Abstract
How kinetochore proteins are organized to connect chromosomes to spindle microtubules, and whether any structural and organizational themes are common to kinetochores from distantly related organisms, are key unanswered questions. Here, we used affinity chromatography and mass spectrometry to generate a map of kinetochore protein interactions. The budding yeast CENP-C homologue Mif2p specifically copurified with histones H2A, H2B, and H4, and with the histone H3-like CENP-A homologue Cse4p, strongly suggesting that Cse4p replaces histone H3 in a specialized centromeric nucleosome. A novel four-protein Mtw1 complex, the Nnf1p subunit of which has homology to the vertebrate kinetochore protein CENP-H, also copurified with Mif2p and a variety of central kinetochore proteins. We show that Mif2 is a critical in vivo target of the Aurora kinase Ipl1p. Chromatin immunoprecipitation studies demonstrated the biological relevance of these associations. We propose that a molecular core consisting of CENP-A, -C, -H, and Ndc80/HEC has been conserved from yeast to humans to link centromeres to spindle microtubules.
Keywords: microtubule; spindle; mitosis; nucleosome; Aurora kinase
Introduction
In eukaryotes, chromosome segregation requires a physical linkage between centromeric DNA and spindle microtubules. The kinetochore serves as the microtubule attachment site and is an important signaling module that controls cell cycle progression via the spindle checkpoint. Although many new kinetochore proteins have been discovered recently (for reviews see Cheeseman et al., 2002a; Cleveland et al., 2003), the physical interactions which establish connectivity between centromeric DNA and spindle microtubules are still ill defined both in yeast and in metazoans.
A variety of strategies have been used to investigate the organization of the budding yeast kinetochore including two-hybrid analyses (Ortiz et al., 1999; Measday et al., 2002; Shang et al., 2003), biochemical purifications of kinetochore subcomplexes (Cheeseman et al., 2001, 2002b; Janke et al., 2001, 2002; Wigge and Kilmartin, 2001; Li et al., 2002), and chromatin immunoprecipitation (ChIP) analysis, which can establish the dependency requirements for the recruitment of a kinetochore protein to centromeric DNA (Meluh and Koshland, 1997; He et al., 2001; Measday et al., 2002; Pot et al., 2003).
We have described previously our purification of the kinetochore proteins that comprise the yeast central and outer kinetochore (Cheeseman et al., 2002b). Here, we examined the DNA binding components of the inner kinetochore. In addition to defining a new kinetochore subcomplex, this analysis has provided insights into how the various subcomplexes interact with each other and suggests a model for a higher order kinetochore structure, which has been conserved throughout evolution.
Results and discussion
Purification of inner kinetochore proteins
Previous studies have identified four different groups of centromeric DNA binding proteins in budding yeast: the four subunit CBF3 complex (Lechner and Carbon, 1991) (Ndc10p, Ctf13p, Cep3p, and Skp1p), a specialized centromeric nucleosome containing the histone H3 variant Cse4p (Stoler et al., 1995; Meluh et al., 1998), and the proteins Cbf1p (Mellor et al., 1990) and Mif2p (Meluh and Koshland, 1995). We sought to purify these proteins using a tandem affinity purification (TAP) strategy (Cheeseman et al., 2002b). Throughout this work, 300 mM KCl was used in our purifications rather than the up to 600 mM KCl we used previously. We found that this lower salt concentration was sufficiently stringent to maintain specificity while at the same time preserving some associations between subcomplexes.
The essential CBF3 complex is a fundamental determinant of budding yeast kinetochore structure. In its absence all known kinetochore proteins fail to associate with centromeric DNA (for review see Cheeseman et al., 2002a). Interestingly, purification of the CBF3 complex using tagged Cep3p resulted in the recovery of the CBF3 subunits Ctf13 and Skp1p (Fig. 1), but not Ndc10p, which is present in the complex when it is isolated by DNA-affinity chromatography (Lechner and Carbon, 1991). Previous studies have demonstrated a distinct assembly pathway for the CBF3 complex with Ndc10p joining a Skp1p–Ctf13p–Cep3p subcomplex as the final assembly step (Russell et al., 1999). Although no other inner or central kinetochore proteins copurified with Cep3p, two proteins related to kinetochore function were detected in trace amounts. These proteins include Hir1p, which together with the chromatin assembly factor CAF-1 has a role in building functional kinetochores (Sharp et al., 2002), and Cdc4p, a subunit of the SCF ubiquitin ligase. The purification of Cbf1p, which binds to the CDEI centromeric element and shows limited homology to metazoan CENP-B, did not reveal associations with other kinetochore proteins under our experimental conditions.
Figure 1.
