Abstract
As the sole site of nucleocytoplasmic transport, the nuclear pore complex (NPC) has a vital cellular role. Nonetheless, much remains to be learned about many fundamental aspects of NPC function. To further understand the structure and function of the mammalian NPC, we have completed a proteomic analysis to identify and classify all of its protein components. We used mass spectrometry to identify all proteins present in a biochemically purified NPC fraction. Based on previous characterization, sequence homology, and subcellular localization, 29 of these proteins were classified as nucleoporins, and a further 18 were classified as NPC-associated proteins. Among the 29 nucleoporins were six previously undiscovered nucleoporins and a novel family of WD repeat nucleoporins. One of these WD repeat nucleoporins is ALADIN, the gene mutated in triple-A (or Allgrove) syndrome. Our analysis defines the proteome of the mammalian NPC for the first time and paves the way for a more detailed characterization of NPC structure and function.
Keywords: nuclear pore complex; nucleoporins; nucleocytoplasmic transport; proteomics; WD repeats
Introduction
Nucleocytoplasmic transport is mediated by nuclear pore complexes (NPCs)* (Allen et al., 2000) which span the nuclear envelope (NE) lumen, inserting into pores formed by the fusion of inner and outer nuclear membranes. NPCs are large multiprotein complexes with octagonal symmetry about their axis and imperfect mirror symmetry about a plane parallel with the NE. They provide a diffusion channel for small molecules and also mediate the active transport of large substrates. As the sole sites of nucleocytoplasmic transport, NPCs play a role in numerous pathways, including cell cycle progression, control of gene expression, and oncogenesis (Lain et al., 1999; Jeffries and Capobianco, 2000; Takizawa and Morgan, 2000). In addition, NPCs have other functions in nuclear organization (Smith and de Lange, 1999; Galy et al., 2000; Belgareh et al., 2001; Ishii et al., 2002).
A proteomic analysis revealed that the yeast NPC is composed of 29 nucleoporins (Rout et al., 2000). To date, 24 nucleoporins have been identified in mammals, with up to 25 remaining to be discovered (Vasu and Forbes, 2001). A subset of known nucleoporins contain functionally significant phenylalanine-glycine (FG) repeat domains, which bind directly to receptors that transport substrates through the NPC (Ryan and Wente, 2000). Although several models have been proposed (Rout et al., 2000; Macara, 2001; Ribbeck and Görlich, 2001), the mechanism by which these FG repeat domains mediate active transport is poorly understood.
Structural comparisons indicate that the vertebrate NPC is larger than its yeast counterpart (1450 × 800 Å compared with 960 × 350 Å) (Akey and Radermacher, 1993; Yang et al., 1998). Mass measurements also suggest that the vertebrate NPC, with an estimated maximum mass of 125 MDa, is significantly larger than the 55–72 MDa yeast NPC (Reichelt et al., 1990; Rout and Blobel, 1993; Yang et al., 1998). However, the combined mass of all yeast nucleoporins is calculated to be only 44 MDa, suggesting that experimentally determined measurements are upper estimates, with NPC-associated proteins and transport complexes contributing significantly to the measured mass (Rout et al., 2000). Given the structural differences between vertebrate and yeast NPCs, a comparison of their proteomes could be of particular value in understanding both conserved and species-specific functions of the NPC.
We used mass spectrometry (MS) to characterize a highly purified nucleoporin fraction from rat liver nuclei. Surprisingly, we found that the mammalian NPC is composed of essentially the same number of nucleoporins as its yeast counterpart. Among the nucleoporins we identified are six novel nucleoporins and a new subfamily of nucleoporins characterized by the presence of WD repeats. Comparison of yeast and mammalian nucleoporins suggests that basic NPC functions are conserved, but that both organisms have also evolved specialized functions.
Results
Enrichment of nucleoporins from rat liver nuclei
To determine the composition of the mammalian NPC, we developed a fractionation procedure to highly enrich nucleoporins from rat liver nuclei. This procedure is a modification of one previously described (Dwyer and Blobel, 1976) and entails the sequential solubilization of nuclear substructures. Until the final step, NPCs remain associated with the lamina and can be separated from solubilized proteins by centrifugation through a sucrose cushion. We have used EM, SDS-PAGE, and immunoblotting to confirm that NPCs remain intact and nucleoporins are not lost during fractionation.
The first step is a digestion with DNase and RNase in the presence of low concentrations of divalent cations. This solubilizes most intranuclear material, although some electron-dense aggregates remain associated with the inner nuclear membrane of the pelleted NEs (Fig. 1 A, arrows). A subsequent extraction with heparin clearly solubilizes these chromatin remnants (Fig. 1, compare A with B). In agreement with the EM data, SDS-PAGE analysis reveals that histones are partially solubilized by DNase/RNase and even more dramatically solubilized by heparin (Fig. 1 E). After the removal of chromatin, the nuclear membranes and their associated proteins are extracted by incubation in Triton X-100 and SDS, leaving NPCs embedded in the lamina. By negative staining (Fig. 1 C) it can be seen that the NPCs retain their characteristic eightfold symmetry and have a central transporter, indicating that they are largely intact. The final step is an incubation with the zwitterionic detergent Empigen BB, which selectively solubilizes the NPC as monomeric nucleoporins. The intact lamina (Fig. 1 D) is cleared from this solution of highly enriched nucleoporins by high-speed centrifugation. When no longer connected to the NPC, the lamina filaments appear to retract, resulting in gaps in the lamina network (Fig. 1 D). By SDS-PAGE it can be seen that the major proteins remaining in the Empigen BB pellet are the lamins (Fig. 1 E).
Figure 1.
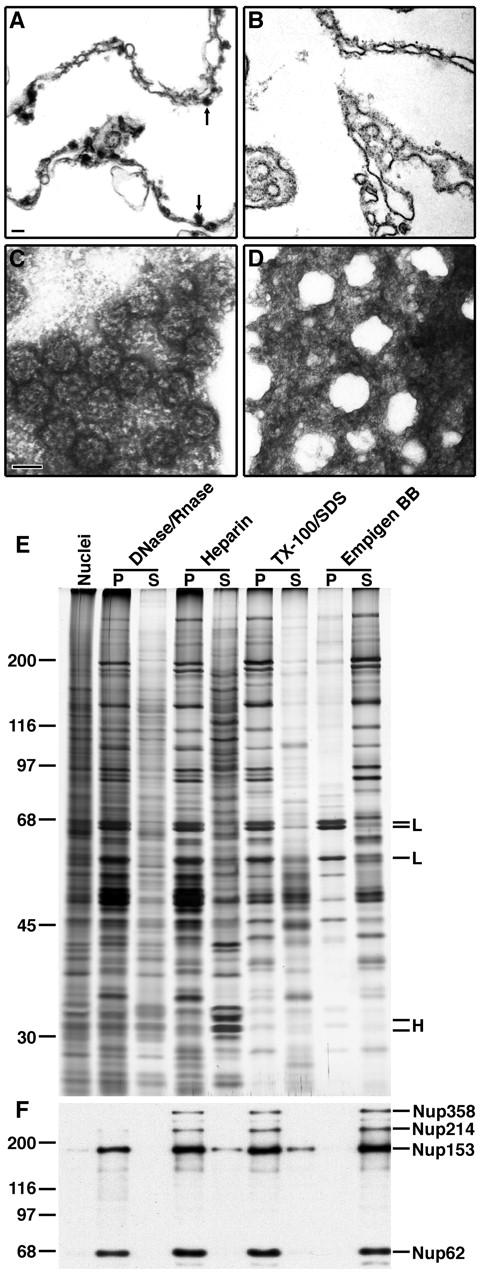
Fractionation of rat liver nuclei. (A–D) EM analysis of the fractionation of rat liver nuclei. Bar, 100 nm. (A) Thin-section EM of pelleted nuclear envelopes following digestion with DNase/RNase. Arrowheads indicate electron-dense aggregates associated with the inner nuclear membrane. (B) Thin-section EM of nuclear envelopes after extraction with heparin. (C) Negative staining EM of the pellet after extraction with Triton X-100/SDS. (D) Negative staining EM of the pellet after extraction with Empigen BB. (E) Silver stained SDS-PAGE analysis of supernatant (S) and pellet (P) from each step of the fractionation. Molecular weight markers are shown on the left. Histones (H) and lamins (L) are denoted on the right. (F) Immunoblot analysis of the pellet and supernatant from each step of the fractionation using mAb414 which recognizes the nucleoporins Nup358, Nup214, Nup153, and Nup62 (Davis and Blobel, 1986). Molecular weight markers are indicated on the left and nucleoporins are indicated on the right.
Using a panel of anti-nucleoporin antibodies (mAb414 is shown as a representative example), we found that all tested nucleoporins fractionate in the Empigen BB supernatant (Fig. 1 F). Importantly, these include nucleoporins from distinct domains of the NPC, including the pore membrane (POM121, gp210), the central transporter (Nup62), and peripheral cytoplasmic (Nup214, Nup358) and nucleoplasmic (Nup153, Tpr) structures. The presence of nucleoporins from these diverse NPC structures further demonstrates that our purified NPCs are intact, strongly arguing that nucleoporins are not quantitatively lost during fractionation.
