Abstract
Novel mutations in the RSW1 and KNOPF genes were identified in a large-scale screen for mutations that affect cell expansion in early Arabidopsis embryos. Embryos from both types of mutants were radially swollen with greatly reduced levels of crystalline cellulose, the principal structural component of the cell wall. Because RSW1 was previously shown to encode a catalytic subunit of cellulose synthase, the similar morphology of knf and rsw1-2 embryos suggests that the radially swollen phenotype of knf mutants is largely due to their cellulose deficiency. Map-based cloning of the KNF gene and enzyme assays of knf embryos demonstrated that KNF encodes α-glucosidase I, the enzyme that catalyzes the first step in N-linked glycan processing. The strongly reduced cellulose content of knf mutants indicates that N-linked glycans are required for cellulose biosynthesis. Because cellulose synthase catalytic subunits do not appear to be N glycosylated, the N-glycan requirement apparently resides in other component(s) of the cellulose synthase machinery. Remarkably, cellular processes other than extracellular matrix biosynthesis and the formation of protein storage vacuoles appear unaffected in knf embryos. Thus in Arabidopsis cells, like yeast, N-glycan trimming is apparently required for the function of only a small subset of N-glycoproteins.
Keywords: cellulose; cell elongation; glycosylation; Arabidopsis; embryo
Introduction
Plant cell elongation is mediated by vacuole-generated turgor pressure, and directed by anisotropic (unequal) yielding of the extracellular matrix, or cell wall (Ray et al., 1972). The cell wall is a complex assembly of carbohydrate polymers and proteins of which cellulose microfibrils, hydrogen-bonded chains of β-1,4–linked glucose, are the major load-bearing component (Carpita and Gibeaut, 1993). In an elegant early demonstration of the importance of cellulose microfibrils in the control of cell elongation, Green (1962) showed that treatment of Nitella cells with the microtubule (MT)* inhibitor colchicine caused randomized orientation of cellulose microfibrils and concomitant isodiametric swelling of the cell. This and subsequent experiments have lent support to the cellulose microfibril “hoop reinforcement” model: inelastic cellulose microfibrils wrapped in a helicoidal pattern around the cell constrain cell expansion unless they are specifically released by a poorly understood active loosening process (Cosgrove, 1999). Recent characterization of mutants that affect the synthesis or organization of cellulose has provided genetic evidence for this model (Arioli et al., 1998; Fagard et al., 2000; Lane et al., 2001).
Cellulose is the principal load-bearing component of plant cell walls but relatively little is known about the proteins involved in cellulose synthesis (Delmer, 1999). The plasma membrane enzyme, cellulose synthase, has been recalcitrant to purification and biochemical approaches have not as yet identified any components of the enzyme complex. A family of genes encoding the putative catalytic subunit of higher plant cellulose synthase (CESA) was identified by genomic methods on the basis of weak homology to bacterial cellulose synthases (Pear et al., 1996). The molecular characterization of mutants with defects in cell wall biogenesis confirmed the participation of the CESA proteins and has also led to the identification of KORRIGAN (KOR), a putative endo-1,4-β-glucanase that is also required for the synthesis of cellulose microfibrils (Nicol et al., 1998; Lane et al., 2001). Both a cellulose synthase catalytic subunit and an endo-1,4-β-glucanase have previously been shown to be required for cellulose synthesis in bacteria (Matthysse et al., 1995). The endo-1,4-β-glucanase has been proposed to be required for either cellulose chain editing or termination, or for cleavage of cellobiose oligosaccharides from a hypothetical lipid-linked precursor (for review see Delmer, 1999).
Some evidence for the importance of N glycosylation in cellulose biosynthesis has come from recent analysis of the embryo-defective mutant cyt1, which is radially swollen beginning early in embryogenesis and has abnormal cell wall structure (Nickle and Meinke, 1998). Lukowitz et al. (2001) showed that the CYT1 gene encodes mannose-1-phosphate guanylyltransferase, an enzyme required for production of GDP-d-mannose, GDP-l-fucose, ascorbic acid, glycosylphosphatidylinositol membrane anchors, and the core N-glycan. cyt1 embryos had strongly reduced levels of cellulose compared with wild type (wt), and growth of Arabidopsis seedlings on sublethal concentrations of the N glycosylation inhibitor, tunicamycin, caused radial swelling of roots similar to that observed in rsw1 (Arioli et al., 1998). Lukowitz et al. (2001) proposed that the cellulose deficiency of cyt1 embryos might be due to the defect in N glycosylation, and they noted that CESA proteins contain multiple putative N-glycan attachment motifs that could be required for protein folding or function (Hammond and Helenius, 1995; Campbell and Braam, 1999).
To identify cell elongation mutants that might result in embryo lethality, we conducted a large-scale genetic screen for mutations that affect the morphology of early Arabidopsis embryos. There was a new, nonconditional, mutant allele of the RSW1 gene (rsw1-2) among the mutations identified, as well as novel alleles of a locus called KNOPF (KNF), for which mutations were previously recovered in a large-scale screen for seedlings with body pattern defects (Mayer et al., 1991). Although not characterized in detail, knopf mutants were proposed to affect the shape, but not the pattern, of the embryo and seedling (Mayer et al., 1991). Here, we present a detailed analysis of the rsw1-2 and knf embryo phenotypes and the KNF gene product. Both rsw1-2 and knf embryos are radially swollen and have strongly reduced cellulose content. We demonstrate that KNF encodes α-glucosidase I, an enzyme that trims the terminal glucose of N-linked glycans. We present evidence that lack of N-glycan trimming does not affect the stability of cellulose synthase catalytic subunits, which contain the conserved N glycosylation motif but are apparently not N glycosylated. Our work indicates that a component or substrate of the cellulose synthase machinery is N glycosylated, and proper glucose trimming of this glycoprotein is required for its folding or function.
Results
A genetic screen for mutations affecting Arabidopsis embryo morphogenesis
Numerous mutants that affect the morphology of the Arabidopsis seedling have been obtained, and in many cases the defect in development can be traced back to an early stage of embryogenesis (Mayer et al., 1991). In addition, there exists a large collection of mutations that result in embryo lethality (Meinke, 1994). However, no systematic genetic screen for mutations that affect embryo morphology specifically during embryogenesis has previously been conducted. To obtain potentially lethal mutations that affect cell expansion we conducted a genetic screen for mutants with altered morphology at the heart to torpedo stage of development.
Our screen was performed by scoring for homozygous mutant embryos segregating in the siliques of mother plants heterozygous for the corresponding embryo mutation. Fruits of ∼12,000 ethylmethane sulfonate–mutagenized M2 plants of the Landsberg erecta ecotype were opened and inspected for ovules containing mutant embryos. Approximately 6,000 M2 plants were found to be segregating at least one embryo-defective mutation. Cleared embryos from these plants were then examined by light microscopy using Nomarski optics. An additional genetic screen was performed by directly examining dry seed from 5,000 individual fast neutron– or X-ray–mutagenized M2 families. In total, ∼40 mutant lines that affect cell elongation in the embryo were obtained (unpublished data).
A strong mutant allele of RSW1
The rsw1-2 mutant (line 12-30) was identified based on its radially swollen phenotype during embryogenesis. The embryo-defective mutation in line 12-30 was mapped to a small interval that also contained RSW1 (unpublished data). A complementation cross was then performed between a homozygous rsw1-1 plant and a plant heterozygous for the mutation in 12-30. 48% (n = 123) of the resulting F1 seedlings displayed the rsw1-1 phenotype of a swollen root tip at the restrictive temperature of 31°C. This segregation matched the 1:1 ratio expected for allelic mutations; thus, the mutation in line 12-30 was designated rsw1-2. The rsw1-2 allele displayed normal penetrance and transmission, as selfed rsw1-2/RSW1-2 plants segregated 23% mutant embryos (n = 1,067). Genomic DNA was prepared from homozygous rsw1-2 seedlings, and the RSW1 gene was sequenced. The RSW1 coding region of rsw1-2 had a single guanine to adenine nucleotide change at bp 1891 of the RSW1 coding sequence (unpublished data). This nucleotide change resulted in a glycine to serine change at amino acid residue 631 of RSW1, a residue that is conserved in all predicted CESA proteins of Arabidopsis and is near the proposed CESA catalytic site (Delmer, 1999).
Wt, rsw1-1, and rsw1-2 seedlings are shown in Fig. 1. Radial swelling of the rsw1-1 root in plants grown at the nonpermissive temperature occurs primarily in the epidermal and cortical layers of the root, and many long root hairs are visible (Fig. 1 D). In contrast, rsw1-2 displays a much more severe phenotype, which is apparent in all tissues of the seedling at all growth temperatures (Fig. 1, E and F). Cotyledons are turgid, elongated petioles are not apparent, and the hypocotyl and root of rsw1-2 seedlings are a swollen mass (Fig. 1 F). Based on the seedling phenotype, rsw1-2 is a stronger mutant allele than rsw1-1.