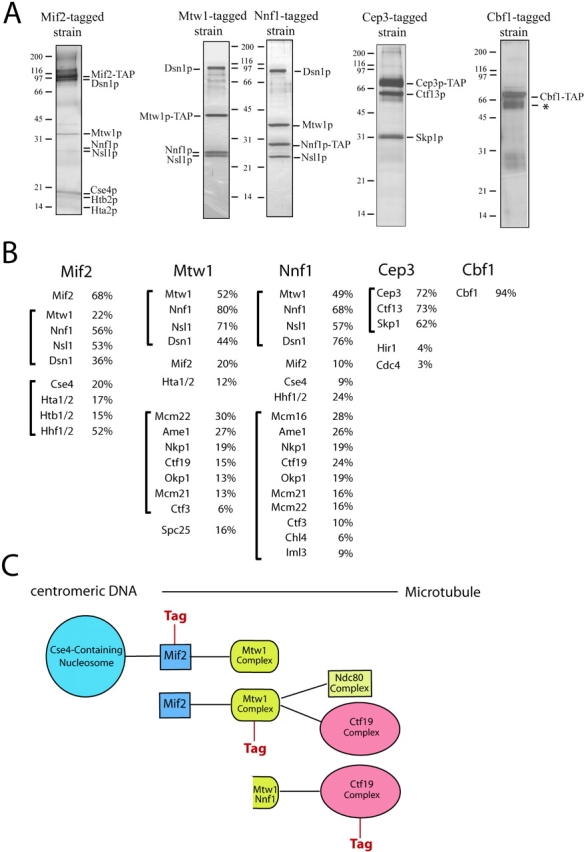
Purification of inner kinetochore proteins. (A) Silver-stained SDS-PAGE gels showing the purification of Mif2p, Cbf1p, the Mtw1 complex, and the CBF3 complex from yeast protein extracts. An asterisk denotes background bands corresponding to the heat shock proteins Ssa1 and Ssa2. (B) Percent sequence coverage obtained from mass spectrometric analysis of each purification shown in A. Brackets show proteins organized into subcomplexes. (C) Schematic diagram showing the associations between the different subcomplexes suggested by their copurification.
Mif2p shows sequence homology to CENP-C, an essential vertebrate kinetochore protein (Meluh and Koshland, 1995, 1997). Strikingly, when we purified a TAP-tagged version of Mif2p from yeast extracts, we found that Mif2p specifically copurified with two distinct sets of proteins (Fig. 1). The first set is composed of the histone H3-related CENP-A homologue Cse4p, and histones H2A, H2B, and H4. The presence of CENP-A proteins is a fundamental feature of all centromeres and they are believed to form a specialized nucleosome found exclusively at centromeric loci (Meluh et al., 1998). As judged by mass spectrometry (Fig. 1 B) and by Western blotting (Fig. S1, available at http://www.jcb.org/cgi/content/full/10.1083/jcb.200305100/DC1), histone H3 was absent from our purification, strongly suggesting that Cse4p completely replaces histone H3 in a centromeric nucleosome. Our purification also implies that a critical role of this specialized nucleosome is to serve as a scaffold for the recruitment of Mif2p/CENP-C.
The second set of proteins that copurifies with Mif2p consists of the kinetochore protein Mtw1p (Goshima and Yanagida, 2000) and the proteins Nnf1p, Dsn1p, and Nsl1p. Recently, a role in chromosome segregation has been demonstrated for the latter three proteins (Euskirchen, 2002). We also found that Nnf1p, Dsn1p, and Nsl1p specifically associate with centromeric DNA in ChIP experiments (Fig. S2, available at http://www.jcb.org/cgi/content/full/10.1083/jcb.200305100/DC1) establishing them as bona fide kinetochore components. Together, these data provide a physical basis for their genetic interactions with Mtw1p (Euskirchen, 2002). As judged by the relative band intensity on Coomassie-stained gels, Cse4p and the histones, as well as Mtw1p, Nnf1p, Dsn1p, and Nsl1p, all copurified substoichiometrically with Mif2p.
In addition to its associations with Mif2p, Mtw1p associates weakly with the Ctf19 complex (Cheeseman et al., 2002b). To examine Mtw1p more closely, we tagged and purified both Mtw1p and Nnf1p. The silver-stained gel (Fig. 1 A) of these purifications shows that an identical set of polypeptides, dominated by four prominent bands corresponding to Mtw1p, Nnf1p, Dsn1p, and Nsl1p, copurified in both cases. Densitometric analysis of a Coomassie-stained gel indicated that these proteins are present in an equimolar ratio, suggesting that they form a distinct kinetochore subcomplex. Therefore, we will refer to this group of proteins as the Mtw1 complex. In addition to these four proteins, mass spectrometric analysis of the Mtw1p and Nnf1p purifications revealed the presence of substoichiometric amounts of Mif2p, confirming its association with the Mtw1 complex. Although small amounts of histones were recovered, the full complement of proteins comprising the Cse4p nucleosome was absent from either sample, suggesting that the Mtw1 complex is more distal from centromeric DNA than Mif2p. Interestingly, mass spectrometric analysis also detected the presence of 9 (Mcm22p, Ame1p, Nkp1p, Ctf19p, Okp1p, Mcm21p, Ctf3p, Chl4p, and Iml3p) of the 11 subunits of the central kinetochore Ctf19 complex that we described previously (Cheeseman et al., 2002b). In addition, we detected the Spc25p subunit of the Ndc80 complex (Janke et al., 2001; Wigge and Kilmartin, 2001) as copurifying with Mtw1p, suggesting a physical association between the Mtw1 complex and a subunit of the central kinetochore Ndc80 complex.
