterial fusion protein in PBS was loaded on a glutathione Sepharose 4B column and purified using reduced glutathione according to the manufacturer's instructions (Amersham Biosciences), and dialyzed against PBS.
HeLa cell extracts were prepared by resuspending cell pellets from 6 near-confluent 10-cm plates (∼6 × 107 cells) in 1 ml PBS using a Dounce homogenizer. Xenopus egg extracts were prepared as described by Murray (1991). Protein concentrations of the extracts prepared varied between 2–5 mg/ml for HeLa cell extracts and 40–100 mg/ml for Xenopus egg extracts. In each copurification experiment, 200 μg GST-centrin-3 or GST alone was added to 1 mg HeLa extract or 10 mg Xenopus egg extract, diluted to 1 ml total volume in PBS, and incubated for 1 h at 4°C. GST fusion protein and associated interactors were then recovered by incubating the mixture with 100 μl glutathione Sepharose beads for 30 min at 4°C. After extensive washes with PBS, bound protein was eluted with 10 mM reduced glutathione, pH 8.0, followed by TCA precipitation and boiling for 5 min in gel loading buffer containing SDS and mercaptoethanol. Recovery of the GST fusion protein was confirmed by SDS-PAGE and Coomassie staining, and the copurification of PCM-1 tested by immunoblotting.
Acknowledgments
A. Dammermann would like to dedicate this work to the memory of Dr. Rainer and Stephanie Dammermann.
We thank our colleagues F. Gardiner, L. Haren, and X. Fant (University of Edinburgh) for technical help and for critically reading this manuscript. We thank Drs. C. Rabouille, K. Sawin, W.C. Earnshaw, M. Heck, and members of their groups (University of Edinburgh) for their help throughout this work, and Drs. M. Bornens, J. Salisbury, H. Joshi, G. Chan, R. Heald, D. Cleveland, and A. Bird for the gift of antibodies and plasmids.
This work was supported by a Wellcome 4-year studentship to A. Dammermann, and a Wellcome senior research fellowship to A. Merdes.
References
- Bacallao, R., C. Antony, C. Dotti, E. Karsenti, E.H.K. Stelzer, and K. Simons. 1989. The subcellular organization of Madin-Darby canine kidney cells during the formation of a polarized epithelium. J. Cell Biol. 109:2817–2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balczon, R., L. Bao, and W.E. Zimmer. 1994. PCM-1, a 228-kD centrosome autoantigen with a distinct cell cycle distribution. J. Cell Biol. 124:783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balczon, R., C.E. Varden, and T.A. Schroer. 1999. Role for microtubules in centrosome doubling in chinese hamster ovary cells. Cell Motil. Cytoskeleton. 42:60–72. [DOI] [PubMed] [Google Scholar]
- Balczon, R., C. Simerly, D. Takahashi, and G. Schatten. 2002. Arrest of cell cycle progression during first interphase in murine zygotes microinjected with anti-PCM-1 antibodies. Cell Motil. Cytoskeleton. 52:183–192. [DOI] [PubMed] [Google Scholar]
- Baron, A.T., V.J. Suman, E. Nemeth, and J.L. Salisbury. 1994. The pericentriolar lattice of PtK2 cells exhibits temperature and calcium-modulated behavior. J. Cell Sci. 107:2993–3003. [DOI] [PubMed] [Google Scholar]
- Bouckson-Castaing, V., M. Moudjou, D.J. Ferguson, S. Mucklow, Y. Belkaid, G. Milon, and P.R. Crocker. 1996. Molecular characterization of ninein, a new coiled-coil protein of the centrosome. J. Cell Sci. 109:179–190. [DOI] [PubMed] [Google Scholar]
- Clark, I.B., and D.I. Meyer. 1999. Overexpression of normal and mutant Arp1α (centractin) differentially affects microtubule organization during mitosis and interphase. J. Cell Sci. 112:3507–3518. [DOI] [PubMed] [Google Scholar]
- Dictenberg, J.B., W. Zimmerman, C.A. Sparks, A. Young, C. Vidair, Y. Zheng, W. Carrington, F.S. Fay, and S.J. Doxsey. 1998. Pericentrin and γ-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J. Cell Biol. 141:163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey, S. 2001. Re-evaluating centrosome function. Nat. Rev. Mol. Cell Biol. 2:688–698. [DOI] [PubMed] [Google Scholar]