Figure 1.

Wt, rsw1-2, and knf seedlings. Wt (A and B), rsw1-1 (C and D), rsw1-2 (E and F), and knf (G and H) seedlings after 5 d growth on MS 1% sucrose plates. Bars: (A, C, E, and G) 0.5 mm; (B, D, F, and H) 100 μm.
knf mutants
Our genetic screens yielded 10 lines with allelic mutations that caused a radial swelling phenotype during embryogenesis similar to that of rsw1-2. Because of the similarity of the embryo phenotype of these allelic lines to knf embryos (Mayer et al., 1991), plants heterozygous for our reference line 9-199 were crossed to plants heterozygous for knf allele U18-7. 10% (n = 279) of the F1 embryos from this cross displayed the knf phenotype, indicating that the mutation in line 9-199 was allelic to knf. Therefore, we designated the mutations in the 10 lines knf-11 to knf-20. knf mutants segregate 11.9% (n = 1,023) in the siliques of heterozygous plants. This segregation is less than half of the 25% expected for a recessive mutation with normal transmission and penetrance. Reciprocal crosses demonstrated that this segregation distortion is due to reduced transmission of knf through pollen, whereas knf eggs have normal fitness (unpublished data). Because all of our alleles showed an indistinguishable phenotype during embryogenesis, our reference allele knf-14 (line 9-199) was used for all analyses. In sterile culture conditions, knf seeds swell slightly, but seedlings are not able to elongate or grow to any extent. A knf seedling that has been dissected out of the seed coat after 5 d on agar-solidified mineral medium is shown in Fig. 1, G and H.
RSW1 and KNF are required for the orientation of cell elongation during embryogenesis
Wt, rsw1-2, and knf embryos are shown at the mid to late heart stage of development in Fig. 2, A, E, and I, respectively. At this stage, the radial pattern of the embryonic hypocotyl and root is well established: the hypocotyl has one epidermal layer (Fig. 2 A, e), two layers of ground tissue (Fig. 2 A, g), and several layers of vascular primordia (Fig. 2 A, v) (Jürgens and Mayer, 1994). The cells of the columella that later form the root meristem and root cap are arranged in two tiers, characteristic of the late heart stage (Fig. 2 A, col). A late heart stage rsw1-2 embryo (Fig. 2 E) is slightly radially swollen compared with wt, as cells of the epidermis and ground tissue (e and g, respectively) are wider than those of its wt counterpart. Despite this change in the shape of cells, the organization of cell layers of the hypocotyl and columella is identical to wt (compare Fig. 2 A with Fig. 1 E). In the mid heart stage knf embryo (Fig. 2 I), the epidermis and two ground layers are present (e and g, respectively), but the cells that make up these layers are wider than those of rsw1-2 and much wider than wt. The columella cells (Fig. 2, A, E, and I, col, bracket) of this knf mid heart stage embryo show some irregularly arranged divisions (Fig. 2 I, arrowhead). This trend of normal differentiation of cell layers, but abnormal width of the cells within these layers, continues throughout the rest of embryo development. At the torpedo stage, rsw1-2 embryos (Fig. 2 F) have a subtle defect in shape, as the rsw1-2 hypocotyl is wider than that of wt (Fig. 2 B). Torpedo stage knf embryos (Fig. 2 J) are larger than at the heart stage, but in overall shape resemble a swollen heart stage embryo more than a torpedo stage embryo.
Figure 2.
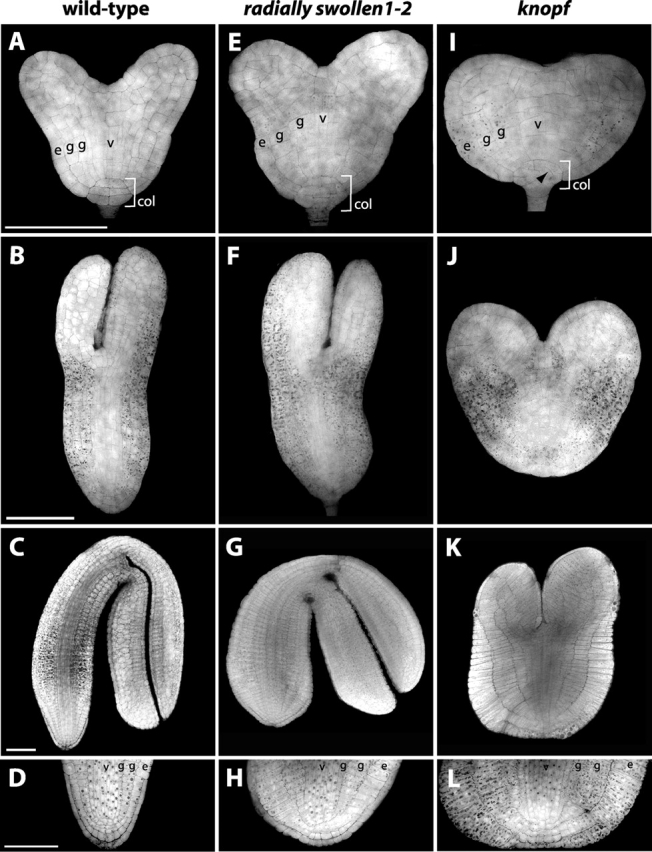
Mutations in RSW1 and KNF affect cell shape and embryo morphogenesis. Whole mount confocal images of wt, rsw1-2, and knf embryos stained with Alexafluor 488 Hydrazide are shown at the following developmental stages: heart (A, E, and I), torpedo (B, F, and J), bent cotyledon (C, G, and K), and root tip of bent cotyledon (D, H, and L). For each developmental stage, embryos are shown at the same magnification. Bars: (A) 10 μm; (B) 50 μm; (C) 50 μm; (D) 10 μm. e, epidermis; c, columella; g, ground tissue; v, vascular tissue.
The radial swelling of rsw1-2 and knf embryos becomes most extreme at the late bent cotyledon stage of embryogenesis, as shown in Fig. 2, C, G, and K. The hypocotyl and cotyledons of the wt embryo (Fig. 2 C) have elongated and folded over so that the tips of the cotyledons are next to the root meristem. The cotyledons of the typical rsw1-2 embryo (Fig. 2 G) have also elongated enough to bend over to the length of the hypocotyl, but are radially swollen compared with the wt embryo. Individual cells have elongated in a lateral direction more than the corresponding cells of wt. Cell numbers as well as cell layers remain essentially the same in rsw1-2 as in wt. By contrast, the overall morphology of the late bent cotyledon stage knf embryo (Fig. 2 K) resembles an extremely swollen heart-shaped embryo. This is due to the altered direction of cell elongation in cells of the embryo. An inspection of cell numbers in the epidermal and ground layers of the hypocotyl reveals that the knf embryo has the same number of cells in these tissues (∼30) as in wt and rsw1-2. However, the cells of the epidermis and outer ground layer especially have become extremely elongated in the radial direction. The epidermis is most dramatically affected, as cells in this layer are close to 50 μm wide (compare the 50 μm scale bar in Fig. 2 C with the knf epidermal cells in Fig. 2 K). Even the inner ground cell layer is approximately twice the width of the corresponding wt cells (Fig. 2, compare K with C). Cells of the cotyledons are also affected and are much more laterally elongated compared with wt.
Despite the extreme defect in cell and embryo shape in rsw1-2 and especially knf, the oriented divisions that are required to generate the different tissue layers of the embryo usually occur, as both rsw1-2 and knf embryos have clearly recognizable shoot and root meristems as well as epidermal, ground, and vascular tissues. The root meristems of wt, rsw1-2, and knf embryos are shown in Fig. 2, D, H, and L. All pattern elements of the wt root meristem are visible in both rsw1-2 and knf embryos, although their placement may differ slightly in knf due to the extreme radial elongation of the outer cell layers (compare wt, Fig. 2 D, with rsw1-2, H, and knf, L). Thus, although mutations in RSW1 and KNF affect cell elongation and, in turn, morphogenesis of the Arabidopsis embryo, they have little or no effect on the orientation of cell division or the formation of the different tissue types of the embryo.
rsw1-2 and knf embryos have thin epidermal cell walls
Because of the similarity of the rsw1-2 and knf phenotypes, it seemed likely that a cell wall defect was responsible for the altered cell elongation in knf embryos. However, to exclude a substantial role for MTs in the knf phenotype, we examined the arrangement of cortical MTs in epidermal cells of wt and knf embryos using a β-tubulin–GFP fusion (Fig. 3, G and H). Although less intense GFP fluorescence was observed in knf embryos compared with wt, the cortical MTs of knf embryos were primarily oriented in parallel arrays within single cells (Fig. 3 H) similar to wt (Fig. 3 G).