Identification and classification of proteins in the Empigen BB supernatant
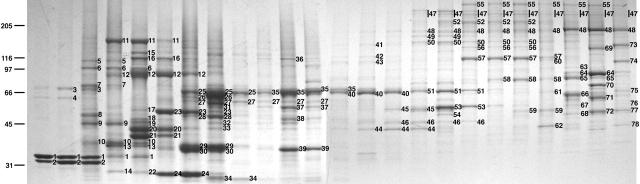
Owing to the complexity of the SDS-PAGE profile of the Empigen BB supernatant, particularly in the 50–80-kD range (Fig. 2), we further separated these proteins by C4 reverse phase chromatography prior to SDS-PAGE (Fig. 3). We used a combination of single-step MS and tandem mass spectrometry (MS/MS) to identify proteins and/or ESTs in bands excised from both unseparated and C4-separated Empigen BB supernatant. C4 reverse phase separation facilitated the identification of less abundant proteins, as up to 100× more material could be analyzed. Reverse phase separation also simplified identification by decreasing the number of proteins in individual SDS-PAGE bands. Parallel analysis of bands from unseparated Empigen BB supernatant ensured that we identified proteins that could have been lost during chromatography, particularly late-eluting proteins that were recovered less efficiently from the C4 column. Proteins identified in unseparated Empigen BB supernatant (Fig. 2) are shown in Table I. Additional proteins identified only from the C4-separated fractions can be found in Table SI (available at http:www.jcb.org/cgi/content/full/jcb.200206106/DC1). A total of 94 proteins were identified. Based on information in the literature and public databases, we initially classified 23 as nucleoporins, 18 as NPC-associated, 42 as non-NPC proteins, and 11 as uncharacterized (Table II).
Figure 2.
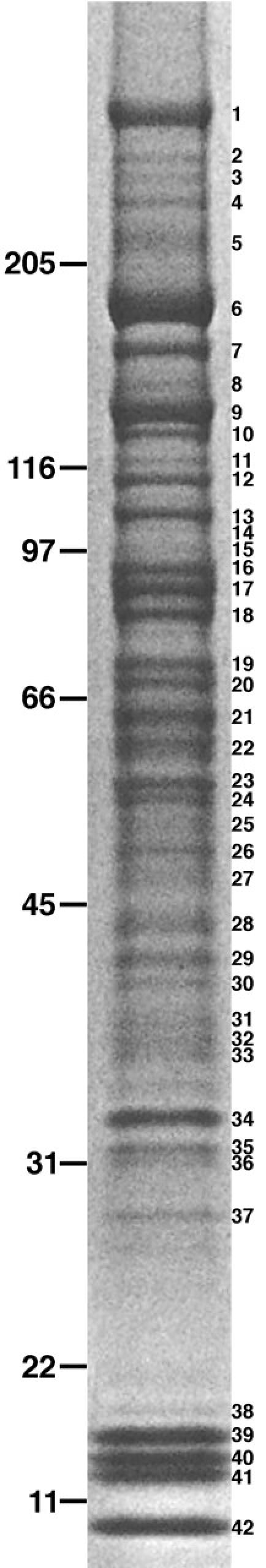
Identification of bands from unseparated Empigen BB supernatant. 10 U of Empigen BB supernatant were separated by SDS-PAGE and stained with Coomassie. Molecular weight markers are shown on the left and bands excised for mass spectrometric analysis are indicated on the right. Proteins identified in these bands are shown in Table I.
Figure 3.
Chromatographic separation of the Empigen BB supernatant. ∼1,000 U of C4-separated Empigen BB supernatant fractions were separated by SDS-PAGE and stained with Coomassie. Molecular weight markers are shown on the left and bands excised for mass spectrometric analysis are indicated to the right of each band. Proteins identified in these bands are shown in Tables I and SI, available at http:www.jcb.org/cgi/content/full/jcb.200206106/DC1 (histones are not shown on this gel).
Table I. Identification of proteins from unseparated Empigen BB supernatant.
| No. | Proteins identified | Accessiona | kDb | C4 band |
|---|---|---|---|---|
| 1 | Nup358/RanBP2 | NP_006258 | 358 | 55 |
| 2 | Tpr | NP_003283 | 266 | 47 |
| 3 | Tpr Nup358c |
NP_003283 NP_006258 |
266 358 |
47 55 |
| 4 | Tpr Nup358c |
NP_003283 NP_006258 |
266 358 |
47 55 |
| 5 | Nup214/CAN Nup358c |
NP_005076 NP_006258 |
214 358 |
52 55 |
| 6 | gp210 Nup205 |
P11654 (rat) BAA13214 |
204 228 |
48 48 |
| 7 | Nup188 Nup153 |
BAA11486 NP_005115 |
196d
154 |
– 11 |
| 8 | POM121 SMC1 |
A40670 (rat) NP_006297 |
121 143 |
50 50 |
| 9 | Nup155 Nup160/Nup120 |
CAA07553 BAA12110 |
155 159d |
73 – |
| 10 | Nup133 | NP_060700 | 129 | 69 |
| 11 | Matrin3 | NP_061322 | 95 | 16 |
| 12 | Nup96 | NP_005378 | 96 | 57 |
| 13 | Nup107 | NP_065134 | 106 | 74 |
| 14 | Nup98 PSF |
NP_005378 NP_005057 |
90 76 |
12 6 |
| 15 | Nup98 | NP_005378 | 90 | 12 |
| 16 | Importin-β | NP_002256 | 97 | – |
| 17 | Nup93 | NP_055484 | 93 | 64 |
| 18 | Nup88/Nup84 RanGAP1 SUMO-1 |
NP_002523 NP_002874 NP_003343 |
84 64 11 |
58 65 65 |
| 19 | LAP1C (long isoform) LAP1C (short isoform) hnRNP M Lamin A |
AAA69914 (rat) AAA69915 (rat) NP_005959 CAA27173 |
57 52 78 77 |
70 70 35 25 |
| 20 | Hsc70 hnRNP M TAP |
NP_006588 NP_005959 NP_006353 |
71 78 70 |
51 35 40 |
| 21 | Nup62 RanGAP1 FLJ12549 |
NP_057637 NP_002874 AAM76706 |
53 64 75 |
66 66 75 |
| 22 | Nup58 (rat) Lamin C ALADIN/AAAS/Adracalin |
AAC52789 CAA27174 NP_056480 |
59 65 60 |
71 27 – |
| 23 | Nup54 | NP_059122 | 55 | 53 |
| 24 | Nup50/NPAP60 LAP2 |
NP_009103 NP_037019 (rat) |
50 50 |
23 59/67 |
| 25 | UDP Glucosyl Transferase (UGT)e | – | – | 46/77 |
| 26 | Nup45 (rat) UGTe |
AAC82318 – |
52 – |
72 46/77 |
| 27 | UGTe | – | – | 46/77 |
| 28 | NLP1/hCG-1 p42 Actine |
NP_031368 AAM76708 – |
45 42 – |
44 20 62 |
| 29 | RAE1/Gle2 HSD3B1 HSD3B2 |
NP_003601 NP_000853 NP_000189 |
41 42 42 |
21 78 78 |
| 30 | Histone macroH2Ae | – | – | – |
| 31 | Sec13-like (Sec13L) | AAM76707 | 40 | 13 |
| 32 | p37 Sec13-related (Sec13R) |
AAM76705 NP_109598 |
37 36 |
29 30 |
| 33 | MP-44 | AAM76704 | 35 | 39 |
| 34 | Histone H1e | – | – | 1 |
| 35 | Histone H1e | – | – | 2 |
| 36 | p30 | AAM76703 | 30d | – |
| 37 | Ran | AAB24940 | 24 | 24 |
| 38 | Ubc9 | AAH00744 | 20 | unpublished data |
| 39 | Histone H3e | – | – | unpublished data |
| 40 | Histone H2Be | – | – | unpublished data |
| 41 | Histone H2Ae | – | – | unpublished data |
| 42 | Histone H4e | – | – | unpublished data |
Unless otherwise stated, accession numbers are for human proteins.
Predicted from amino acid sequence.
Proteolysis product or alternative isoform.
Estimated molecular weight. Exact 5' end of sequence unknown.
Exact isoform not determined although, in most cases, it appears that several distinct isoforms may be present in a single band.