Strikingly, comparing the overlapping sets of copurifying proteins defines a path of connectivity from centromeric DNA to the protein complexes of the central and outer kinetochore (Fig. 1 C). For example, whereas the Mif2p sample copurified with the centromeric nucleosome and the Mtw1 complex, purification of the Mtw1 complex resulted in coisolation of Mif2p and components of the Ctf19 and Ndc80 complexes, but not of the complete centromeric nucleosome, indicating that we had moved one step away from centromeric DNA. Similarly, our previous purification of the Ctf19 complex yielded a small amount of Mtw1p and Nnf1p (Cheeseman et al., 2002b; unpublished data).
Nnf1 displays homology to metazoan CENP-H
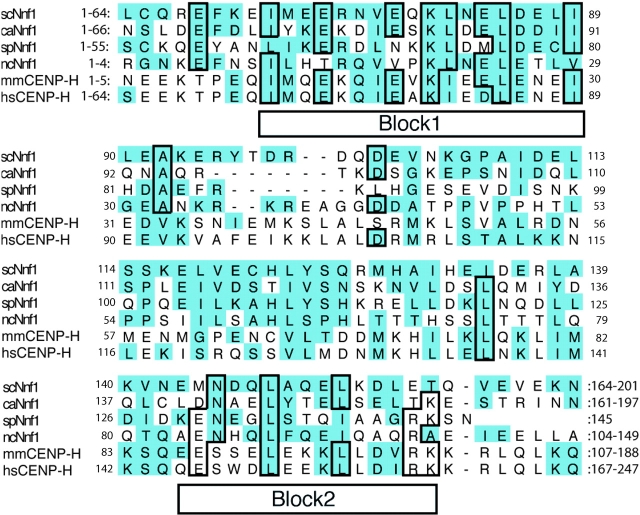
Recently a human homologue of Mtw1p, hMis12, was shown to be essential for faithful chromosome segregation (Goshima et al., 2003). Using the Block Maker (Henikoff et al., 1995) and motif alignment and search tools (MASTs), we identified significant homology between the Mtw1 complex subunit Nnf1p and CENP-H (Fig. 2). CENP-H is a constitutive component of the metazoan inner kinetochore that colocalizes with CENP-A and -C (Sugata et al., 2000; Fukagawa et al., 2001). The homology between the Saccharomyces cerevisiae Nnf1p and human CENP-H protein sequences (17.4% identity and 49% homology over the entire protein) is similar to the level of homology observed between Mtw1p and hMis12 (19% identity and 49% homology). In addition, the overall P value from the MAST was 2.4 × 10−7, similar to the P value of 4.3 × 10−6 obtained from the Mtw1/hMis12 search. We also note that all of the fungal Nnf1 proteins, as well as the vertebrate CENP-H members, are roughly the same size (∼200 aa) and feature a central coiled-coil domain (residues 60–100 and 120–175 in scNnf1, residues 50–200 in hsCENP-H). Thus, at least two subunits of the Mtw1 complex have been conserved throughout evolution.
Figure 2.
Nnf1p is homologous to vertebrate CENP-H. Sequence alignments of four fungal Nnf1p sequences and two vertebrate CENP-H sequences. The conserved blocks identified by Block Maker are indicated. Homologous residues are shaded; and identical residues are boxed (mm = Mus musculus; hs = Homo sapiens; sc = Saccharomyces cerevisiae; ca = Candida albicans; sp = Schizosaccharomyces pombe; and nc = Neuorospora crassa).
Interdependencies in centromere association of kinetochore proteins
To further explore the structural organization of the inner kinetochore and to independently test the biological importance of the subcomplex associations revealed by the copurification experiments, we next performed ChIP analysis of these inner kinetochore proteins in a variety of mutant backgrounds (Fig. 3). We used the TAP-tagged proteins described above and isolated these proteins with Ig G, which binds to the protein A region of the tag. Although it has been shown that Mif2p depends on Ndc10p for association with the centromere (Meluh and Koshland, 1997), we also found that its localization to CEN DNA is substantially diminished in cse4-1 mutants at the restrictive temperature (Fig. 3 A). In contrast, ndc80-1 mutants had no effect on the centromeric association of Mif2p. The dependency between Cse4p and Mif2p agrees with the observation that vertebrate CENP-C is mislocalized in a CENP-A knockout (Howman et al., 2000), but is recruited along with CENP-A to ectopic sites when CENP-A is overexpressed (Van Hooser et al., 2001). Intriguingly, the Mif2p signal was strongly reduced in both mtw1-1 and nnf1-17 mutants at the restrictive temperature (Fig. 3 A) supporting the observation that vertebrate CENP-H is required for the targeting of CENP-C to centromeres (Fukagawa et al., 2001). In the reciprocal experiment, we observed a substantial reduction of the Mtw1p signal in a mif2-3 mutant at the restrictive temperature. Therefore, we conclude that Mif2p and the Mtw1p complex are interdependent for full association with the centromere, consistent with their close association revealed in our affinity purification experiments.
Figure 3.