Figure 3.
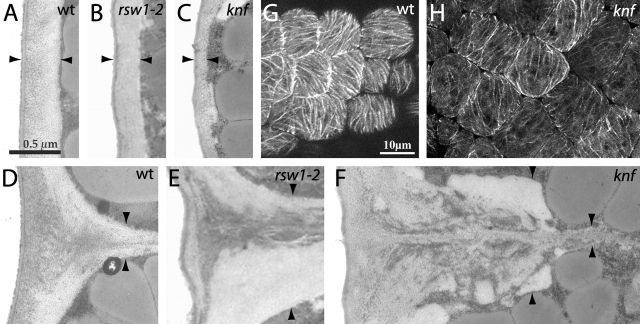
Epidermal cell walls and cortical MTs of wt, rsw1-2, and knf embryos. Transmission electron micrographs of epidermal cell walls at the midpoint of the hypocotyl of wt (A and D), rsw1-2 (B and E), and knf (C and F) bent cotyledon stage embryos, all shown at equal magnification. Cortical MTs of wt (G) and knf (H) bent cotyledon stage hypocotyl epidermal cells visualized with β-tubulin–GFP translational fusion, shown at equal magnification. Arrowheads are pointing to the cell wall boundary. Bars: (A) 0.5 μm; (G) 10 μm.
Transmission electron micrographs of typical hypocotyl epidermal walls of wt, rsw1-2, and knf embryos at the late bent cotyledon stage of embryogenesis are shown in Fig. 3, A–F. Compared with wt (Fig. 3 A), rsw1-2 walls are decreased in thickness (Fig. 3 B), and knf epidermal walls are even thinner (Fig. 3 C). The average thickness of walls of individual epidermal cells is shown in Table I. Whereas wt epidermal walls had an average thickness of 438 nm, those of rsw1-2 had an average thickness of 307 nm, and knf walls showed an average thickness of 129 nm. The decreasing thickness of epidermal walls in rsw1-2 and knf embryos correlates with the increased radial swelling phenotype of rsw1-2 and knf embryos.
Table I. Thickness of hypocotyl epidermal cell wall.
| Strain | Width in nma |
|---|---|
| wt | 438 (96) n = 16 |
| rsw1-2 | 307 (55) n = 25 |
| knf | 129 (79) n = 18 |
Standard deviation listed in parentheses.
Wall thickness measured for individual cells at midpoint of hypocotyl.
In addition to their effect on the thickness of epidermal walls, the rsw1-2 and knf mutations also caused alterations in cell wall ultrastructure. The junction of two epidermal cell walls in wt, rsw1-2, and knf is shown in Fig. 3, D–F. The wt wall junction in Fig. 3 D shows a smooth shape with an even organization of electron-dense staining, characteristic of the pectin-rich middle lamella. The thickness of the cell junction is increased in rsw1-2 (Fig. 3 E), and the area of electron-dense staining of the middle lamella is increased as well as disorganized compared with wt. A two-cell junction from a knf epidermis (Fig. 3 F) shows a lack of normal wall morphology, as the outer part of the junction is disorganized and lumpy, and the thickness of the wall changes abruptly. Electron-dense staining is heterogeneous and diffuse, not focused in the center of the wall as in wt. Cellular organelles such as chloroplasts, mitochondria, and nuclei appear unchanged by the knf mutation, with the exception that protein storage bodies are fewer in number, have an abnormal crystalline structure, and form later in development than in wt (unpublished data).
rsw1-2 and knf cell walls are deficient in cellulose
To more specifically characterize the cell wall defect in rsw1-2 and knf embryos, cell wall components were quantified by chemical analysis. The results of these analyses are summarized in Table II. The most dramatic change observed was the large decrease in crystalline cellulose in rsw1-2 and knf embryos. Crystalline cellulose in wt embryos was measured at 200 nmol glucose/mg embryo dry wt, whereas rsw1-2 embryos contained 45.4 nmol/mg (22% of wt) and knf embryos contained 27.5 nmol/mg (13% of wt). More modest changes in total uronic acid, the principle component of pectins, were also observed. Wt embryos contained 64.8 nmol/mg dry wt, rsw1-2 embryos contained 74.6 nmol/mg (115% of wt), and knf embryos showed 89.4 nmol/mg (138% of wt).
Table II. Cell wall composition of wt, knf, and rsw1-2 embryosa .
| wt | knf | rsw1-2 | |
|---|---|---|---|
| Cell wall polymer | |||
| Celluloseb | 200 (5) | 27.5 (5) | 45.4 (2) |
| Pectinc | 64.8 (6) | 89.4 (8) | 74.6 (6) |
| Neutral sugars | |||
| Rhamnose | 26.5 (2) | 24.5 (6) | ND |
| Fucose | 12.8 (1) | 7.6 (1) | ND |
| Arabinose | 356.4 (13) | 373.4 (21) | ND |
| Xylose | 106.4 (10) | 94.5 (5) | ND |
| Mannose | 23.1 (1) | 26.7 (4) | ND |
| Galactose | 85.3 (6) | 80.9 (3) | ND |
| Average dry weight per embryo | 5.5 μg | 2.5 μg | 5.0 μg |
Values expressed as nmol sugar/mg dry weight of embryo. Value is the average of at least three measurements, with standard deviation in parentheses.
Cellulose measured as acetic/nitric acid–insoluble glucose.
Pectin measured as total uronic acid after trifluoroacetic acid hydrolysis.
To determine if the knf mutation affected the hemicellulosic polymers of the cell wall, neutral sugars of wt and knf embryos were measured as alditol acetates (Table II). With the exception of fucose, no significant changes were observed. Fucose in wt embryos was measured at 12.8 nmol/mg dry wt, whereas levels of fucose in knf decreased to 7.6 nmol/mg (60% of wt). Neutral sugars in rsw1-2 were not quantified, as Peng et al. (2000) have previously shown that the rsw1-1 mutation does not affect levels of neutral sugars.
Map-based cloning of KNF
The KNF gene was located on the lower arm of chromosome I by bulk segregant analysis of wt F2 progeny from a selfed knf/KNF (Landsberg erecta ecotype/Columbia ecotype) F1 plant, and subsequently mapped between the SSLP markers K09 and nga111 (Lukowitz et al., 2000). In total, 98 F2 plants with recombinations in the K09–nga111 interval were obtained, and the interval containing KNF was narrowed down using additional SSLP markers. KNF was mapped to a final genetic interval of eight recombination breakpoints, which corresponded to a 160-kb region containing 29 predicted genes (Fig. 4 A). Out of 960 recombinant chromosomes, only one recombination between marker K12 and KNF was recovered, indicating very close linkage between the two. Subsequently, Southern blot analysis demonstrated that gene At1g67490, 10 kb from marker K12, was rearranged in the fast neutron–generated allele knf-19 (unpublished data). This predicted gene corresponded to six anonymous ESTs (Asamizu et al., 2000). Sequencing of the EST clones revealed that two (GenBank/EMBL/DDBJ accession nos. AV549758 and AV537739) contained an open reading frame of 2,559 bp. A comparison of genomic and cDNA sequences for At1g67490 is shown in Fig. 4 B.
Figure 4.
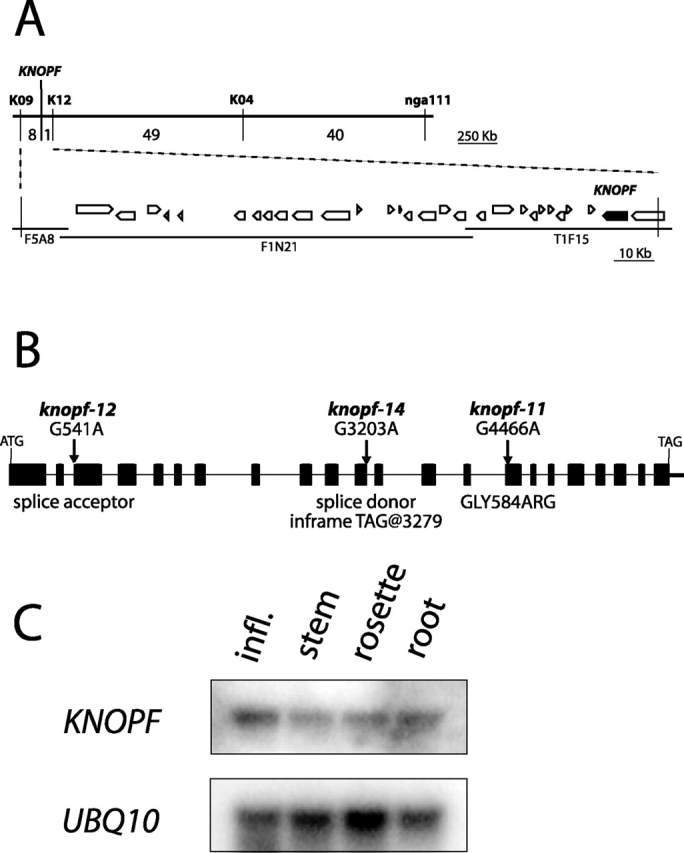
KNF cloning, gene structure, and gene expression. (A) Identification of the KNF gene based on its map position at ∼100 cM on chromosome I. Upper line is the genetic map of the interval containing KNF. PCR-based SSLP markers used for recombination mapping are listed above the line, and the number of recombinations obtained between each marker (out of a total of 948) are listed below the top line. KNF was found to lie between markers K09 and K12, with a single recombination event between the KNF gene and marker K12. Lower lines represent three overlapping BAC clones (F5A8, F1N21, and T1F15), which cover the ∼160-kb physical map between markers K09 and K12, with predicted open reading frames shown as open pointed boxes, and the KNF gene shown as a filled-in box. The names of the BAC clones are listed below the line representing each clone. (B) Gene structure of KNF, with exons represented as boxes and introns as a thin line. The nucleotide changes in three knf alleles are shown above arrows indicating the site of the mutation. The result of the nucleic acid change is shown below the gene model. (C) KNF is expressed throughout the plant. Northern blot analysis of KNF expression in inflorescence, stem, rosette, and root tissue. UBIQUITIN10 expression is shown as a loading control.