Table II. Summary of identified proteins.
| Nucleoporins | NPC-associated proteins | Non-NPC proteins | Uncharacterized proteins |
|---|---|---|---|
| Nup358 | Importin-α1 | Nuclear matrix/chromatin-associated proteins | Abundant |
| Tpr | Importin-β1 | LAP1C | FLJ12549 |
| Nup214 | TAP | LAP2 | Sec13L |
| gp210 | hnRNP F | MAN1 | MP-44 |
| Nup205 | hnRNP H | Matrin3 | ALADIN/AAAS |
| Nup188 | hnRNP M | SMARCC1 | p42 |
| Nup153 | Ran | SMARCC2 | p37 |
| Nup160 | RanGAP1 | SMARCD2 | p30 |
| Nup155 | RCC1 | SMARCE1 | Nonabundant |
| Nup133 | Hsc70 | DDX5 | c1orf28 |
| POM121 | Lamin A | HDAC1 | CoAA |
| Nup107 | Lamin B | HDAC2 | HP1-BP74 |
| Nup98 | Lamin C | RbAp46 | KIAA1551 |
| Nup96 | Sec13R | RbAp48 | |
| Nup93 | Actin | MTA2 | |
| Nup88 | Vimentin | SMC1 | |
| Nup62 | Ubc9 | SMC3 | |
| Nup58 | SENP2 | RAD21 | |
| Nup54 | TIF1β | ||
| Nup50 | RUVBL1 | ||
| Nup45 | RUVBL2 | ||
| NLP1 | Histone H1 | ||
| RAE1 | Histone H2A | ||
| Histone H2B | |||
| Histone H3 | |||
| Histone H4 | |||
| Histone macroH2A | |||
| Fibrillarin | |||
| SAF-B | |||
| DEK | |||
| PSF | |||
| Splicing factors | |||
| SAP145 | |||
| TDP-43 | |||
| SF3a | |||
| SART-1 | |||
| Cytoskeletal proteins | |||
| α-tubulin | |||
| β-tubulin | |||
| Chaperones | |||
| Hsp27 | |||
| DnaJ proteins | |||
| Enzymes | |||
| HSD3B1 | |||
| HSD3B2 | |||
| UGT enzymes | |||
| Catalase |
The 23 proteins classified as nucleoporins (Table II, column 1) were all identified in unseparated Empigen BB supernatant and are, therefore, major components of the NPC fraction. Only ∼1/2 of these nucleoporins were identified in rat databases, with the remaining proteins identified by searching the more complete mouse and human databases. This approach proved feasible because of the relatively high sequence conservation between rat, mouse, and human proteins and because of the large number of MS/MS measurements performed on peptides from each sample. We identified all previously described vertebrate nucleoporins with the exception of Gle1, whose association with the NPC may be dynamic (Watkins et al., 1998).
Of the 18 proteins classified as NPC-associated proteins (Table II, column 2), most were factors involved in nucleocytoplasmic transport, such as importin-α1, importin-β1, TAP, hnRNPs, Ran, RanGAP1, RCC1, and Hsc70. Lamins A, B, and C are components of the lamina which is intimately associated with the NPC and remains associated with it until the final step of the fractionation. The function of the other NPC-associated proteins in transport is not well understood, although all have been shown to localize, at least in part, to the NPC (Cai et al., 1997; Saitoh et al., 1997; Fontoura et al., 1999; Hofmann et al., 2001; Vasu et al., 2001; Hang and Dasso, 2002; Zhang et al., 2002). Proteins classified as non-NPC proteins (Table II, column 3) are known to function and/or localize at sites other than the NPC. Most non-NPC proteins were minor components since they were identified only in the C4-separated fractions. A number of these non-NPC proteins have been reported to be nuclear matrix- or chromatin-associated (Table II) and may, therefore, be connected directly or indirectly to the NPC/lamina. Other proteins may be contaminants, such as abundant transport substrates caught during translocation, or abundant liver or ER-associated enzymes. The final subset of proteins included those whose function and/or localization was unknown (Table II, column 4). These were further divided into two subgroups: abundant proteins that were identified in unseparated Empigen BB supernatant and non-abundant proteins that were identified only in the C4-separated fractions. p42, p37, and p30 are preliminary names for proteins identified for the first time in this study.
To determine if any nucleoporins remained insoluble after the final fractionation step, we also identified proteins in the Empigen BB pellet (Fig. S1, available at http:www.jcb.org/cgi/content/full/jcb.200206106/DC1 and Table SII). Lamins A, B, and C were the major components of the Empigen BB pellet. Some nucleoporins (Nup214, gp210, Nup153, and Nup98) were also present, though only Nup98 was a major component. The uncharacterized proteins, p30 and MP-44, were also identified in the Empigen BB supernatant and pellet and will be discussed below.
Sequence analysis of uncharacterized proteins
Sequence analysis of the uncharacterized proteins revealed that three of the abundant proteins (FLJ12549, Sec13L, and MP-44) have homology to known nucleoporins. FLJ12549 has low-sequence homology to the Schizosaccharomyces pombe homologue of Nup85p (19% identity and 37% similarity over its central ∼400 amino acids). Although it displays no significant homology to Saccharomyces cerevisiae Nup85p, this low level of conservation is frequently seen in nucleoporins and may still indicate functional conservation. The central ∼150 amino acids of MP-44 are 21% identical (40% similar) to S. cerevisiae Nup53p. Although this homology does not extend to the COOH-terminal amphipathic α-helix of Nup53p (Marelli et al., 1998, 2001), there are predicted amphipathic regions in MP-44 which may be functionally homologous. Like Nup53p, MP-44 has a small number of scattered FG repeats but not the large domains seen in other nucleoporins. Sec13L is related to yeast Sec13p (30% identity and 47% similarity) but is more homologous to Seh1p (34% identity, 54% similarity). This unusually high degree of sequence conservation in a nucleoporin is largely due to the presence of six WD repeat motifs. The homology between Sec13L and Seh1p also extends to regions outside these repeats, making Sec13L the most likely candidate for the human homologue of Seh1p. Sec13R is the human homologue of Sec13p in both sequence (50% identity, 66% similarity) and function (Shaywitz et al., 1995). The remaining four abundant proteins (ALADIN, also called Adracalin; p42; p37; and p30) are not homologous to any known nucleoporin and have no characterized homologues that could suggest a function. However, we did note that ALADIN, p42, and p37 all contain WD repeats (4, 5, and 4, respectively). Of the nonabundant proteins, none have homology to nucleoporins, although two have conserved sequence motifs. CoAA has two RNA recognition motif domains and HP1-BP74 has a domain found in linker histones H1 and H5 (Le Douarin et al., 1996; Iwasaki et al., 2001).
Subcellular localization of uncharacterized proteins
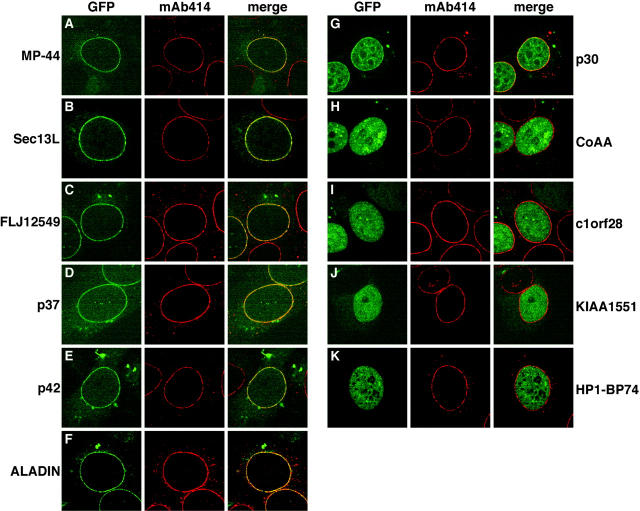
The uncharacterized proteins were further studied by examining the subcellular localization of transiently expressed GFP-tagged fusion proteins. Six of the abundant novel proteins (MP-44, Sec13L, FLJ12549, p37, p42, and ALADIN) showed punctate nuclear rim localization typical of nucleoporins (Fig. 4, A–F, left). This was particularly apparent in views of the nuclear surface (unpublished data). In each case the GFP signal colocalized with that of anti-nucleoporin antibody mAb414 (which recognizes Nup358, Nup214, Nup153, and Nup62), further confirming the NPC localization of these proteins (Fig. 4, A–F, middle and right). On the basis of their subcellular localization and, in some cases, homology to known nucleoporins, we propose that the five uncharacterized proteins (MP-44, Sec13L, FLJ12549, p37, and p42) be named Nup35, Seh1, Nup75, Nup37, and Nup43 (to avoid confusion with the unrelated yeast nucleoporin Nup42p). The remaining abundant novel protein (p30) was mostly nuclear with a significant enrichment at the nuclear periphery and around the nucleolus (Fig. 4 G, left). However, the GFP signal at the nuclear periphery did not colocalize with mAb414 (Fig. 4 G, middle and right) indicating that p30 is not a nucleoporin. We speculate that this protein may be involved in linking peripheral nuclear structures, such as the nuclear lamina, with the nuclear interior. A more detailed study of the localization of p30 will provide further insight into the functions of this novel protein.
Figure 4.
Subcellular localization of uncharacterized proteins. GFP-tagged fusion proteins were transiently transfected into HeLa cells and their localization visualized by confocal microscopy 48 h posttransfection (green, left). Transfected cells were also labeled with mAb414 (red, middle). Merged images showing the extent of colocalization are shown in the right panel.