Analysis of kinetochore subcomplex organization by ChIP. Agarose gels (left column) showing ChIP analysis of TAP-tagged kinetochore proteins in various mutant backgrounds. CEN3 DNA was amplified by PCR from either total chromatin solution (Total, serial dilutions 1:32, 1:64, and 1:128), an immunoprecipitate (IP), or a mock-treated control (−). Control reactions show no amplification of noncentromeric DNA (PGK1) in the immunoprecipitates. Quantitation of the ChIP results (right column): the amount of CEN3 DNA in the immunoprecipitate at the permissive (25°C) and restrictive temperature (37°C) is expressed as the percentage of the DNA in the lysate. Western blotting demonstrated that there was no significant change in the level of the tagged proteins 3 h after shifting to the restrictive temperature (Fig. S3, available at http://www.jcb.org/cgi/content/full/10.1083/jcb.200305100/DC1).
Next, we determined whether there are additional kinetochore proteins that depend on the Mtw1 complex for centromeric localization. We found that the Ctf19 complex subunits Ctf19p (Fig. 3) and Chl4p (not depicted) persist at centromeres in mtw1-1 and nnf1-17 mutants at the restrictive temperature. However, the Ctf19p signal was strongly reduced in the mif2-3 mutant, demonstrating complexity in the centromere localization dependencies of Ctf19 complex subunits. The association between subunits of the Mtw1 complex and the Ndc80 complex identified in our purifications is supported by the observation that the centromere association of the Ndc80 subunit Spc24p was strongly reduced in an nnf1-17 mutant, whereas in the reciprocal experiment Mtw1p or Nnf1p did not depend on Ndc80p for their association with the centromere. A summary of the localization dependencies observed in the ChIP experiments is presented in Table I. The associations and dependencies detected between Cse4p, Mif2p, the Mtw1 complex, and the Ndc80 complex in comparison with results obtained in metazoans, suggest that a structural CENP-A, -C, -H, and Ndc80/HEC core mediates the connectivity between centromeric DNA and proteins of the central and outer kinetochore, and has been conserved from yeast to metazoans.
Table I. Summary of the dependencies in centromeric association determined by chromatin immunoprecipitation.
| cse4-1 | mif2-3 | mtw1-1 | nnf1-17 | ndc80-1 | |
|---|---|---|---|---|---|
| Mif2-TAP | − | n.a | − | − | + |
| Mtw1-TAP | − | − | n.a. | n.d. | + |
| Ctf19-TAP | n.d. | − | + | + | n.d. |
| Spc24-TAP | n.d. | n.d. | − | − | n.a. |
−, reduced centromeric association; +, persistent centromeric association; n.a., not applicable; n.d., not done.
Phosphoregulation of inner kinetochore proteins
We have described previously the phosphoregulation of the central and outer kinetochore by the Aurora kinase Ipl1p (Cheeseman et al., 2002b). To determine whether inner kinetochore proteins are also regulated by phosphorylation, we mapped in vivo phosphorylation sites in the kinetochore proteins described in this paper. Mass spectrometric analysis identified two phosphorylation sites in Mif2p (serine 54 and 325). Interestingly, serine 54 lies within a sequence that directly matches the previously established Ipl1p consensus site (Cheeseman et al., 2002b). We also detected phosphorylation of the Mtw1 complex subunit Dsn1p at serine 250, and phosphorylation of the CBF3 component Cep3p at serine 574 (Fig. 4 A). To determine whether any of these proteins are substrates for Ipl1p in vitro, we purified the Mif2, Mtw1, and CBF3 complexes and treated the samples with λ-phosphatase to remove the endogenous phosphate groups. When we incubated the isolated complexes in the presence of Ipl1p purified from Escherichia coli and γ-[32P]ATP, we found that Mif2p, as well as the Mtw1 complex subunit Dsn1p, were readily phosphorylated by Ipl1p in vitro (Fig. 4 B).
Figure 4.