To confirm the identity of KNF, gene At1g67490 was PCR-amplified from genomic DNA of homozygous knf-11, knf-12, and knf-14 embryos. Sequencing of the PCR products revealed that each of these ethylmethane sulfonate–induced alleles contained a single base pair substitution in the At1g67490 genomic sequence (Fig. 4 B). On the basis of sequenced mutations in three separate alleles as well as a restriction fragment length polymorphism in a fourth allele, we concluded that gene At1g67490 corresponds to KNF. Molecular identification of the KNF gene revealed that it was identical to GCS1, a gene recently identified based on its role in seed development (Boisson et al., 2001). gcs1 mutants were shown to affect N-glycan trimming, the formation of complex glycans, and the accumulation of seed storage proteins.
Northern analysis of KNF gene expression in different tissues and at different stages of development showed that KNF is expressed throughout plant development (Fig. 4 C). KNF mRNA is present at approximately the same low level in inflorescences, stems, and rosettes of 5-wk-old greenhouse-grown plants and in roots of 3-wk-old plants grown in sterile culture on agar-solidified mineral medium. This observation is consistent with the role of KNF in providing a basic metabolic function (see below).
KNF encodes α-glucosidase I
Fig. 5 shows a comparison of the predicted KNF protein with human and yeast α-glucosidase I. The similarity between the three proteins extends over the entire protein sequence: KNF and human α-glucosidase I share 32.7% identity, whereas KNF and the yeast α-glucosidase I Cwh41p are 19.7% identical. The mutations in three knf alleles are shown in Fig. 5, and all occur in conserved regions of the protein. Loss of the splice donor site in knf-14 introduces an in-frame stop codon at bp 3279 of the KNF genomic sequence (Fig. 4 B). Because this occurs ∼100–amino acid residues before the proposed active site of the enzyme (Fig. 5, brackets), this allele, which was used for all phenotypic analysis, is presumably a null.
Figure 5.

KNF is similar to α-glucosidase I enzymes from human and yeast. Sequence alignment of the predicted 852–amino acid KNF protein with human α-glucosidase I (GenBank/EMBL/DDBJ accession no. CAA60683) and the yeast α-glucosidase I Cwh41p (AAC49157). Identical residues are shaded in black. The single predicted transmembrane domain between the cytoplasmic NH2 terminus and ER-lumenal COOH terminus is marked by a broken line, the predicted substrate-binding domain (Romaniouk and Vijay, 1997) is marked by horizontal brackets, and the mutations in different knf alleles are marked with stars. KNF is 32.7% identical to human α-glucosidase I and 19.7% identical to yeast α-glucosidase I. The KNOPF sequence is available from GenBank/EMBL/DDBJ under accession no. AY055858.
The lack of α-glucosidase I activity in knf embryos should result in a loss of trimming of N-linked glycans. To test this, protein extracts from wt and knf embryos were probed with an antibody to protein disulfide isomerase (PDI), an ER-residing N glycoprotein (Shimoni et al., 1995). The absence of α-glucosidase I activity should lead to a higher molecular weight of PDI in knf than in wt extracts, due to a lack of trimming of the three terminal glucose residues that normally occurs in the ER. As predicted, PDI has a slightly reduced mobility in the protein extracts from knf embryos as compared with wt embryos (Fig. 6 A).
Figure 6.
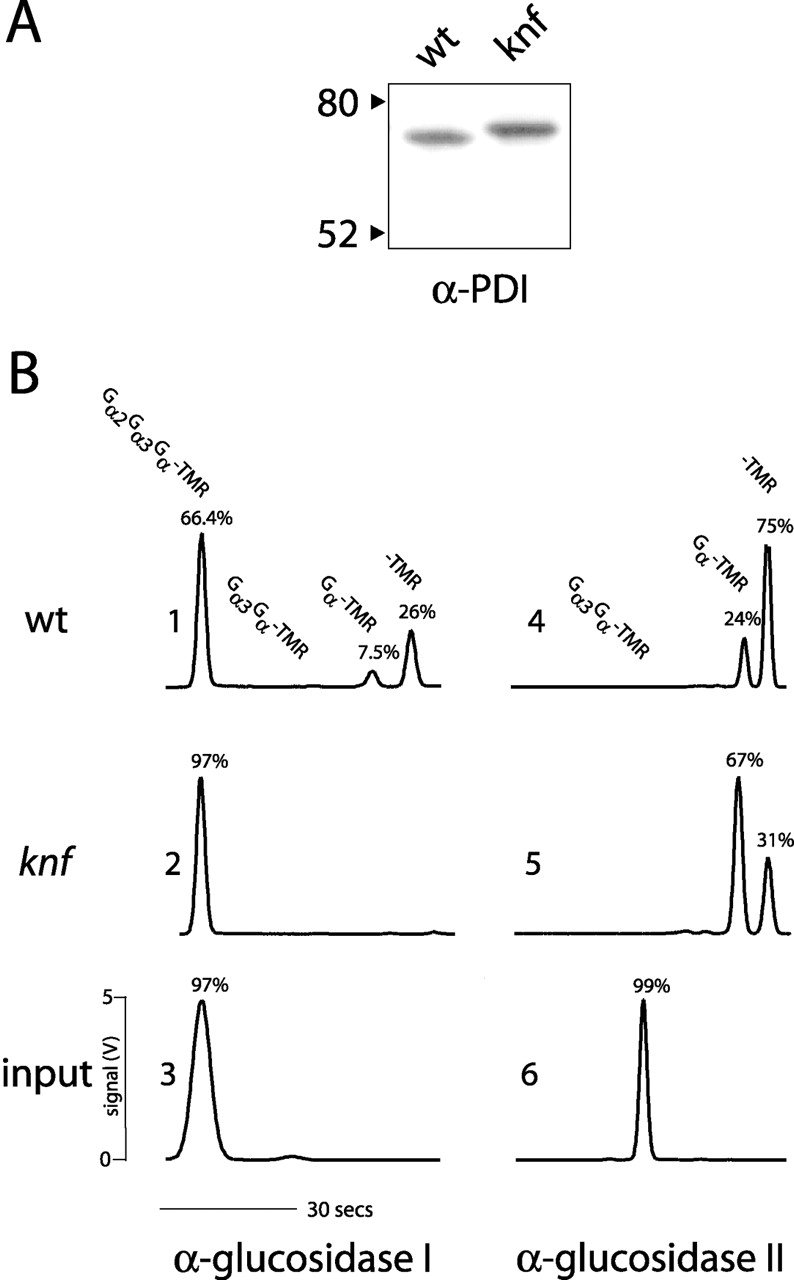
KNF encodes α-glucosidase I. (A) The ER resident glycoprotein, PDI, shows reduced mobility in extracts from knf embryos. 10 μg of total protein was probed with PDI antisera. Left, molecular mass standards. (B) knf embryo extracts lack α-glucosidase I activity. Wt and knf embryo extracts were incubated for 24 h with the rhodamine-conjugated triglucoside substrate Gα2Gα3Gα-TMR to test for α-glucosidase I activity, or with the rhodamine-conjugated diglucoside substrate Gα3Gα-TMR to test for glucosidase II activity. Reaction products were separated and measured by capillary electrophoresis laser-induced fluorescence detection at 580 nm. Left column, α-glucosidase I activity; right column, α-glucosidase II activity; y-axis, signal (V); x-axis, migration time (s).