The nonabundant uncharacterized proteins (CoAA, c1orf28, KIAA1551, and HP1-BP74) were all nuclear with no obvious enrichment at the nuclear periphery (Fig. 4, H–K, left). In double labeling experiments with mAb414 (Fig. 4, H–K, middle and right), we saw no overlap between these proteins and the NPC. Therefore, these proteins are unlikely to be nucleoporins. Because many nonabundant proteins in the Empigen BB supernatant are nuclear matrix- or chromatin-associated, CoAA, c1orf28, KIAA1551, and HP1-BP74 may be similarly localized.
Mass estimation of the NPC
We estimated the relative abundance of nucleoporins in the Empigen BB supernatant by quantitation of SDS-PAGE band intensities (see Materials and methods). From their relative abundance we estimated the copy number per NPC of each nucleoporin, based on the assumption that nucleoporins would be present at a copy number of 8 or multiples of 8, owing to the rotational symmetry of the NPC (Table III). From the nucleoporin copy number, we estimated NPC mass. This approach has certain limitations, such as nonquantitative dye-binding, stain saturation, and incomplete recovery of nucleoporins. Although these limitations were controlled for using different staining methods and different loading conditions, the results are an approximation. Overall, the estimated relative abundance of individual nucleoporins correlated well with that determined for their yeast homologues (Table III; Rout et al., 2000) and resulted in an NPC mass estimate of ∼60 MDa. The mass of the vertebrate NPC was previously estimated by STEM analysis of manually isolated Xenopus oocyte nuclear envelopes (Reichelt et al., 1990). The mass of 125 MDa reported in this study is a maximum since Xenopus oocyte nuclear envelopes were isolated using mild conditions and probably retain many transport factors and cargo. By comparison, the yeast NPC has an estimated mass of 55–72 MDa (Rout and Blobel, 1993; Yang et al., 1998) but a calculated mass of only 44 MDa (Rout et al., 2000). In vertebrates, the closely apposed lamina could also contribute to experimental mass measurements, as would posttranslational modification of nucleoporins by glycosylation and/or phosphorylation. Tissue and species differences may also contribute to differences between the estimated masses of the rat liver and Xenopus oocyte NPCs. Overall, our analysis indicates that the mammalian NPC is composed of ∼30 distinct nucleoporins that are present in similar ratios to their yeast counterparts.
Table III. Relative abundance of mammalian nucleoporins.
| Nucleoporin | Relative abundancea | Yeast homologue(s) | Relative abundancea , b |
|---|---|---|---|
| Nup358 | 8 | – | – |
| Tpr | 16 | Mlp1p, Mlp2p | ND |
| Nup214 | 8 | Nup159p | 8 |
| gp210 | 16 | – | – |
| Nup205 | 16 | Nup192p | 16 |
| Nup153 | 8 | Nup1pc | 8 |
| Nup188 | 8 | Nup188p | 16 |
| POM121 | 8 | – | – |
| Nup155 | 32 | Nup157p, Nup170p | 32 |
| Nup160 | 8 | Nup120p | 16 |
| Nup133 | 16 | Nup133p | 16 |
| Nup96 | 16 | C-Nup145p | 16 |
| Nup107 | 32 | Nup84p | 16 |
| Nup98 | 8 | N-Nup145p, Nup116p, Nup100p | 32 |
| Nup93 | 32–48 | Nic96p | 32 |
| Nup88 | 32 | Nup82p | 8–16 |
| Nup62 | 16 | Nsp1p | 32 |
| Nup75 (FLJ12549) | 16 | Nup85p | 16 |
| Nup58 | 48 | Nup49p | 16 |
| ALADIN | 8 | – | – |
| Nup54 | 32–48 | Nup57p | 16 |
| Nup50 | 32 | Nup2pc | ND |
| Nup45 | 32 | (Nup49p) | (16) |
| NLP1 | 16 | Nup42p | 8 |
| Nup43 (p42) | 16 | – | – |
| RAE1 | 48 | Gle2p | 16–32 |
| Seh1 (Sec13L) | 16–32 | Seh1p | 16 |
| Nup37 (p37) | 16–32 | – | – |
| Sec13R | 16–32 | Sec13p | ND |
| Nup35 (MP-44) | 16–32 | Nup53p, Nup59p | 32 |
Copies per NPC.
Rout et al., 2000.
Direct sequence homology is not necessarily apparent. Rather, conserved domains, interactions, and localization imply functional similarities.
Discussion
Although the vertebrate NPC was first described several decades ago and its structure has been relatively well characterized (Allen et al., 2000), its protein composition has not yet been completely defined and a detailed understanding of many of its cellular functions has proved elusive. Estimates of the number of distinct NPC proteins have been as high as 100 but have recently fallen to a more conservative 40–50 (Vasu and Forbes, 2001). Our analysis of biochemically isolated and enriched nucleoporins from rat liver nuclei shows that the vertebrate NPC is comprised of ∼30 distinct proteins. Although we cannot exclude the possibility that some nucleoporins were not retained or not identified in our analysis, we believe that this number is small. Our EM and immunoblot data, combined with the results of our MS analysis, all indicate that the NPC remained intact during fractionation and that nucleoporins were not lost prior to or during the final extraction. We have maximized the number of identified proteins by further fractionating the Empigen BB supernatant to identify less abundant protein components. Large-scale MS/MS analysis facilitated the identification of proteins encoded by partial cDNA sequences or encoded by cDNAs from other species.
Nucleoporins and NPC-associated proteins
We identified the component proteins of a biochemically purified nucleoporin fraction. Of the 94 proteins identified, 29 (∼1/3) were classified as nucleoporins (Table IV). Six of these nucleoporins have not been previously described. A further 18 proteins were classified as NPC-associated based on previous functional characterization. However, this distinction between nucleoporins and NPC-associated proteins is not necessarily clear cut. Therefore, it may not be meaningful to make such a clear distinction between NPC components when trying to understand the structure and function of the NPC. Because an in-depth characterization of all the novel proteins identified here was beyond the scope of this study, we have classified novel proteins as nucleoporins based on their apparently exclusive localization to the NPC. Future work will reveal more on the functions of these proteins and clarify their roles at the NPC.
Table IV. Summary of vertebrate nucleoporins.
| Motifsb
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nup | Nup subcomplexes | Localizationa | FG | WD | Gly | RBD | ZF | CC | LZ | AH | EF | Disease |
| Nup358 | ? | C | Y | Y | Y | Y | ||||||
| Tpr | Nup98 | N | Y | Y | Oncogenic fusions | |||||||
| Nup214 | Nup88 Nup62 |
C | Y | Y | Y | Y | Y | Oncogenic fusions | ||||
| gp210 | POM121 | PM | Y | Y | PBC autoantibodies | |||||||
| Nup205 | Nup188/Nup93 | ? | Y | |||||||||
| Nup153 | Nup107 complexc | N | Y | Y | Y | |||||||
| Nup188 | Nup205/Nup93 | ? | ||||||||||
| POM121 | gp210 | PM | Y | Y | ||||||||
| Nup155 | ? | C,N | ||||||||||
| Nup160 | Nup107 complexc
Nup98 Nup153 |
N | ||||||||||
| Nup133 | Nup107 complexc
Nup98 Nup153 |
C,N | ||||||||||
| Nup96 | Nup107 complexc
Nup98 Nup153 |
N | ||||||||||
| Nup107 | Nup107 complexc
Nup98 Nup153 |
C,N | Y | |||||||||
| Nup98 | RAE1 Nup107 complexc Tpr |
N | Y | Y | Oncogenic fusions | |||||||
| Nup93 | Nup62 Nup205/Nup188 |
N | Y | |||||||||
| Nup88 | Nup214 | C | Y | Upregulated in some tumors | ||||||||
| Nup62 | Nup62 complexd
Nup214 |
C,N | Y | Y | Y | PBC autoantibodies | ||||||
| Nup75 | ? | ? | ||||||||||
| Nup58 | Nup62 complexd | C,N | Y | Y | Y | PBC autoantibodies | ||||||
| ALADIN | ? | ? | Y | AAAS | ||||||||
| Nup54 | Nup62 complexd | C,N | Y | Y | Y | PBC autoantibodies | ||||||
| Nup50 | Nup153 | N | Y | Y | ||||||||
| Nup45 | Nup62 complexd | C,N | Y | Y | Y | PBC autoantibodies | ||||||
| NLPI | ? | C | Y | Y | ||||||||
| Nup43 | ? | ? | Y | Y | ||||||||
| RAE1 | Nup98 | N | Y | |||||||||
| Seh1 | ? | ? | Y | |||||||||
| Nup37 | Nup107 complexc
Nup98 Nup153 |
? | Y | |||||||||
| Nup35 | ? | ? | Y | |||||||||
Unless specifically discussed in the text, data are summarized from previously published reports (Allen et al., 2000; Ryan and Wente, 2000; Vasu and Forbes, 2001).
C, cytoplasmic face; N, nucleoplasmic face; PM, pore membrane.
FG, FG repeats; WD, WD repeats; Gly, glycosylated; RBD, Ran-binding domain; ZF, zinc fingers; CC, coiled coil; LZ, leucine zipper; AH, amphipathic helix; EF, EF hand.
Nup107 complex: Nup160, Nup133, Nup107, Nup96, Nup37, Sec13.
Nup62 complex: Nup62, Nup58, Nup54, Nup45.