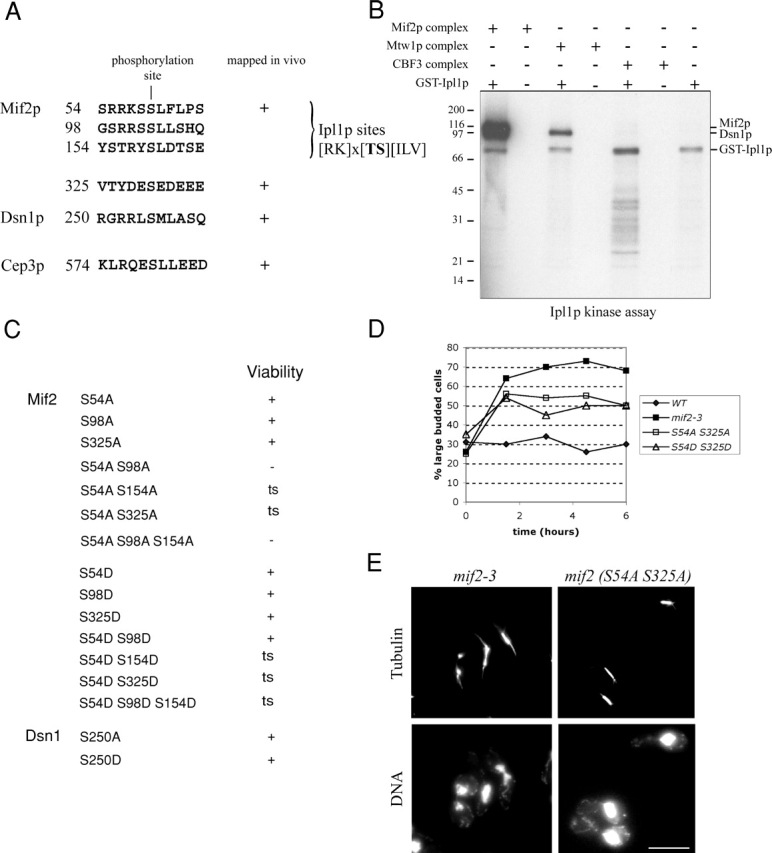
Phosphoregulation of inner kinetochore proteins. (A) Phosphorylation sites identified in inner kinetochore proteins by mass spectrometry. (B) Mif2p and Dsn1p are direct targets of Ipl1p in vitro. Mif2, Mtw1, and CBF3 inner kinetochore complexes were purified and dephosphorylated with λ-phosphatase. In vitro phosphorylation with E. coli GST-Ipl1p was analyzed by autoradiography. Comparison with a Coomassie-stained gel (not depicted) demonstrates that Mif2p and Dsn1p are direct targets of Ipl1p. (C) Mutational analysis of the phosphorylation sites in inner kinetochore proteins. Growth on rich medium (YPD) is indicated (ts, temperature sensitive). (D) mif2 temperature-sensitive phosphorylation mutants display a metaphase arrest: cells were grown to mid-log phase and shifted to the restrictive temperature of 37°C at t = 0. The percentage of large-budded cells was determined in fixed and sonicated samples at the indicated time points. (E) Analysis of mif2 mutants 3 h after shift to 37°C. mif2-3 mutants display large-budded cells with broken down or weakened spindles (left column). The mif2 (S54A S325A) mutant shows large-budded cells with a short spindle and a single DNA mass. Bar, 5 μm.
To test the importance of the phosphorylation sites identified above, we systematically mutated the target residues, alone or in combination, to either alanine (to eliminate phosphorylation at that site) or aspartate (to mimic the constitutive phosphorylation at that site). Because serine 54 of Mif2p corresponded to an Ipl1p target site, and because Mif2p was strongly phosphorylated by Ipl1p in vitro, we also included two additional predicted Ipl1p sites within the Mif2 sequence (serine 98 and 154) in our mutational analysis. This analysis failed to produce an observable growth phenotype for dsn1 (Fig. 4 C) and cep3 mutants (not depicted). Similarly, individually mutating each of the phosphorylation sites in Mif2p showed no effect. However, mutating both in vivo phosphorylation sites to alanine (S54A S325A) resulted in temperature sensitivity at 37°C. Moreover, mutating serine 54 to alanine in combination with a second predicted Ipl1p site (mif2 S54A S98A) resulted in lethality, suggesting that phosphorylation of Mif2p on either serine 54 or 98 is essential for viability. In contrast, the corresponding aspartate double mutant (mif2 S54D S98D) was alive at 25°C but displayed very slow growth at 37°C. To determine the consequences of disrupting Mif2p phosphorylation on spindle function, we next examined the phenotypes associated with the different mif2 phosphorylation site mutants. A previously described temperature-sensitive allele of mif2 (mif2-3; Meluh and Koshland, 1995) causes a cell cycle arrest with greater than 70% large-budded cells and shows a variety of mitotic defects including broken down spindles with abnormal DNA segregation. Mif2 (S54A S325A) and mif2 (S54D S325D) cultures shifted to 37°C display an accumulation of large-budded cells that reached 55% for mif2 (S54A S325A) compared with around 30% for wild-type cells (Fig. 4 D). In contrast to mif2-3, mif2 (S54A S325A) or mif2 (S54A S154A) mutants did not show spindle defects, but 80% of the large-budded cells had a short spindle and a single DNA mass (Fig. 4 E, right) indicative of a metaphase arrest. Western blotting with an affinity-purified Mif2 antibody indicated that there was no significant change in the protein levels 3 h after shifting to the restrictive temperature (unpublished data).
We conclude that phosphorylation plays a critical role in Mif2 function. Mif2p is likely regulated by both Ipl1p and by at least one other, as yet unidentified, kinase because serine 325 lies within an acidic sequence, which is not characteristic of an Ipl1p site. The analysis presented here brings the total number of in vivo Ipl1p targets at the kinetochore to eight (Ipl1p, Dam1p, Spc34p, Ask1p, Ndc80p, Sli15p, Mif2p, and Dsn1p). Interestingly, these targets are distributed throughout the inner, central, and outer kinetochore, suggesting that Ipl1p acts at multiple levels to regulate kinetochore structure and function. Intriguingly, based on the phenotypes observed for the mif2 phosphorylation site mutants, Mif2p phosphorylation by Ipl1p appears to play a distinct role from the phosphorylation of the Dam1 complex (Cheeseman et al., 2002b). This suggests that phosphorylation of Mif2p may not be part of the Dam1p pathway required for the establishment of chromosome biorientation (Cheeseman et al., 2002b), but instead contributes to a distinct aspect of kinetochore regulation.