An in vivo assay was used to directly measure α-glucosidase I activity in knf extracts (Fig. 6 B). Extracts of wt and knf embryos were incubated with the rhodamine-conjugated triglucoside substrate α-d-glucose-(1–2)-α-d-glucose-(1–3)-α-d-glucose-O-(CH2)8COOCH3-tetramethylrhodamine (Glcα2Glcα3Glcα-TMR; Scaman et al., 1996). This substrate has been demonstrated to be specific for α-(1,2) glucosidase I, which cleaves the terminal α-1,2–linked glucose to generate a diglucose product with a terminal α-(1,3)-glucose that is, in turn, a substrate for α-(1,3)-glucosidase II (Le et al., 1999). Reaction products were separated by capillary electrophoresis and quantified using laser-induced fluorescence (Le et al., 1999). As shown in Fig. 6 B, panel 1, extracts of wt embryos efficiently cleaved the terminal α-1,2–linked glucose of the triglucoside substrate. The diglucose product Gα3Gα-TMR is not visible because it is subsequently cleaved by α-glucosidase II to monoglucose (Gα-TMR) and rhodamine linker arm only (TMR). Extracts of knf embryos incubated with triglucose substrate (Fig. 6 B, panel 2) had no detectable α-glucosidase I activity. As a positive control for general enzyme activity in knf embryos, wt (Fig. 6 B, panel 4) and knf (panel 5) extracts were also incubated with the diglucose substrate Gα3Gα-TMR. In both extracts, α-glucosidase II completely digested the diglucose substrate to the monoglucoside or the linker arm only.
CESA proteins are not affected by lack of N-glycan trimming
We have demonstrated that KNF encodes α-glucosidase I, an N-glycan trimming enzyme that is not directly involved in cellulose synthesis. Thus, the most likely explanation for the lack of cellulose in knf embryos is that one or more proteins required for cellulose biosynthesis are N glycosylated, and the proper trimming of these glycans is essential for the synthesis of an active protein. Cellulose synthase catalytic subunits such as RSW1 (Arioli et al., 1998) have five N glycosylation motifs. To directly test whether cellulose synthase proteins are modified by the attachment of N-glycans, we probed native and peptide:N-glycosidase F (PNGase)–treated protein extracts with CESA antibody 60836. Most plant complex glycans are not a substrate for PNGase because they are substituted at the proximal GlcNAc with α-1,3-fucose. To circumvent this problem, we used the cgl1 mutant, which totally lacks complex glycan epitopes and is phenotypically normal. In this mutant background, the majority of N-glycans have the structure Man5GlcNAc2, and are thus suitable substrates for PNGase (von Schaewen et al., 1993).
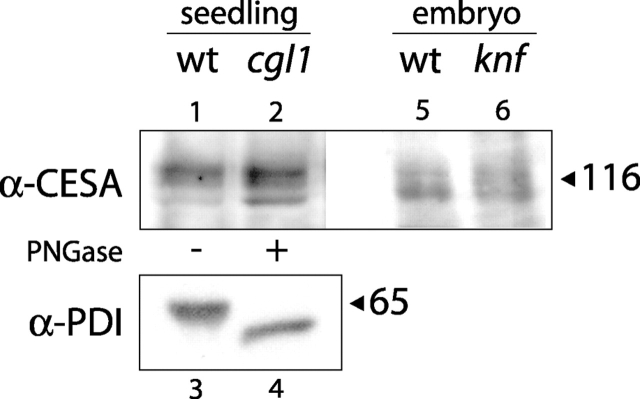
As shown in Fig. 7, lane 1, antibody 60836 recognizes three proteins in the ∼120-kD size range (the predicted size range for CESA proteins) in protein extracts from 2-d-old wt seedlings. The presence of three CESA isoforms is in good agreement with other evidence that three CESA genes are expressed in young seedlings (RSW1/CESA1, this study and Arioli et al., 1998; PRC1/CESA6, Fagard et al., 2000; IXR1/CESA3, Scheible et al., 2001). These same three bands migrate with equivalent molecular weight in PNGase-treated extracts from cgl1 seedlings (Fig. 7, lane 2). The lack of shift in band size after PNGase treatment suggests that these CESA isoforms are not modified by the attachment of N-glycans. To verify that CESA proteins are present in knf embryos, we probed protein extracts of wt and knf embryos with antibody 60836. As shown in Fig. 7, both wt (lane 5) and knf (lane 6) embryos contained three cross-reacting bands in the ∼120-kD size range. Thus, the stability of CESA proteins is not affected by loss of KNF α-glucosidase I activity.
Figure 7.
Cellulose synthase catalytic subunits are not N glycosylated. No size shift in CESA proteins was observed between native wt (lane 1) and PNGase-treated cgl1 (lane 2) protein extracts from 2-d-old seedlings. As a control for deglycosylation, the same extracts were probed with PDI antiserum (lane 3 vs. 4). CESA proteins are present in equal amounts in wt (lane 5) and knf (lane 6) extracts from late stage embryos. 75 μg total protein was probed with CESA antiserum 60836 or PDI antiserum. Size standards are indicated on the right.
Discussion
We have isolated and characterized mutations in the RSW1 and KNF genes, which alter cell elongation and embryo morphogenesis in Arabidopsis. Both of these mutations cause large reductions in crystalline cellulose. Characterization of the nonconditional rsw1-2 phenotype extends previous studies of a conditional mutant allele of RSW1, which encodes a catalytic subunit of cellulose synthase (Arioli et al., 1998). Map-based cloning of the KNF gene revealed that it encodes α-glucosidase I, which catalyzes the first step in N-glycan trimming. Because α-glucosidase I has no direct role in cellulose biosynthesis, the decrease in cellulose in knf mutants is due to a secondary effect of lack of N-glycan trimming on other protein(s) required for biosynthesis of cellulose.
Mutant alleles of KNF (called gcs1) were recently described by Boisson et al. (2001). Analysis of the structure of N-linked glycans in gcs1 mutants indicated a complete deficiency of α-1,3-fucose– and β-1,2-xylose–containing complex N-linked glycans. Instead, gcs1 mutants accumulate the N-glycan Glc3Man7GlcNAc2. In addition, the gcs1 mutation was shown to affect accumulation of seed storage proteins. Our work extends that of Boisson et al. (2001) by providing direct evidence for a defect in α-glucosidase I activity, an explanation of the morphological phenotype of knf/gcs1 mutants, and analysis of the effects of the lesion on cell wall structure and function.
Reduced cellulose underlies the swollen morphology of rsw1-2 and knf embryos
Cellulose is thought to be the main load-bearing polymer of the plant cell wall (Carpita and Gibeaut, 1993). The arrangement of cellulose microfibrils in helicoidal hoops around the circumference of cells, and the cross-linking of these microfibrils by xyloglucans, reinforces cells so that expansion occurs perpendicular to the direction of hoop reinforcement (Cosgrove, 1999; Kerstens et al., 2001). The epidermis in particular has been proposed to bear much of the stress experienced by plant tissues, and thus is critical for the control of morphogenesis (for review see Green, 1980).
rsw1-2 and knf embryos have four- and sevenfold reduced levels of cellulose, respectively. The direction of cell expansion is altered in the epidermal and ground layers of rsw1-2 embryos and in all cell layers of knf embryos. We have shown that the average thickness of the epidermal layer is reduced by one third in rsw1-2 embryos and by two thirds in knf embryos. Taken together, these data suggest a simple explanation for the altered shape of rsw1-2 and knf embryos, especially when the importance of the epidermis in controlling plant shape is taken into consideration. Fig. 8 illustrates the change in shape between the heart and torpedo stage of wt and knf embryos. In wt embryos, growth primarily occurs in a longitudinal direction so that the torpedo stage embryo is approximately four times as long as the heart stage embryo. This anisotropic growth requires that the cell walls of the embryo, and especially the epidermis, are differentially reinforced. The reduced amount of cellulose reinforcement in knf embryos, especially in the epidermis, means that growth cannot occur in one direction more than another, and thus overall shape is maintained while volume increases. The fact that rsw1-2 embryos are swollen but are able to elongate considerably more than knf is most likely due to the fact that these embryos have more cellulose early in development than knf embryos, as evidenced by the relatively normal rsw1-2 phenotype at the heart and torpedo stages. In the absence of anisotropic reinforcement, objects retain their relative dimensions as they increase in volume. An initial difference in cellulose also suggests an earlier requirement for KNF enzyme activity compared with RSW1.
Figure 8.