It was surprising that only certain transport receptors, importin-β, and TAP, were abundant in the Empigen BB supernatant. Importin-α1 was also identified as a minor component. Other transport receptors may be less tightly bound to the NPC or may mediate the transport of less abundant substrates. In addition, RanGAP1 remains associated with the cytoplasmic fibrils and is expected to promote the dissociation of export complexes in transit through the NPC (Floer and Blobel, 1999). The presence of hnRNPs F, H, and M in the Empigen BB supernatant may reflect the extent to which these hnRNPs form protein–protein contacts with the NPC during mRNA export. As TAP is the receptor responsible for the majority of mRNA export (Reed and Hurt, 2002), it will be interesting to see if there is any link between this receptor and hnRNPs F, H, and M. Ran, RanGAP1, and RCC1 are all known to associate with the NPC (Wilken et al., 1995; Wu et al., 1995; Yokoyama et al., 1995; Matunis et al., 1996; Mahajan et al., 1997; Nakielny et al., 1999; Yaseen and Blobel, 1999; Fontoura et al. 2000). It is logical that proteins involved in nuclear transport are localized, at least in part, to the NPC, as this is where they function. In addition, the binding of RanGAP1 and RCC1 to opposite sides of the NPC maintains an asymmetric distribution of RanGTP/RanGDP that specifies the directionality of transport (Mattaj and Englmeier, 1998).
Comparison of yeast and vertebrate NPCs
Even though yeast and vertebrate NPCs share a conserved basic framework, the vertebrate NPC is significantly larger and has several additional domains (Akey and Radermacher, 1993; Yang et al., 1998). These include more intricate peripheral rings/filaments, a larger lumenal spoke complex domain, and a larger, hourglass-shaped central transporter. Interestingly, many nucleoporins that localize to these additional structures have no yeast orthologue, for example Nup358, gp210, and POM121 (Greber et al., 1990; Hallberg et al., 1993; Wilken et al., 1995; Wu et al., 1995). Other nucleoporins, including Nup214, Nup98, and Nup62, undergo certain vertebrate-specific modifications such as glycosylation (Davis and Blobel, 1987; Finlay et al., 1987; Kraemer et al., 1994; Powers et al., 1995). Glycosylation has been shown to alter protein structure (Shogren et al., 1989) and, although it would cause only a small increase in mass, the effect of glycosylation on NPC size may be disproportionate. Our estimates also suggest that Nup58, Nup54, and Nup45 (putative components of the central transporter; Guan et al., 1995) may be more abundant in the vertebrate NPC than are their yeast homologues. These factors may all potentially contribute to the greater dimensions and complexity of the vertebrate NPC.
In addition to the aforementioned structural differences, the vertebrate NPC also performs certain specialized functions. The most obvious of these occurs during mitosis when the vertebrate NE breaks down and the NPC disassembles into subcomplexes. NE breakdown is thought to be initiated by phosphorylation and several nucleoporins, including Nup62, Nup98, Nup214, and gp210, are phosphorylated in a cell cycle–dependent manner (Macaulay et al., 1995; Favreau et al., 1996; Miller et al., 1999). Because yeast undergo a closed mitosis, mitotic phosphorylation as a trigger for NE breakdown may be unnecessary. However, a closed mitosis creates other problems, including a requirement for the spindle to somehow invade the nucleus. Two yeast nucleoporins (Ndc1p and Cdc31p) are shared components of both NPCs and the spindle pole body and may facilitate this process (Chial et al., 1998; Rout et al., 2000). Mammals have no known homologue of Ndc1p and the closest human homologue of Cdc31p, Centrin3, apparently does not localize to the NPC (Middendorp et al., 1997). Another vertebrate NPC specialization is the interaction between NPCs and the lamina, an interaction mediated, at least in part, by Nup153 (Smythe et al., 2000). Therefore, it is not surprising that some Nup153 remains in the Empigen BB pellet, and it may explain why a substantial amount of Nup98 also does. Yeast, in contrast, has no homologous nuclear lamina.
In several cases, yeast have two or more related nucleoporins that correspond to a single mammalian nucleoporin. Specifically, the yeast nucleoporins Nup157p/Nup170p, Nup53p/Nup59p, and Nup100p/Nup116p/N-Nup145p have homology to the mammalian nucleoporins Nup155, Nup35, and Nup98, respectively. Although a search of human genome sequences reveals putative gene duplication events for both Nup35 and Nup155, these duplications are pseudogenes or partial transpositions and are, therefore, unlikely to be functional. It is possible that one mammalian nucleoporin can perform the functions of all its yeast orthologues. Nup98, for example, contains domains specific to both N-Nup145p, and Nup116p (Ryan and Wente, 2000). Alternative splicing, an event very rare in yeast genes (Graveley, 2001) but seen in some mammalian nucleoporins (Hu and Gerace, 1998; Fontoura et al., 1999) may further increase vertebrate nucleoporin diversity in a manner analogous to the gene duplications seen in yeast.
Evolutionary conservation of the NPC
Given their structural and functional differences, it is surprising that vertebrate and yeast NPCs are composed of a similar number of distinct proteins. As discussed above, vertebrate-specific nucleoporins are probably the major factor giving rise to species differences in NPC size and complexity. However, two-thirds of the nucleoporins are conserved between the two species and probably mediate conserved functions. Although sequence homology between yeast and mammalian nucleoporins is frequently low, their interactions and localization suggests conservation of function. In addition, some NPC-associated proteins are conserved from yeast to humans. Sec13 is part of a nucleoporin subcomplex in both yeast and mammals (Siniossoglou et al., 1996; Fontoura et al., 1999; Vasu et al., 2001). A SUMO-deconjugating enzyme also associates with NPCs in both species (Schwienhorst et al., 2000; Hang and Dasso, 2002; Zhang et al., 2002). In vertebrates, the SUMO E2 conjugating enzyme, Ubc9, and the E3 ligase, Nup358, also associate with NPCs (Saitoh et al., 1997; Pichler et al., 2002) although it is unknown if E2 or E3 enzymes associate with the yeast NPC. However, it is interesting that at least some components of the SUMO modification/demodification pathway associate with NPCs in both higher and lower eukaryotes. This implies that SUMO modification of proteins may play a conserved role in nucleocytoplasmic transport.
Similar to the yeast NPC, our analysis revealed no motor proteins, ATPases or other evidence of mechanochemical activity. The most highly conserved features are FG repeat domains, present in ∼1/3 of the vertebrate nucleoporins. This implies that yeast and vertebrate NPCs function similarly and is consistent with a central role for the FG repeat domains. Like the yeast NPC, the vertebrate NPC has a surprisingly simple protein composition for a structure of its size and mass. Compared to the ribosome, which has a mass of ∼4 MDa and consists of ∼80 distinct proteins (Wool et al., 1995), the vertebrate NPC has a mass of at least 60 MDa and yet consists of only ∼30 distinct proteins. This surprisingly simple protein composition is a result of the high copy number of nucleoporins within the NPC (owing to the high degree of symmetry of the NPC) and also the high molecular mass of many nucleoporins.
A novel subfamily of nucleoporins
It is intriguing to note that, of the six novel nucleoporins identified in this study, four of these (Nup37, Nup43, Seh1, and ALADIN) are members of the WD repeat family of proteins. Together with RAE1 and Sec13, these proteins define a novel WD subfamily of NPC proteins. WD repeats are thought to mediate the assembly of large multiprotein complexes (Smith et al., 1999). In the NPC, WD repeats may mediate the assembly of subdomains of the NPC, or facilitate the interaction of transport complexes or other, novel multiprotein complexes with the NPC. It is likely that evolutionarily conserved WD nucleoporins, such as Seh1, Sec13, and RAE1, are involved in conserved NPC functions. However, Nup37, Nup43, and ALADIN have no identifiable yeast orthologues, suggesting that these nucleoporins have functions specific to the higher eukaryotic NPC.
Nup43 and Nup37 have no characterized orthologues in other organisms and no significant sequence homology outside the WD repeats. Nup37 was previously described as cofractionating with a nucleoporin complex containing Nup107, Nup96, and Sec13 (Fontoura et al., 1999), further supporting its classification as a nucleoporin. A complex homologous to the Nup107 complex has been described in yeast and also contains Sec13p and the Sec13p-homologue Seh1p (Siniossoglou et al., 1996). We do not yet know whether human Seh1 is also present in the Nup107 complex or whether Nup37 is a functional orthologue of Seh1p.
ALADIN also has no sequence homology outside its WD repeats and its role at the NPC is unknown. ALADIN's function may prove to be particularly interesting since mutations in this protein are associated with triple-A syndrome (AAAS) (Tullio-Pelet et al., 2000). Although AAAS displays some heterogeneity, it is characterized by three major symptoms: adrenocorticotropic hormone–resistant adrenal insufficiency, achalasia (failure to relax) of the esophageal sphincter muscle and alacrima (deficient tear production) (Orrell and Clark, 2002). Neurological defects are also usually present although it is unclear whether these are primary or secondary defects of AAAS. Several truncations, point mutations and splice site mutations have been identified in AAAS patients (Tullio-Pelet et al., 2000; Handschug et al., 2001; Sandrini et al., 2001; Schmittmann-Ohters et al., 2001; Goizet et al., 2002) and we are currently examining the effects of these mutations on the localization and function of ALADIN. Further characterization of ALADIN could provide valuable insights into both NPC function and the pathogenesis of AAAS.