- Doxsey, S.J., P. Stein, L. Evans, P.D. Calarco, and M. Kirschner. 1994. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell. 76:639–650. [DOI] [PubMed] [Google Scholar]
- Echeverri, C.J., B.M. Paschal, K.T. Vaughan, and R.B. Vallee. 1996. Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J. Cell Biol. 132:617–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., J. Harborth, W. Lendeckel, A. Yalcin, K. Weber, and T. Tuschl. 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 411:494–498. [DOI] [PubMed] [Google Scholar]
- Felix, M.A., C. Antony, M. Wright, and B. Maro. 1994. Centrosome assembly in vitro: role of γ-tubulin recruitment in Xenopus sperm aster formation. J. Cell Biol. 124:19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannak, E., M. Kirkham, A.A. Hyman, and K. Oegema. 2001. Aurora-A kinase is required for centrosome maturation in Caenorhabditis elegans. J. Cell Biol. 155:1109–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman, J.J., J. Mahr, K. McNally, K. Okawa, A. Iwamatsu, S. Thomas, S. Cheesman, J. Heuser, R.D. Vale, and F.J. McNally. 1998. Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell. 93:277–287. [DOI] [PubMed] [Google Scholar]
- Heald, R., R. Tournebize, A. Habermann, E. Karsenti, and A. Hyman. 1997. Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J. Cell Biol. 138:615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalt, A., and M. Schliwa. 1993. Molecular components of the centrosome. Trends Cell Biol. 3:118–128. [DOI] [PubMed] [Google Scholar]
- Keating, T.J., J.G. Peloquin, V.I. Rodionov, D. Momcilovic, and G.G. Borisy. 1997. Microtubule release from the centrosome. Proc. Natl. Acad. Sci. USA. 94:5078–5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov, A., and C.L. Rieder. 1999. The sudden recruitment of γ-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J. Cell Biol. 146:585–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klotz, C., M.C. Dabauvalle, M. Paintrand, T. Weber, M. Bornens, and E. Karsenti. 1990. Parthenogenesis in Xenopus eggs requires centrosomal integrity. J. Cell Biol. 110:405–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, A., H. Sasaki, A. Yuba-Kubo, S. Tsukita, and N. Shiina. 1999. Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J. Cell Biol. 147:969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laoukili, J., E. Perret, S. Middendorp, O. Houcine, C. Guennou, F. Marano, M. Bornens, and F. Tournier. 2000. Differential expression and cellular distribution of centrin isoforms during human ciliated cell differentiation in vitro. J. Cell Sci. 113:1355–1364. [DOI] [PubMed] [Google Scholar]
- Li, Q., D. Hansen, A. Killilea, H.C. Joshi, R.E. Palazzo, and R. Balczon. 2001. Kendrin/pericentrin-B, a centrosome protein with homology to pericentrin that complexes with PCM-1. J. Cell Sci. 114:797–809. [DOI] [PubMed] [Google Scholar]
- McKean, P.G., S. Vaughan, and K. Gull. 2001. The extended tubulin superfamily. J. Cell Sci. 114:2723–2733. [DOI] [PubMed] [Google Scholar]
- Merdes, A., R. Heald, K. Samejima, W.C. Earnshaw, and D.W. Cleveland. 2000. Formation of spindle poles by dynein/dynactin-dependent transport of NuMA. J. Cell Biol. 149:851–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middendorp, S., A. Paoletti, E. Schiebel, and M. Bornens. 1997. Identification of a new mammalian centrin gene, more closely related to Saccharomyces cerevisiae CDC31 gene. Proc. Natl. Acad. Sci. USA. 94:9141–9146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen, M.M. 1999. Microtubule release and capture in epithelial cells. Biol. Cell. 91:331–341. [PubMed] [Google Scholar]
- Mogensen, M.M., J.B. Tucker, and H. Stebbings. 1989. Microtubule polarities indicate that nucleation and capture of microtubules occurs at cell surfaces in Drosophila. J. Cell Biol. 108:1445–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen, M.M., A. Malik, M. Piel, V. Bouckson-Castaing, and M. Bornens. 2000. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J. Cell Sci. 113:3013–3023. [DOI] [PubMed] [Google Scholar]