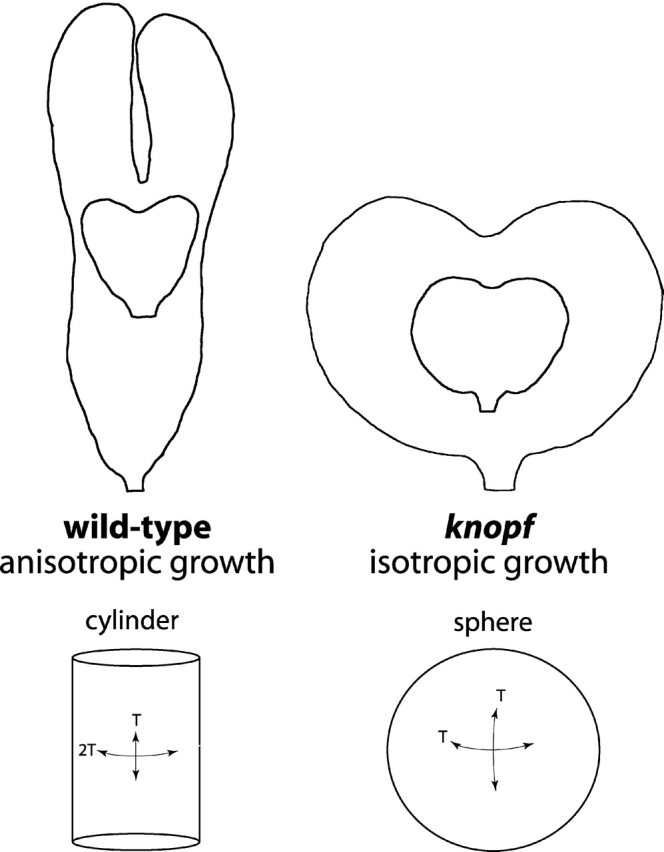
A biophysical model for morphogenesis of wt and knf embryos. Growth of a wt embryo requires differential reinforcement of the epidermis by cellulose and can be modeled as a cylinder. Because of decreased cellulose reinforcement of the epidermis, growth of a knf embryo resembles the enlargement of a sphere. Top, tracings of wt (left) and knf (right) heart and torpedo stage embryos shown at the same scale. Bottom, the relative tension (T) experienced in different directions by the wall of a hollow cylinder or sphere.
N-glycan trimming is required for cellulose biosynthesis
In the cgl1 mutant of Arabidopsis, complex glycans are replaced by the high mannose glycan Man5GlcNAc2 (von Schaewen et al., 1993). However, cgl1 plants are phenotypically normal. In contrast, knf/gcs1 mutants, which also lack complex glycans and predominantly accumulate the N-glycan Glc3Man7GlcNAc2 (Boisson et al., 2001), produce almost no cellulose. The difference in the N-glycan structures of these two mutants demonstrates that correct trimming of only the glucose-substituted branch of the core N-glycan is crucial for cellulose synthesis.
Different hypotheses to explain the effect of glucose trimming on cellulose biosynthesis can be envisioned. First, one or more of the proteins required for cellulose biosynthesis may be N glycosylated, and trimming of these N-glycans may be required for enzyme activity. For instance, glycosylation by high mannose glycans is required for in vitro enzyme activity of the xyloglucan endotransglycosylases TCH4 and Meri-5 (Campbell and Braam, 1999). Second, in animals and fungi, α-glucosidase I and II action is required to generate the monoglucosylated substrate recognized by the ER-localized lectin-like chaperone calnexin (Hammond and Helenius, 1995). Lack of α-glucosidase I activity in knf mutants might affect the folding or stability of enzymes required for cellulose synthesis.
Three lines of evidence suggest that CESA proteins themselves are not affected by the knf mutation. First, no size shift of CESA proteins after PNGase treatment of seedling extracts is observed. Second, CESA proteins are present in equal amounts in wt and knf embryos. Third, based on the CESA topology proposed by Delmer (1999), only one N glycosylation motif is located in an extracellular loop of the enzyme, and this motif is too close to the plasma membrane to serve as a substrate for the oligosaccharyltransferase that transfers the core N-glycan to nascent proteins (Nilsson and von Heijne, 1993). A second possibility, noted by Lukowitz et al. (2001) is that an N-glycosylated protein may be required as a primer for cellulose biosynthesis. In view of the fact that the cyt1 mutant appears to be completely deficient in high mannose glycosylation, and the knf mutant appears to be completely deficient in α-glucosidase I activity, the existence of residual cellulose in both mutants renders this hypothesis unlikely. It seems more likely that N glycosylation has a stimulatory, rather than an obligate, role in cellulose synthesis.
Thus, our results suggest a requirement for N glycosylation of one or more components of the cellulose synthase complex other than the catalytic subunit itself. One possible candidate is KORRIGAN, a secreted endo-1,4-β-glucanase that has eight N glycosylation signature motifs in its predicted extracellular domain (Nicol et al., 1998). Analysis of rsw2 mutants, which are allelic to KOR, demonstrated that KOR activity is required for cellulose synthesis (Lane et al., 2001). In addition, a recent study by Molhoj et al. (2001) has shown that N-glycans are required for enzyme activity of a heterologously expressed Brassica endo-1,4-β-glucanase homologous to KOR.
Conclusion
The mechanism of cellulose biosynthesis is poorly understood. Our results demonstrate that one or more proteins required for cellulose synthesis, but most likely not the cellulose synthase catalytic subunits, must be glycosylated to function properly. In addition, the observation that defects in cellulose biosynthesis can be identified during embryogenesis should allow the isolation, by mutant analysis, of additional genes required for cellulose synthesis. From a broad perspective, it is interesting to note that although a large number of proteins are modified by the addition of N-glycans in Arabidopsis and yeast, these N-glycans are apparently essential for the function of only a small number of proteins. In yeast, defects in N-glycan processing affect synthesis of β-1,6-glucan by decreasing stability of the β-1,6-glucan synthase Kre6p, but do not change the secretion and activity of other glycoproteins such as invertase, pro-α-factor, and carboxypeptidase Y (Abeijon and Chen, 1998; Simons et al., 1998). The characterization of knf and gcs1 alleles of α-glucosidase I in Arabidopsis shows that N-glycan trimming is also essential for plant cell wall synthesis and formation of protein storage vacuoles. We have not observed any other cellular defects in knf mutants. In view of the fact that knf embryos are able to develop thousands of cells, the lethality of the knf mutation (like that of rsw1) can reasonably be attributed to the reduction in cellulose deposition. It seems likely that knf embryo lethality coincides with the requirement for osmotic stabilization of the embryo during seed desiccation.
Materials and methods
Plant material and growth conditions
All mutant lines, generated in the Landsberg erecta ecotype, were propagated as heterozygotes and backcrossed at least twice before analysis. Plants were grown in a greenhouse in 16 h photoperiods at an average temperature of 25°C. Seedlings were grown on 0.5× MS media (GIBCO BRL), 1% sucrose, 0.8% agar plates in 24 h light (∼100 μmol m−2 s−1) at 22°C or 31°C. Plasmid pEGAD-TUB1, which expresses a GFP–β1-tubulin fusion protein, was obtained from S. Cutler and D. Ehrhardt (Carnegie Institution), and was introduced into the Landsberg erecta ecotype by agrobacterium transformation (Cutler et al., 2000) to produce line SGGFP4. Line 12-30 was a gift from W. Lukowitz (Carnegie Institution). knf U18-7 seed was obtained from the Arabidopsis Biological Resource Center (ABRC) at Ohio State University. rsw1-1 seed was provided by R. Williamson (Australian National University, Canberra, Australia). cgl1-1 seed was provided by M. Chrispeels (University of California, San Diego, CA).
Light and EM
The genetic screen for embryo mutations was performed by dissecting ovules into Hoyer's solution (7.5 g gum arabic, 100 g chloral hydrate, 5 ml glycerol, 30 ml water) overnight and viewing cleared ovules with Nomarski optics. Embryos for EM were dissected out of the ovule and fixed in 4% paraformaldehyde and 0.25% glutaraldehyde. Embryos were postfixed in 1% osmium tetroxide, stained en bloc in 1% uranyl acetate, dehydrated through an ethanol series, and infiltrated with Spurr's resin, according to the manufacturer's instructions (Polysciences, Inc.). Silver-gold sections (60–90 nm thick) were stained with 2% uranyl acetate and lead citrate and viewed with a Philips 410 transmission electron microscope.