Conclusions/perspectives
We have completed a proteomic study of the mammalian NPC. We have found that the mammalian NPC is composed of ∼30 distinct protein components and that many of these components are conserved from yeast to mammals. These findings indicate that the overall structure and function of the NPC is conserved. Nonetheless, there are also nucleoporins unique to both yeast and mammals, suggesting a degree of organism-specific specialization. We identified six novel mammalian nucleoporins and defined a new subfamily of nucleoporins characterized by the presence of WD repeats, one of which is linked to the human disease known as AAAS. A more detailed knowledge of the composition of the mammalian NPC paves the way to a better understanding of its assembly, interactions and functions.
Materials and methods
Preparation and chromatographic separation of an enriched nucleoporin fraction
Rat liver nuclei were prepared as previously described (Blobel and Potter, 1966) and stored at −80°C in 100-U (3 × 108 nuclei) aliquots. Nuclei were thawed and pelleted at 800 g for 1 min. Pelleted nuclei were resuspended with constant vortexing at a final concentration of 100 U/ml−1 by drop-wise addition of buffer A (0.1 mM MgCl2, 1 mM DTT, 0.5 mM PMSF, 1 μg/ml−1 leupeptin/pepstatin/aprotinin) supplemented with 5 μg/ml−1 DNase I and 5 μg/ml−1 RNase A. After resuspension, nuclei were immediately diluted to 20 U/ml−1 by addition of buffer B (buffer A + 10% sucrose, 20 mM triethanolamine, pH 8.5) with constant vortexing. After digestion at room temperature for 15 min, the suspension was underlayed with 4 ml ice-cold buffer C (buffer A + 30% sucrose, 20 mM triethanolamine, pH 7.5) and centrifuged at 3,500 g for 10 min in a swinging bucket rotor (Sorvall SH-3000). The pellet was resuspended in ice-cold buffer D (buffer A + 10% sucrose, 20 mM triethanolamine, pH 7.5) at a final concentration of 100 U/ml−1. The suspension was diluted to 67 U/ml−1 with buffer D + 0.3 mg/ml−1 heparin, and then immediately underlayed and pelleted as above. The heparin pellet was resuspended in ice-cold buffer D (100 U/ml−1), diluted to 67 U/ml−1 with buffer D + 3% Triton X-100, 0.075% SDS, and then pelleted as above. The resultant pellet (the NPC-lamina fraction) was resuspended in buffer D + 0.3% Empigen BB (final concentration of 100 U/ml−1). After incubation on ice for 10 min, the insoluble lamina was separated from soluble nucleoporins by centrifugation in a microfuge at 16,000 g for 15 min.
For reverse-phase separation, the Empigen BB supernatant was TCA-precipitated and resuspended in 70% A: 30% B (A: 60% formic acid; B: 60% formic acid, 33% acetonitrile) at ∼1,000 U/ml−1. After centrifugation to remove debris, the supernatant was loaded onto a C4 column (Perkin Elmer) equilibrated with 70% A: 30% B. SDS-PAGE analysis found no proteins in the flowthrough. Bound proteins were eluted with a 4-h gradient from 70% A: 30% B to 100% B, followed by 1.5 h at 100% B (flow rate of 0.25 ml/min−1). 45 × 1-ml fractions were collected every 4 min from 2 h (35% A: 65% B). Fractions and flowthrough were diluted in distilled water and precipitated with 20% TCA in the presence of 0.01% deoxycholate (final dilution factor of 1:5) on ice for 1 h. The pellet was washed twice with −20°C acetone, once overnight and once for 15 min. After air drying, the pellet was dissolved in SDS sample buffer and analyzed by 4–20% Tris-Glycine SDS-PAGE (Novex, Invitrogen). Because late-eluting proteins tended to elute inefficiently over a wide range, later fractions were pooled prior to SDS-PAGE.
EM
The DNase/RNase and heparin pellets were prepared for EM analysis as previously described (Pain et al., 1990). The Triton X-100 and Empigen BB pellets were resuspended in buffer D and sonicated briefly to produce fragments that could be viewed as single layers. After sonication, the samples were pelleted and washed once with distilled water before adhering to glow-discharged Formvar carbon-coated copper grids and staining with 2% uranyl acetate.
Mass spectrometric protein identification
Individual bands were excised from an SDS-PAGE gel (Novex; Invitrogen) and processed for analysis as described previously (Krutchinsky et al., 2000). Proteins were identified using a combination of peptide mapping (single-stage MS) and MS/MS. All mass spectra were obtained using in-house assembled matrix-assisted laser desorption/ionization (MALDI)–quadrupole-quadrupole time of flight (QqTOF) (Krutchinsky et al., 2000) and MALDI-ion trap (Krutchinsky et al., 2001) instruments.
Peptide mapping data obtained by MALDI-QqTOF-MS were compared with theoretical maps computed from protein sequences present in the NCBI nonredundant (NR) database, using the program ProFound (Zhang and Chait, 2000). Protein identities were confirmed using MS/MS of selected peptide ion peaks on the same instrument, using the program PepFrag (Fenyö et al., 1998). To identify proteins whose sequences were not present in the NR database, we used MS/MS data to search the NCBI expressed sequence tag database using PepFrag.
To maximize the number of identified proteins, we repeated the isolation and identification of proteins using a newly constructed MALDI-ion trap capable of high throughput MS/MS measurements (Krutchinsky et al., 2001). MALDI-ion trap-MS/MS data were analyzed using Mascot (Perkins et al., 1999) and Sonar (http://canada.proteometrics.com/PROWL/sonar.html) to search rat, mouse, and human NR, and expressed sequence tag databases. Large numbers of peptides from each protein band were analyzed by MS/MS to increase the probability of identifying rat proteins from mouse and human databases.
Molecular biology
A partial sequence of p30 was obtained by sequencing IMAGE EST 3936128. Comparison with genomic sequences suggests that the first nucleotide of this EST is the third nucleotide of a putative start methionine. The size of this predicted protein agrees with that determined experimentally by SDS-PAGE. However, 5′-RACE to determine the complete 5′ sequence was unsuccessful, probably owing to the high GC content. p37 was amplified directly from a fetal brain cDNA library (CLONTECH Laboratories, Inc.) using primers deduced from EST sequences. The 3′ sequence of p43 was obtained by sequencing IMAGE EST 3918958 and the 5′ sequence was deduced by 5′-RACE from a fetal brain cDNA library (Marathon-Ready; CLONTECH Laboratories, Inc.). To assess subcellular localization, full-length cDNAs (or, in the case of p30, all of the known cDNA sequence) were cloned into pEGFP-C1 (CLONTECH Laboratories, Inc.) in frame with an NH2-terminal EGFP tag and transfected into HeLa cells as described below.
Transfections
HeLa cells were maintained in DME supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, and 10 mM Hepes (Invitrogen). For transfection, cells were grown on coverslips to ∼50% confluency and transfected with 1 μg DNA using lipofectAMINE-PLUS (Invitrogen) according to the manufacturer's protocol. 48 h posttransfection, cells were fixed in 2% formaldehyde in PBS (30 min at room temperature) then permeabilized in acetone at −20°C for 2 min. NPCs were visualized with mAb414 followed by Alexa 594-conjugated anti–mouse (Molecular Probes). Confocal images were collected using the Ultraview confocal microscope and acquisition software (Perkin-Elmer) mounted on a Zeiss Axiovert and equipped with a Coolpix CCD camera.
Estimation of the relative abundance of nucleoporins
2, 5, 10, and 20 U of Empigen BB supernatant were separated by 4–20% Tris-Glycine SDS-PAGE (Novex; Invitrogen) and stained with zinc or Coomassie. Using NIH Image, we calculated the relative abundance of bands within each load/staining condition. For each band an average relative abundance was calculated using values from the different conditions. Certain values were excluded to ensure that loading and/or stain saturation did not distort the value. The relative abundance was then corrected for molecular weight.
Online supplemental material
Online supplemental materials are available at http:www.jcb.org/cgi/content/full/jcb.200206106/DC1. Proteins identified only in the C4-separated Empigen BB supernatant (Fig. 3) are shown in Table SI. Bands excised from the Empigen BB pellet are indicated in Fig. S1 and the proteins identified are shown in Table SII.
Supplemental Material
Acknowledgments
We apologize to authors who we could not cite directly owing to space limitations. Many thanks go to Kathy Wilson, Mike Rout, and all members of the Chait lab for extremely valuable discussion and advice. We also thank Martin Goldberg, Terry Allen, and Spencer Collis for their time and support, Helen Shio for assistance with electron microscopy, and Günter Blobel for his encouragement and support during the early stages of this project.
This work was funded by the American Cancer Society (RSG-01-064-01-CSM; M.J. Matunis) and the National Institutes of Health (RR00862 and CA89810; B.T. Chait).