- Moritz, M., M.B. Braunfeld, J.C. Fung, J.W. Sedat, B.M. Alberts, and D. Agard. 1995. Three-dimensional structural characterization of centrosomes from early Drosophila embryos. J. Cell Biol. 130:1149–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moudjou, M., N. Bordes, M. Paintrand, and M. Bornens. 1996. γ-Tubulin in mammalian cells: the centrosomal and the cytosolic forms. J. Cell Sci. 109:875–887. [DOI] [PubMed] [Google Scholar]
- Murray, A.W. 1991. Cell cycle extracts. In Methods in Cell Biology, vol. 36. B.K. Kay and H.B. Peng, editors. Academic Press, Inc., San Diego, CA. 581–605. [PubMed]
- Ohta, T., R. Essner, J.H. Ryu, R.E. Palazzo, Y. Uetake, and R. Kuriyama. 2002. Characterization of Cep135, a novel coiled-coil centrosomal protein involved in microtubule organization in mammalian cells. J. Cell Biol. 156:87–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou, Y.Y., G.J. Mack, M. Zhang, and J.B. Rattner. 2002. CEP110 and ninein are located in a specific domain of the centrosome associated with centrosome maturation. J. Cell Sci. 115:1825–1835. [DOI] [PubMed] [Google Scholar]
- Paoletti, A., M. Moudjou, M. Paintrand, J.L. Salisbury, and M. Bornens. 1996. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J. Cell Sci. 109:3089–3102. [DOI] [PubMed] [Google Scholar]
- Piel, M., P. Meyer, A. Khodjakov, C.L. Rieder, and M. Bornens. 2000. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 149:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit, A., S.H. Tynan, R. Vallee, and S.J. Doxsey. 1999. Direct interaction of pericentrin with cytoplasmic dynein light intermediate chain contributes to mitotic spindle organization. J. Cell Biol. 147:481–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne, N.J., S.R. Gill, D.M. Eckley, C.L. Crego, D.A. Compton, and T.A. Schroer. 1999. Dynactin is required for microtubule anchoring at centrosomes. J. Cell Biol. 147:321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner, J.B. 1992. Ultrastructure of centrosome domains and identification of their protein components. In The Centrosome. V.I. Kalnins, editor. Academic Press, Inc., San Diego, CA. 45–69.
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Sanders, M.A., and J.L. Salisbury. 1994. Centrin plays an essential role in microtubule severing during flagellar excision in Chlamydomonas reinhardtii. J. Cell Biol. 124:795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnackenberg, B.J., A. Khodjakov, C.L. Rieder, and R.E. Palazzo. 1998. The disassembly and reassembly of functional centrosomes in vitro. Proc. Natl. Acad. Sci. USA. 95:9295–9300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, P.A., C.P. Toret, A.N. Salic, M.M. Rolls, and T. Rapoport. 2002. A novel centrosome-associated protein with affinity for microtubules. J. Cell Sci. 115:3389–3402. [DOI] [PubMed] [Google Scholar]
- Tynan, S.H., A. Purohit, S.J. Doxsey, and R.B. Vallee. 2000. Light intermediate chain 1 defines a functional subfraction of cytoplasmic dynein which binds to pericentrin. J. Biol. Chem. 275:32763–32768. [DOI] [PubMed] [Google Scholar]
- Wittmann, T., and T. Hyman. 1999. Recombinant p50/dynamitin as a tool to examine the role of dynactin in intracellular processes. Methods Cell Biol. 61:137–143. [DOI] [PubMed] [Google Scholar]
- Young, A., J.B. Dictenberg, A. Purohit, R. Tuft, and S.J. Doxsey. 2000. Cytoplasmic dynein-mediated assembly of pericentrin and γ tubulin onto centrosomes. Mol. Biol. Cell. 11:2047–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, Y., M.L. Wong, B. Alberts, and T. Mitchison. 1995. Nucleation of microtubule assembly by a γ-tubulin-containing ring complex. Nature. 378:578–583. [DOI] [PubMed] [Google Scholar]