Confocal microscopy
For visualization of embryo morphology with confocal microscopy, torpedo stage and earlier stage ovules were dissected from siliques into 70% ethanol, and bent cotyledon stage embryos were dissected out of the seed coat into 70% ethanol. Ethanol was replaced with 1:1 chloroform/methanol and the embryos were gently agitated for 30 min. The chloroform/methanol was replaced with 100% methanol for 15 min. Methanol was then replaced by NaPT buffer (50 mM sodium phosphate buffer, pH 7.2, with 0.05% Triton X-100) and tubes were rotated for 30 min. NaPT buffer was replaced by 50 μl of 150 μg/ml Alexafluor 488 Hydrazide (Molecular Probes) in NaPT buffer, and tubes were put in a light-tight box on a horizontal rotating shaker for 2 h. Stained embryos/ovules were then rinsed three times for 5 min with water and embedded in Hoyer's solution (6 g gum arabic, 40 g chloral hydrate, 4 g glycerol, 10 ml water; Bougourd et al., 2000) on a microscope slide. Embryos were visualized immediately using a Bio-Rad Laboratories MRC 1024 confocal microscope at 488 nm excitation and 520 nm absorbance, with Kalman averaging n = 20. Images were processed using NIH Image for the Macintosh (http://rsbweb.nih.gov/), Adobe Photoshop® 6.0, and Adobe Illustrator® 9.0 (Adobe Systems, Inc.).
Positional cloning and molecular biology
Rough and fine mapping of the KNF gene were performed as described in Lukowitz et al., (2000). Primer sequences for SSLP markers are available at http://www.arabidopsis.org/. The KNF gene was directly sequenced from PCR products amplified from genomic DNA prepared from homozygous mutant embryos. The full-length KNF cDNA clone, pSG8, was obtained as an EST (GenBank/EMBL/DDBJ accession no. AV549758) from the Kazusa Arabidopsis cDNA sequencing project.
Northern analysis of KNF gene expression was performed as follows: total RNA was extracted from inflorescences (including young siliques), stems, and rosette leaves of 5-wk-old greenhouse-grown plants of the Columbia ecotype and from roots of 3-wk-old plants grown on 0.5× MS 1% sucrose plates using Trizol reagent (Life Technologies). PolyA+ RNA was isolated using PolyAtract beads (Promega). PolyA+ RNA (2.5 μg) was transferred to nylon membrane and probed with a 32P-labeled SacI fragment of the KNF cDNA (bp 1–736 of AY055858) and the full-length UBQ10 EST clone 170I13T7 (Callis et al., 1995) using standard protocols (Ausubel et al., 1995). Blots were exposed on a phosphor screen for 1 wk (KNF) or 3 h (UBQ10) and visualized using a Typhoon 8600 variable mode imager (Molecular Dynamics). EST clone 170I13T7 was obtained from the ABRC.
Western analysis
For α-PDI Western blot, green bent cotyledon stage wt and knf embryos were dissected from the seed coat into extraction buffer (2% SDS, 100 mM TrisCl, pH 8.0, 10 mM MgCl2, 18% sucrose, 40 mM β-mercaptoethanol) and ground in a tiny mortar and pestle. For CESA Western blots, embryos were dissected into grinding buffer (50 mM sodium phosphate buffer, pH 8.0, 300 mM NaCl) plus 1× Calbiochem-Novabiochem protease inhibitor cocktail set I. Gel loading buffer (2×) contained 9.5 M urea. Proteins were quantified using the Bradford protein assay kit (Bio-Rad Laboratories), separated by 7.5% SDS-PAGE, and electro-blotted using semidry transfer to nitrocellulose (PDI) or PVDF (CESA) membrane. Rabbit PDI antiserum (Rose Biotechnology) was used at a dilution of 1:2,500, and rabbit CESA antiserum 60836 (generated against the peptide NELPRLVYVSREKRPGC and affinity purified against the same peptide by Quality Controlled Biochemicals) was used at a dilution of 1:1,000. Primary antibodies were visualized by incubation with goat anti–rabbit IgG secondary antibodies conjugated to horseradish peroxidase (Bio-Rad Laboratories) followed by a chemiluminescence reaction (SuperSignal®; Pierce Chemical Co.).
Cell wall analysis
For each sample, 200–300 (0.5–1.0 mg dry wt) late bent cotyledon stage wt, rsw1-2, and knf embryos were dissected out of the seed coat, incubated in 100% ethanol at 65°C for 1 h, and then incubated in 100% acetone at room temperature for 5 min. Cellulose was isolated and solubilized by the method of Updegraff (1960), and glucose content was determined by the method of Scott and Melvin (1953). Total uronic acid was determined by autoclaving ∼0.5 mg dry embryo tissue in 100 μl of 2 M trifluoroacetic acid for 1 h. Trifluoroacetic acid was then evaporated under a nitrogen stream, and embryo residue was dissolved in 100 μl distilled water. Total uronic acid content was determined per Blumenkrantz and Asboe-Hansen (1973). Cell wall neutral sugars were determined as in Reiter et al. (1993). Absolute amounts of neutral sugars were determined by running pure standards using myo-inositol as an internal control.
α-Glucosidase assay
The α-glucosidase assays were performed essentially as described in Le et al. (1999), with the following modifications: ∼100 frozen wt or knf bent cotyledon stage embryos were thawed and gently homogenized with a pestle in an Eppendorf tube in 30 μl lysis buffer (100 mM sodium phosphate buffer, pH 7.0, 1.0% Triton X-100, 1× Roche protease inhibitor). The pestle was rinsed with 10 μl lysis buffer, the tubes were centrifuged at 4,000 g for 30 min, and the supernatant was removed for assay. For the triglucoside (α-glucosidase I) assay, samples were diluted 1:1 with lysis buffer, and 5 μl of diluted sample was incubated with 2.5 μl of 270 μM triglucose-TMR at 37°C. For the diglucoside (glucosidase II) assay, samples were diluted 1:10 with lysis buffer and 5 μl of sample was incubated with 0.5 μl of 1 mM diglucose-TMR at 37°C. Enzyme activity time points were taken by removing 0.5 μl of the extract–substrate mixture, adding 19.5 μl of capillary electrophoresis running buffer (10 mM Na2HPO4, 10 mM phenylboronic acid, 50 mM SDS, 2.5 mM Na2B4O7, 10% methanol, pH 9.4) to quench, and freezing at −20°C. For capillary electrophoresis, 0.5 μl of quenched aliquot was diluted in 9.5 μl running buffer. Samples were injected at 2,000, 3,000, or 5,000 V for 5 s (to achieve comparable signals between samples) and then run at 400 V/cm on a 35-cm capillary. The instrument was locally constructed with a 5.0-mW He-Ne laser (λexcit = 543.5 nm) focused into a postcolumn sheath flow cuvette for laser-induced fluorescence detection at 580 nm. Data were analyzed with IgorPro 3.1 (Wavemetrics).
Acknowledgments
Thanks to K. Sujino (University of Alberta, Alberta, Canada) for conducting preliminary α-glucosidase assays, and to R. Gillmor (Carnegie Institution) for expert technical assistance. S. Cutler and D. Ehrhardt generously provided the GFP–β1-tubulin construct, and J. Griffits (Carnegie Institution) is acknowledged for plant transformation. W. Lukowitz, S. Cutler (Carnegie Institution), and D. Bonetta (Carnegie Institution) provided advice and insights throughout this work. Line 12-30 was a gift from W. Lukowitz. J. Dumais (Stanford University, Stanford, CA) is acknowledged for illuminating conversations on the physical properties of the cell wall, as is R. Torres-Ruiz (Technical University of Munich, Munich, Germany) for suggesting knf allelism. W. Lukowitz and D. Bergmann (Carnegie Institution) provided helpful comments on the manuscript.
C.S. Gillmor was partially supported by a U.S. Department of Energy/National Science Foundation/U.S. Department of Agriculture tri-agency training grant. J. Lorieau was supported by an Alberta Heritage Foundation for Medical Research Summer Studentship award. This work was supported in part by a grant from the U.S. Department of Energy (DE-FG02-97ER20133) to C.R. Somerville, and by a grant from the Natural Sciences and Engineering Research Council of Canada to M.M. Palcic.
Footnotes
Abbreviations used in this paper: CESA, cellulose synthase; MT, microtubule; PDI, protein disulfide isomerase; PNGase, peptide:N-glycosidase F; TMR, tetramethylrhodamine; wt, wild type.
References
- Abeijon, C., and L.Y. Chen. 1998. The role of glucosidase I (Cwh41p) in the biosynthesis of cell wall β-1,6-glucan is indirect. Mol. Biol. Cell. 9:2729–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arioli, T., L. Peng, A.S. Betzner, J. Burn, W. Wittke, W. Herth, C. Camilleri, H. Höfte, J. Plazinski, R. Birch, et al. 1998. Molecular analysis of cellulose biosynthesis in Arabidopsis. Science. 279:717–720. [DOI] [PubMed] [Google Scholar]
- Asamizu, E., Y. Nakamura, S. Sato, and S. Tabata. 2000. A large scale analysis of cDNAs in Arabidopsis thaliana: generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res. 7:175–180. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidmann, J.A. Smith, and K. Struhl. 1995. Current Protocols in Molecular Biology. Vol. 1. John Wiley & Sons Inc., New York. 500 pp.