The online version of this article contains supplemental material.
Footnotes
Abbreviations used in this paper: AAAS, triple-A syndrome; FG, phenylalanine-glycine; MALDI, matrix-assisted laser desorption/ionization; MS, mass spectrometry; NE, nuclear envelope; NPC, nuclear pore complex; NR, nonredundant; QqTOF, quadrupole-quadrupole time of flight.
References
- Akey, C.W., and M. Radermacher. 1993. Architecture of the Xenopus nuclear pore complex revealed by three-dimensional cryo-electron microscopy. J. Cell Biol. 122:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen, T.D., J.M. Cronshaw, S. Bagley, E. Kiseleva, and M.W. Goldberg. 2000. The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J. Cell Sci. 113:1651–1659. [DOI] [PubMed] [Google Scholar]
- Belgareh, N., G. Rabut, S.W. Bai, M. van Overbeek, J. Beaudouin, N. Daigle, O.V. Zatsepina, F. Pasteau, V. Labas, M. Fromont-Racine, et al. 2001. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. J. Cell Biol. 154:1147–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel, G., and V.R. Potter. 1966. Nuclei from rat liver: isolation method that combines purity with high yield. Science. 154:1662–1665. [DOI] [PubMed] [Google Scholar]
- Cai, S.T., F.L. Zhou, and J.Z. Zhang. 1997. Immunogold labeling electron microscopy showing vimentin filament anchored on nuclear pore complex. Shi Yan Sheng Wu Xue Bao. 30:193–199. [PubMed] [Google Scholar]
- Chial, H.J., M.P. Rout, T.H. Giddings, and M. Winey. 1998. Saccharomyces cerevisiae Ndc1p is a shared component of nuclear pore complexes and spindle pole bodies. J. Cell Biol. 143:1789–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, L.I., and G. Blobel. 1986. Identification and characterization of a nuclear pore complex protein. Cell. 45:699–709. [DOI] [PubMed] [Google Scholar]
- Davis, L.I., and G. Blobel. 1987. Nuclear pore complex contains a family of glycoproteins that includes p62: glycosylation through a previously unidentified cellular pathway. Proc. Natl. Acad. Sci. USA. 84:7552–7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer, N., and G. Blobel. 1976. A modified procedure for the isolation of a pore complex-lamina fraction from rat liver nuclei. J. Cell Biol. 70:581–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favreau, C., H.J. Worman, R.W. Wozniak, T. Frappier, and J.-C. Courvalin. 1996. Cell cycle-dependent phosphorylation of nucleoporins and nuclear pore membrane protein gp210. Biochemistry. 35:8035–8044. [DOI] [PubMed] [Google Scholar]
- Fenyö, D., J. Qin, and B.T. Chait. 1998. Protein identification using mass spectrometric information. Electrophoresis. 19:998–1005. [DOI] [PubMed] [Google Scholar]
- Finlay, D.R., D.D. Newmeyer, T.M. Price, and D.J. Forbes. 1987. Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J. Cell Biol. 104:189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floer, M., and G. Blobel. 1999. Putative reaction intermediates in Crm1-mediated nuclear protein export. J. Biol. Chem. 274:16279–16286. [DOI] [PubMed] [Google Scholar]
- Fontoura, B., G. Blobel, and M.J. Matunis. 1999. A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kD precursor generates Nup98 and the novel nucleoporin, Nup96. J. Cell Biol. 144:1097–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontoura, B.M., G. Blobel, and N.R. Yaseen. 2000. The nucleoporin Nup98 is a site for GDP/GTP exchange on ran and termination of karyopherin β2-mediated nuclear import. J. Biol. Chem. 275:31289–31296. [DOI] [PubMed] [Google Scholar]
- Galy, V., J.-C. Olivo-Marin, H. Scherthan, V. Doye, N. Rascalou, and U. Nehrbass. 2000. Nuclear pore complexes in the organization of silent telomeric chromatin. Nature. 403:108–112. [DOI] [PubMed] [Google Scholar]
- Goizet, C., B. Catargi, F. Tison, A. Tullio-Pelet, S. Hadj-Rabia, F. Pujol, A. Lagueny, S. Lyonnet, and D. Lacombe. 2002. Progressive bulbospinal amyotrophy in Triple A syndrome with AAAS gene mutation. Neurology. 58:962–965. [DOI] [PubMed] [Google Scholar]
- Graveley, B.R. 2001. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 17:100–107. [DOI] [PubMed] [Google Scholar]
- Greber, U.F., A. Senior, and L. Gerace. 1990. A major glycoprotein of the nuclear pore complex is a membrane-spanning polypeptide with a large lumenal domain and a small cytoplasmic tail. EMBO J. 9:1495–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, T., S. Müller, G. Klier, N. Panté, J.M. Blevitt, M. Haner, B. Paschal, U. Aebi, and L. Gerace. 1995. Structural analysis of the p62 complex, an assembly of O-linked glycoproteins that localizes near the central gated channel of the nuclear pore complex. Mol. Biol. Cell. 6:1591–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallberg, E., R.W. Wozniak, and G. Blobel. 1993. An integral membrane protein of the pore membrane domain of the nuclear envelope contains a nucleoporin-like region. J. Cell Biol. 122:513–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handschug, K., S. Sperling, S.J. Yoon, S. Hennig, A.J. Clark, and A. Huebner. 2001. Triple A syndrome is caused by mutations in AAAS, a new WD-repeat protein gene. Hum. Mol. Genet. 10:283–290. [DOI] [PubMed] [Google Scholar]
- Hang, J., and M. Dasso. 2002. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J. Biol. Chem. 277:19961–19966 [DOI] [PubMed] [Google Scholar]
- Hofmann, W., B. Reichart, A. Ewald, E. Muller, I. Schmitt, R.H. Stauber, F. Lottspeich, B.M. Jockusch, U. Scheer, J. Hauber, and M.C. Dabauvalle. 2001. Cofactor requirements for nuclear export of Rev response element (RRE)- and constitutive transport element (CTE)-containing retroviral RNAs. An unexpected role for actin. J. Cell Biol. 152:895–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, T., and L. Gerace. 1998. cDNA cloning and analysis of the expression of nucleoporin p45. Gene. 221:245–253. [DOI] [PubMed] [Google Scholar]
- Iwasaki, T., W.W. Chin, and L. Ko. 2001. Identification and characterization of RRM-containing coactivator activator (CoAA) as TRBP-interacting protein, and its splice variant as a coactivator modulator (CoAM). J. Biol. Chem. 276:33375–33383. [DOI] [PubMed] [Google Scholar]
- Ishii, K., G. Arib, C. Lin, G. Van Houwe, and U.K. Laemmli. 2002. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 109:551–562. [DOI] [PubMed] [Google Scholar]
- Jeffries, S., and A.J. Capobianco. 2000. Neoplastic transformation by Notch requires nuclear localization. Mol. Cell. Biol. 20:3928–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer, D., R.W. Wozniak, G. Blobel, and A. Radu. 1994. The human CAN protein, a putative oncogene product associated with myeloid leukemogenesis, is a nuclear pore complex protein that faces the cytoplasm. Proc. Natl. Acad. Sci. USA. 91:1519–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutchinsky, A.N., W. Zhang, and B.T. Chait. 2000. Rapidly switchable matrix-assisted laser desorption/ionization and electrospray quadrupole-time-of-flight mass spectrometry for protein identification. J. Am. Soc. Mass Spectrom. 11:493–504. [DOI] [PubMed] [Google Scholar]
- Krutchinsky, A.N., M. Kalkum, and B.T. Chait. 2001. Automatic identification of proteins with a MALDI-quadrupole ion trap mass spectrometer. Anal. Chem. 73:5066–5077. [DOI] [PubMed] [Google Scholar]
- Lain, S., D. Xirodimas, and D.P. Lane. 1999. Accumulating active p53 in the nucleus by inhibition of nuclear export: a novel strategy to promote the p53 tumor suppressor function. Exp. Cell Res. 253:315–324. [DOI] [PubMed] [Google Scholar]
- Le Douarin, B., A.L. Nielsen, J.M. Garnier, H. Ichinose, F. Jeanmougin, R. Losson, and P. Chambon. 1996. A possible involvement of TIF1α and TIF1β in the epigenetic control of transcription by nuclear receptors. EMBO J. 15:6701–6715. [PMC free article] [PubMed] [Google Scholar]
- Macara, I.G. 2001. Transport into and out of the nucleus. Microbiol. Mol. Biol. Rev. 65:570–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay, C., E. Meier, and D.J. Forbes. 1995. Differential mitotic phosphorylation of proteins of the nuclear pore complex. J. Biol. Chem. 270:254–262. [DOI] [PubMed] [Google Scholar]
- Mahajan, R., C. Delphin, T. Guan, L. Gerace, and F. Melchior. 1997. A small ubiquitin-related polypeptide involved in targeting RanGAP1 to nuclear pore complex protein RanBP2. Cell. 88:97–107. [DOI] [PubMed] [Google Scholar]