An updated model for the budding yeast kinetochore
Fig. 5 represents an updated model of the budding yeast kinetochore incorporating the information presented in this paper. Interestingly, whereas the CBF3 complex is essential for kinetochore function in budding yeast, our purifications did not reveal significant physical interactions between CBF3 and other kinetochore proteins. No obvious homologues of the CBF3 complex have been detected in Schizosaccharomyces pombe or in metazoans, suggesting that its role may be specialized for the short, nucleotide sequence-constrained centromere unique to budding yeast. In contrast, our purifications show that the interactions establishing connectivity between centromeric DNA and central/outer kinetochore complexes are mediated by conserved proteins consisting of Cse4p/CENP-A, Mif2p/CENP-C, Mtw1p/hMis12, Nnf1p/CENP-H, Nuf2p/hNuf2, and Ndc80/HEC. Fig. 5 B highlights this conserved protein core. It is tempting to speculate that the much larger vertebrate kinetochore is a repetitive expansion of this proposed single core present in budding yeast.
Figure 5.
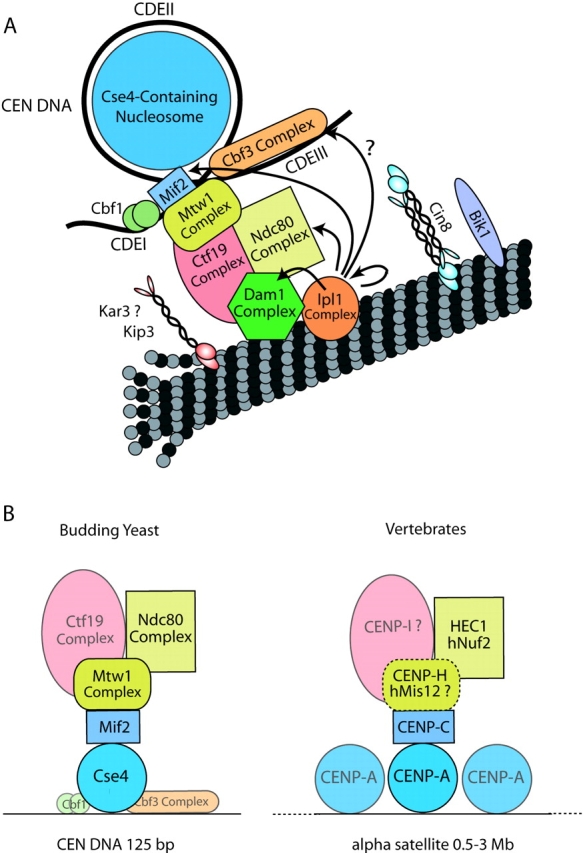
Revised model of budding yeast kinetochore structure. (A) Updated model of the budding yeast kinetochore incorporating the interaction between Mif2p and the centromeric nucleosome, the positioning of the Mtw1 complex and the newly identified Ipl1p target. (B) Illustration of a structural core of conserved proteins based on the copurification and ChIP experiments. A dotted line indicates a putative complex between vertebrate CENP-H and hMis12 that awaits experimental verification.
Materials and methods
Strains and growth conditions
Yeast strains used in this work are listed in Table SI, available at http://www.jcb.org/cgi/content/full/10.1083/jcb.200305100/DC1. COOH-terminal S-TEV-ZZ (TAP) tags and deletions of MIF2, DSN1 and CEP3 were constructed by PCR as described previously (Cheeseman et al., 2002b). Phosphorylation site mutants were generated using QuikchangeTM site directed mutagenesis (Stratagene), cloned into pRS306, and integrated at the URA3 locus. All growth experiments were conducted in YPD (YP + 2% dextrose). Geneticin (G418; GIBCO BRL) was used at 0.4 mg/ml.
Complex purifications
Purification of kinetochore complexes was conducted as described previously (Cheeseman et al., 2001) except that 300 mM KCl was used throughout the study. Identification of proteins and phosphorylation sites by mass spectrometry were performed as described previously (Cheeseman et al., 2001; MacCoss et al., 2002). In vitro phosphorylation of kinetochore complexes by GST-Ipl1p from E. coli was performed as described previously (Cheeseman et al., 2002b).
Sequence characterization
Fungal homologues of Nnf1p were identified using basic local alignment search tool searches of the Candida albicans (http://www-sequence.stanford.edu/group/candida), S. pombe (http://www.sanger.ac.uk/Projects/S_pombe), Neurospora crassa (http://www-genome.wi.mit.edu/cgi-bin/annotation/neurospora/blast_page.cgi?organismName=Neurospora), and Magnaporthe grisea (http://www-genome.wi.mit.edu/cgi-bin/annotation/magnaporthe/blast_page.cgi?organismName=Magnaporthe) web sites. Block Maker (http://www.blocks.fhcrc.org/blockmkr/make_blocks.html) was used to align conserved blocks of the fungal sequences. MAST (http://meme.sdsc.edu/meme/website/mast-intro.html) was used to search the nonredundant database for high-scoring sequences with two generated blocks as input. Coiled coils were identified using the COILS server (http://www.ch.embnet.org/software/COILS_form.html). All programs were used with default settings.