- Boisson, M., V. Gomord, C. Audran, N. Berger, B. Dubreucq, F. Granier, P. Lerouge, L. Faye, M. Caboche, and L. Lepiniec. 2001. Arabidopsis glucosidase I mutants reveal a critical role of N-glycan trimming in seed development. EMBO J. 20:1010–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougourd, S., J. Marrison, and J. Haseloff. 2000. An aniline blue staining procedure for confocal microscopy and 3D imaging of normal and perturbed cellular phenotypes in mature Arabidopsis embryos. Plant J. 24:543–550. [DOI] [PubMed] [Google Scholar]
- Blumenkrantz, N., and G. Asboe-Hansen. 1973. New method for quantitative determination of uronic acids. Anal. Biochem. 54:484–489. [DOI] [PubMed] [Google Scholar]
- Callis, J., T. Carpenter, C.W. Sun, and R.D. Vierstra. 1995. Structure and evolution of genes encoding polyubiquitin and ubiquitin-like proteins in Arabidopsis thaliana ecotype Columbia. Genetics. 139:921–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, P., and J. Braam. 1999. In vitro activities of four xyloglucan endotransglycosylases from Arabidopsis. Plant J. 18:371–382. [DOI] [PubMed] [Google Scholar]
- Carpita, N.M., and D.M. Gibeaut. 1993. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3:1–30. [DOI] [PubMed] [Google Scholar]
- Cosgrove, D.J. 1999. Enzymes and other agents that enhance cell wall extensibility. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50:391–417. [DOI] [PubMed] [Google Scholar]
- Cutler, S.R., D.W. Ehrhardt, J.S. Griffits, and C.R. Somerville. 2000. Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at high frequency. Proc. Natl. Acad. Sci. USA. 97:3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmer, D.P. 1999. Cellulose biosynthesis: exciting times for a difficult field of study. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50:245–276. [DOI] [PubMed] [Google Scholar]
- Fagard, M., T. Desnos, T. Desprez, F. Goubet, G. Refregier, G. Mouille, M. McCann, C. Rayon, S. Vernhettes, and H. Höfte. 2000. PROCUSTE1 encodes a cellulose synthase required for normal cell elongation specifically in roots and dark-grown hypocotyls of Arabidopsis. Plant Cell. 12:2409–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, P.B. 1962. Mechanism for plant cellular morphogenesis. Science. 138:1404–1405. [DOI] [PubMed] [Google Scholar]
- Green, P.B. 1980. Organogenesis—a biophysical view. Annu. Rev. Plant Physiol. 31:51–82. [Google Scholar]
- Hammond, C., and A. Helenius. 1995. Quality control in the secretory pathway. Curr. Opin. Cell Biol. 7:523–529. [DOI] [PubMed] [Google Scholar]
- Jürgens, G., and U. Mayer. 1994. Arabidopsis. Embryos: Color Atlas of Development. J.B.L. Bard, editor. Wolfe, London. 7–20.
- Kerstens, S., W.F. Decraemer, and J.P. Verbelen. 2001. Cell walls at the plant surface behave mechaniclaly like fiber-reinforced composite materials. Plant Physiol. 127:381–385. [PMC free article] [PubMed] [Google Scholar]
- Lane, D.R., A. Wiedermeier, L. Peng, H. Höfte, S. Vernhettes, T. Desprez, C.H. Hocart, R.J. Birch, T.I. Baskin, J.E. Burn, et al. 2001. Temperature sensitive alleles of RSW2 link the KORRIGAN endo-1,4,-β-glucanase to cellulose synthesis and cytokinesis in Arabidopsis. Plant Physiol. 126:278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le, X.C., W. Tan, C.H. Scaman, A. Szpacenko, E. Arriaga, Y. Zhang, N.J. Dovichi, O. Hindsgaul, and M.M. Palcic. 1999. Single cell studies of enzymatic hydrolysis of a tetramethylrhodamine labeled triglucoside in yeast. Glycobiology. 9:219–225. [DOI] [PubMed] [Google Scholar]
- Lukowitz, W., C.S. Gillmor, and W.-R. Scheible. 2000. Positional cloning in Arabidopsis. Why it feels good to have a genome initiative working for you. Plant Physiol. 123:795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz, W., T.C. Nickle, D.W. Meinke, R.L. Last, P.L. Conklin, and C.R. Somerville. 2001. Arabidopsis cyt1 mutants are deficient in a mannose-1-phosphate guanylyltransferase and point to a requirement of N-linked glycosylation for cellulose biosynthesis. Proc. Natl. Acad. Sci. USA. 98:2262–2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse, A.G., D.O.L. Thomas, and A.R. White. 1995. Mechanism of cellulose synthesis in Agrobacterium tumefaciens. J. Bacteriol. 177:1076–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, U., R.A. Torres-Ruiz, T. Berleth, S. Miséra, and G. Jürgens. 1991. Mutations affecting body organization in the Arabidopsis embryo. Nature. 353:402–407. [Google Scholar]
- Meinke, D.W. 1994. Seed development in Arabidopsis thaliana. Arabidopsis. E. Meyerowitz and C.R. Somerville, editors. Cold Spring Harbor Laboratory Press, Plainview, NY. 253–296.
- Molhoj, M., P. Ulvskov, and F. Dal Degan. 2001. Characterization of a functional soluble form of a Brassica napus membrane anchored endo-1,4-β-glucanase heterologously expressed in Pichia pastoris. Plant Physiol. 127:674–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol, F., I. His, A. Jauneau, S. Vernhettes, H. Canut, and H. Höfte. 1998. A plasma membrane-bound putative endo-1,4-β−glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 17:5563–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickle, T.C., and D.W. Meinke. 1998. A cytokinesis-defective mutant of Arabidopsis (cyt1) characterized by embryonic lethality, incomplete cell walls, and excessive callose accumulation. Plant J. 15:321–332. [DOI] [PubMed] [Google Scholar]
- Nilsson, I., and G. von Heijne. 1993. Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J. Biol. Chem. 268:5798–5801. [PubMed] [Google Scholar]
- Pear, J.R., Y. Kawagoe, W.E. Schreckengost, D.P. Delmer, and D.M. Stalker. 1996. Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. USA. 93:12637–12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, L., C.H. Hocart, J.W. Redmond, and R.E. Williamson. 2000. Fractionation of carbohydrates in Arabidopsis root cell walls shows that three radial swelling loci are specifically involved in cellulose production. Planta. 211:406–414. [DOI] [PubMed] [Google Scholar]
- Ray, P.M., P.B. Green, and R. Cleland. 1972. Role of turgor in plant cell growth. Nature. 239:163–164. [Google Scholar]
- Reiter, W.-D., C.C.S. Chapple, and C.R. Somerville. 1993. Altered growth and cell walls in a fucose-deficient mutant of Arabidopsis. Science. 261:1032–1035. [DOI] [PubMed] [Google Scholar]
- Romaniouk, A., and I.K. Vijay. 1997. Structure-function relationships in glucosidase I: amino acids involved in binding the substrate to the enzyme. Glycobiology. 7:399–404. [DOI] [PubMed] [Google Scholar]
- Scaman, C.H., O. Hindsgaul, M.M. Palcic, and O.P. Srivastava. 1996. Synthesis of a-d-Glcp-(1→2)-α-d=Glcp-(1→3)-α-d-Glcp-O-(CH2)8 COOH3 for use in the assay of α-glucose I activity. Carbohydr. Res. 296:203–213. [DOI] [PubMed] [Google Scholar]
- Scheible, W.-R., R. Eshed, T. Richmond, D. Delmer, and C. Somerville. 2001. Modifications of cellulose synthase confer resistance to isoxaben and thiazolidinone herbicides in Arabidopsis ixr1 mutants. Proc. Natl. Acad. Sci. USA. 98:10079–10084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, A.S., and E.H. Melvin. 1953. Determination of dextran with anthrone. Anal. Chem. 25:1656–1661. [Google Scholar]
- Shimoni, Y., X.-Z. Zhu, H. Levanony, G. Segal, and G. Galili. 1995. Purification, characterization, and intracellular localization of glycosylated protein disulfide isomerase from wheat grains. Plant Physiol. 108:327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, J.F., M. Ebersold, and A. Helenius. 1998. Cell wall 1,6-B-glucan synthesis in Saccharomyces cerevisiae depends on ER glucosidases I and II, and the molecular chaperone BiP/Kar2p. EMBO J. 17:396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updegraff, D.M. 1960. Semi-micro determination of cellulose in biological materials. Anal. Biochem. 32:420–424. [DOI] [PubMed] [Google Scholar]
- von Schaewen, A., A. Sturm, J. O'Neill, and M.J. Chrispeels. 1993. Isolation of a mutant Arabidopsis plant that lacks N-acetyl glucosaminyl transferase I and is unable to synthesize Golgi-modified complex N-linked glycans. Plant Physiol. 102:1109–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]