- Marelli, M., J.D. Aitchison, and R.W. Wozniak. 1998. Specific binding of the karyopherin Kap121p to a subunit of the nuclear pore complex contianing Nup53p, Nup59p, and Nup170p. J. Cell Biol. 143:1813–1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marelli, M., C.P. Lusk, H. Chan, J.D. Aitchison, and R.W. Wozniak. 2001. A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. J. Cell Biol. 153:709–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattaj, I.W., and L. Englmeier. 1998. Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem. 67:265–306. [DOI] [PubMed] [Google Scholar]
- Matunis, M.J., E. Coutavas, and G. Blobel. 1996. A novel ubiquitin-like modification modulates the partitioning of the Ran-GTPase-activating protein RanGAP1 between the cytosol and the nuclear pore complex. J. Cell Biol. 135:1457–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middendorp, S., A. Paoletti, E. Schiebel, and M. Bornens. 1997. Identification of a new mammalian centrin gene, more closely related to Saccharomyces cerevisiae CDC31 gene. Proc. Natl. Acad. Sci. USA. 94:9141–9146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, M.W., M.R. Caracciolo, W.K. Berlin, and J.A. Hanover. 1999. Phosphorylation and glycosylation of nucleoporins. Arch. Biochem. Biophys. 367:51–60. [DOI] [PubMed] [Google Scholar]
- Nakielny, S., S. Shaikh, B. Burke, and G. Dreyfuss. 1999. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J. 18:1982–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orrell, R.W., and A.J. Clark. 2002. ALADIN, but where's the Genie? Neurology. 58:847–848. [DOI] [PubMed] [Google Scholar]
- Pain, D., H. Murakami, and G. Blobel. 1990. Identification of a receptor for protein import into mitochondria. Nature. 347:444–449. [DOI] [PubMed] [Google Scholar]
- Perkins, D.N., D.J. Pappin, D.M. Creasy, and J.S. Cottrell. 1999. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 20:3551–3567. [DOI] [PubMed] [Google Scholar]
- Pichler, A., A. Gast, J.S. Seeler, A. Dejean, and F. Melchior. 2002. The nucleoporin RanBP2 has SUMO1 E3 ligase activity. Cell. 108:109–120. [DOI] [PubMed] [Google Scholar]
- Powers, M.A., C. Macaulay, F.R. Masiarz, and D.J. Forbes. 1995. Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J. Cell Biol. 128:721–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, R., and E. Hurt. 2002. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell. 108:523–531. [DOI] [PubMed] [Google Scholar]
- Reichelt, R., A. Holzenburg, E.L. Buhle, Jr., M. Jarnik, A. Engel, and U. Aebi. 1990. Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J. Cell Biol. 110:883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck, K., and D. Görlich. 2001. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 20:1320–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout, M.P., and G. Blobel. 1993. Isolation of the yeast nuclear pore complex. J. Cell Biol. 123:771–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout, M.P., J.D. Aitchison, A. Suprapto, K. Hjertaas, Y. Zhao, and B.T. Chait. 2000. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J. Cell Biol. 148:635–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan, K.J., and S.R. Wente. 2000. The nuclear pore complex: a protein machine bridging the nucleus and cytoplasm. Curr. Opin. Cell Biol. 12:361–371. [DOI] [PubMed] [Google Scholar]
- Saitoh, H., R. Pu, M. Cavenagh, and M. Dasso. 1997. RanBP2 associates with Ubc9p and a modified form of RanGAP1. Proc. Natl. Acad. Sci. USA. 94:3736–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandrini, F., C. Farmakidis, L.S. Kirschner, S.M. Wu, A. Tullio-Pelet, S. Lyonnet, D.L. Metzger, C.J. Bourdony, D. Tiosano, W.Y. Chan, and C.A. Stratakis. 2001. Spectrum of mutations of the AAAS gene in Allgrove syndrome: lack of mutations in six kindreds with isolated resistance to corticotropin. J. Clin. Endocrinol. Metab. 86:5433–5437. [DOI] [PubMed] [Google Scholar]
- Schmittmann-Ohters, K., A. Huebner, A. Richter-Unruh, and B.P. Hauffa. 2001. Clinical and novel molecular findings in a 6.8-year-old Turkish boy with triple A syndrome. Horm. Res. 56:67–72. [DOI] [PubMed] [Google Scholar]
- Schwienhorst, I., E.S. Johnson, and R.J. Dohmen. 2000. SUMO conjugation and deconjugation. Mol. Gen. Genet. 263:771–786. [DOI] [PubMed] [Google Scholar]
- Shaywitz, D.A., L. Orci, M. Ravazzola, A. Swaroop, and C.A. Kaiser. 1995. Human SEC13Rp functions in yeast and is located on transport vesicles budding from the endoplasmic reticulum. J. Cell Biol. 128:769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shogren, R., T.A. Gerken, and N. Jentoft. 1989. Role of glycosylation on the conformation and chain dimensions of O-linked glycoproteins: light-scattering studies of ovine submaxillary mucin. Biochemistry. 28:5525–5536. [DOI] [PubMed] [Google Scholar]
- Siniossoglou, S., C. Wimmer, M. Rieger, V. Doye, H. Tekotte, C. Weise, S. Emig, A. Segref, and E.C. Hurt. 1996. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 84:265–275. [DOI] [PubMed] [Google Scholar]
- Smith, S., and T. de Lange. 1999. Cell cycle dependent localization of the telomeric PARP, tankyrase, to nuclear pore complexes and centrosomes. J. Cell Sci. 112:3649–3656. [DOI] [PubMed] [Google Scholar]
- Smith, T.F., C. Gaitatzes, K. Saxena, and E.J. Neer. 1999. The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24:181–185. [DOI] [PubMed] [Google Scholar]
- Smythe, C., H.E. Jenkins, and C.J. Hutchison. 2000. Incorporation of the nuclear pore basket protein Nup153 into nuclear pore structures is dependent upon lamina assembly: evidence from cell-free extracts of Xenopus eggs. EMBO J. 19:3918–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa, C.G., and D.O. Morgan. 2000. Control of mitosis by changes in the subcellular location of cyclin-B1-Cdk1 and Cdc25C. Curr. Opin. Cell Biol. 12:658–665. [DOI] [PubMed] [Google Scholar]
- Tullio-Pelet, A., R. Salomon, S. Hadj-Rabia, C. Mugnier, M.H. de Laet, B. Chaouachi, F. Bakiri, P. Brottier, L. Cattolico, C. Penet, et al. 2000. Mutant WD-repeat protein in triple-A syndrome. Nat. Genet. 26:332–335. [DOI] [PubMed] [Google Scholar]
- Vasu, S., S. Shah, A. Orjalo, M. Park, W.H. Fischer, and D.J. Forbes. 2001. Novel vertebrate nucleoporins Nup133 and Nup160 play a role in mRNA export. J. Cell Biol. 155:339–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasu, S.K., and D.J. Forbes. 2001. Nuclear pores and nuclear assembly. Curr. Opin. Cell Biol. 13:363–375. [DOI] [PubMed] [Google Scholar]
- Watkins, J.L., R. Murphy, J.L. Emtage, and S.R. Wente. 1998. The human homologue of Saccharomyces cerevisiae Gle1p is required for poly(A)+ RNA export. Proc. Natl. Acad. Sci. USA. 95:6779–6784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilken, N., J.-L. Senécal, U. Scheer, and M.-C. Dabauvalle. 1995. Localization of the Ran-GTP binding protein RanBP2 at the cytoplasmic side of the nuclear pore complex. Eur. J. Cell Biol. 68:211–219. [PubMed] [Google Scholar]
- Wool, I.G., Y.L. Chan, and A. Gluck. 1995. Structure and evolution of mammalian ribosomal proteins. Biochem. Cell Biol. 73:933–947. [DOI] [PubMed] [Google Scholar]
- Wu, J., M.J. Matunis, D. Kraemer, G. Blobel, and E. Coutavas. 1995. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J. Biol. Chem. 270:14209–14213. [DOI] [PubMed] [Google Scholar]
- Yang, Q., M.P. Rout, and C.W. Akey. 1998. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol. Cell. 1:223–234. [DOI] [PubMed] [Google Scholar]
- Yaseen, N.R., and G. Blobel. 1999. GTP hydrolysis links initiation and termination of nuclear import on the nucleoporin Nup358. J. Biol. Chem. 274:26493–26502. [DOI] [PubMed] [Google Scholar]
- Yokoyama, N., N. Hayashi, T. Seki, N. Panté, T. Ohba, K. Nishii, K. Kuma, T. Hayashida, T. Miyata, U. Aebi, et al. 1995. A giant nucleopore protein that binds Ran/TC4. Nature. 376:184–188. [DOI] [PubMed] [Google Scholar]
- Zhang, H., H. Saitoh, and M.J. Matunis. 2002. Enzymes of the SUMO modification pathway localize to the filaments of the nuclear pore complex. Mol. Cell. Biol. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, W., and B.T. Chait. 2000. ProFound: an expert system for protein identification using mass spectrometric peptide mapping information. Anal. Chem. 72:2482–2489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.