ChIP
Immunoprecipitation of formaldehyde cross-linked chromatin was performed as described previously (Enquist-Newman et al., 2001) with the following modifications: immunoprecipitations on TAP-tagged strains were conducted overnight with 0.1 mg/ml rabbit Ig G (Sigma-Aldrich) at 4°C. Immune complexes were subsequently isolated on protein A–Sepharose CL-4B beads (Amersham Biosciences). PCR used BIO-X-ACT polymerase (Bioline) and were typically run for 29 cycles. PCR products were resolved on 2.5% agarose gels and visualized with ethidium bromide. Stained gels were quantified using a Gel-Doc 1000 system (Bio-Rad Laboratories) and ImageQuant software (Molecular Dynamics).
Immunofluorescence microscopy
Indirect immunofluorescence microscopy on intact yeast cells was performed as described previously (Cheeseman et al., 2002b). The YOL134 antitubulin antibody (Accurate Chemical and Scientific Corporation) was used at a dilution of 1:200. Fluorescein-conjugated anti-IgG heavy chain secondary antibodies (Cappel/Organon Technika Inc. or Jackson Laboratory) were used at 1:500. Light microscopy was performed using a microscope (model TE300; Nikon) equipped with a 100×/1.4 Plan-Apo objective and a cooled CCD camera (model Orca-100; Hamamatsu) controlled by Phase-3 software (Phase-3 Imaging Systems).
Online supplemental material
Fig. S1 shows a Western analysis of purified Mif2p complex with a histone H3-specific antibody (provided by P. Kaufman and J. Sharp, University of California, Berkeley, CA). Fig. S2 provides a ChIP analysis of the proteins Nnf1p, Nsl1p, and Dsn1p. Fig. S3 confirms the presence of TAP-tagged proteins in various mutant backgrounds by Western blotting. Table S1 provides a list of the yeast strains used in this paper. Online supplemental material is available at available at http://www.jcb.org/cgi/content/full/10.1083/jcb.200305100/DC1.
Supplemental Material
Acknowledgments
The authors thank Yuko Nakajima, Ching Shang, and Jonathan Wong for discussions; Dan Foltz and Ben Black for critical reading of the manuscript; G. Euskirchen, M. Fitzgerald-Hayes, J. Kilmartin, M. Yanagida, and P. Meluh for strains; and P. Kaufman and J. Sharp for antibodies.
This work was supported by grants from the National Institute of General Medical Sciences to G. Barnes (GM-47842); a grant to T. Davis and the Yeast Research Center from the National Center for Research Resources of the National Institutes of Health (Comprehensive Biology: Exploiting the Yeast Genome; PHS P41 RR11823); a National Science Foundation Graduate Research Fellowship to I.M. Cheeseman; and a fellowship of the Deutsche Forschungsgemeinschaft to S. Westermann.
The online version of this article contains supplemental material.
The present address of I.M. Cheeseman is Ludwig Institute for Cancer Research University of California, San Diego, La Jolla, CA 92093-0660.
Abbreviations used in this paper: ChIP, chromatin immunoprecipitation; MAST, motif alignment and search tool; TAP, tandem affinity purification.
References
- Cheeseman, I.M., C. Brew, M. Wolyniak, A. Desai, S. Anderson, N. Muster, J.R. Yates, T.C. Huffaker, D.G. Drubin, and G. Barnes. 2001. Implication of a novel multiprotein Dam1p complex in outer kinetochore function. J. Cell Biol. 155:1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., D.G. Drubin, and G. Barnes. 2002. a. Simple centromere, complex kinetochore: linking spindle microtubules and centromeric DNA in budding yeast. J. Cell Biol. 157:199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman, I.M., S. Anderson, M. Jwa, E.M. Green, J. Kang, J.R. Yates, C.S.M. Chan, D.G. Drubin, and G. Barnes. 2002. b. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 111:163–172. [DOI] [PubMed] [Google Scholar]
- Cleveland, D.W., Y. Mao, and K.F. Sullivan. 2003. Centromeres and kinetochores: from epigenetics to mitotic checkpoint signalling. Cell. 112:407–421. [DOI] [PubMed] [Google Scholar]
- Enquist-Newman, M., I.M. Cheeseman, D. Van Goor, D.G. Drubin, P. Meluh, and G. Barnes. 2001. Dad1p, third component of the Duo1p/Dam1p complex involved in kinetochore function and mitotic spindle integrity. Mol. Biol. Cell. 12:2601–2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euskirchen, G.M. 2002. Nnf1p, Dsn1p, Mtw1p and Nsl1p: a new group of proteins important for chromosome segregation in Saccharomyces cerevisiae. Eukaryot. Cell. 1:229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukagawa, T., Y. Mikami, A. Nishihashi, V. Regnier, T. Haraguchi, Y. Hiraoka, N. Sugata, K. Todokoro, W. Brown, and T. Ikemura. 2001. CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J. 20:4603–4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima, G., and M. Yanagida. 2000. Establishing biorientation occurs with precocious separation of the sister kinetochores, but not the arms, in the early spindle of budding yeast. Cell. 100:619–633. [DOI] [PubMed] [Google Scholar]
- Goshima, G., T. Kiyomitsu, K. Yoda, and M. Yanagida. 2003. Human centromere chromatin protein hMis12, essential for equal segregation, is independent of CENP-A loading pathway. J. Cell Biol. 160:25–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, X., D.R. Rines, C.W. Espelin, and P.K. Sorger. 2001. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 106:195–206. [DOI] [PubMed] [Google Scholar]
- Henikoff, S., J.G. Henikoff, W.J. Alford, and S. Pietrokovski. 1995. Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene. 163:GC17–GC26. [DOI] [PubMed] [Google Scholar]
- Howman, E.V., K.J. Fowler, A.J. Newson, S. Redward, A.C. MacDonald, P. Kalitsis, and K.H. Choo. 2000. Early disruption of centromeric chromatin organization in centromere protein A (CENP-A) null mice. Proc. Natl. Acad. Sci. USA. 97:1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, C., J. Ortiz, J. Lechner, A. Shevchenko, M.M. Magiera, C. Schramm, and E. Schiebel. 2001. The budding yeast proteins Spc24p and Spc25p interact with Ndc80p and Nuf2p at the kinetochore and are important for kinetochore clustering and checkpoint control. EMBO J. 20:777–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janke, C., J. Ortiz, T.U. Tanaka, J. Lechner, and E. Schiebel. 2002. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 21:181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner, J., and J. Carbon. 1991. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 64:717–725. [DOI] [PubMed] [Google Scholar]
- Li, Y., J. Bachant, A.A. Alcasabas, Y. Wang, J. Qin, and S.J. Elledge. 2002. The mitotic spindle is required for loading of the DASH complex onto the kinetochore. Genes Dev. 16:183–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCoss, M.J., W.H. McDonald, A. Saraf, R. Sadygov, J.M. Clark, J.J. Tasto, K.L. Gould, D. Wolters, M. Washburn, A. Weiss, et al. 2002. Shotgun identification of potein modifications from protein complexes and lens tissue. Proc. Natl. Acad. Sci. USA. 99:7900–7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measday, V., D.W. Hailey, I. Pot, S. Givan, K.M. Hyland, G. Cagney, S. Fields, T.N. Davis, and P. Hieter. 2002. Ctf3p, the Mis6 budding yeast homolog, interacts with Mcm22p and Mcm16p at the yeast outer kinetochore. Genes Dev. 16:101–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor, J., W. Jiang, M. Funk, J. Rathjen, C.A. Barnes, T. Hinz, J.H. Hegemann, and P. Philippsen. 1990. CPF1, a yeast protein which functions in centromeres and promoters. EMBO J. 9:4017–4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P.B., and D. Koshland. 1995. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol. Biol. Cell. 6:793–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P.B., and D. Koshland. 1997. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 11:3401–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh, P.B., P. Yang, L. Glowczewski, D. Koshland, and M.M. Smith. 1998. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 94:607–613. [DOI] [PubMed] [Google Scholar]
- Ortiz, J., O. Stemmann, S. Rank, and J. Lechner. 1999. A putative protein complex consisting of Ctf19, Mcm21, and Okp1 represents a missing link in the budding yeast kinetochore. Genes Dev. 13:1140–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pot, I., V. Measday, B. Snydsman, G. Cagney, S. Fields, T.N. Davis, E.G.D. Miller, and P. Hieter. 2003. Chl4p and Iml3p are two new members of the budding yeast outer kinetochore. Mol. Biol. Cell. 14:460–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, I.D., A.S. Grancell, and P.K. Sorger. 1999. The unstable F-Box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex. J. Cell Biol. 145:933–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang, C., T.Z. Hazburn, I.M. Cheeseman, J. Aranda, S. Fields, D.G. Drubin, and G. Barnes. 2003. Kinetochore protein interactions and their regulation by the Aurora kinase Ipl1p. Mol. Biol. Cell. 14:3342–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, J.A., A.A. Franco, M.A. Osley, and P.D. Kaufman. 2002. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 16:85–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler, S., K.C. Keith, K.E. Curnick, and M. Fitzgerald-Hayes. 1995. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 9:573–586. [DOI] [PubMed] [Google Scholar]
- Sugata, N., S. Li, W.C. Earnshaw, T.J. Yen, K. Yoda, H. Masumoto, E. Munekata, P.E. Warburton, and K. Todokoro. 2000. Human CENP-H multimers colocalize with CENP-A and CENP-C at active centromere-kinetochore complexes. Hum. Mol. Genet. 9:2919–2926. [DOI] [PubMed] [Google Scholar]
- Van Hooser, A.A., I.I. Ouspenski, H.C. Gregson, D.A. Starr, T.J. Yen, M.L. Goldberg, K. Yokomori, W.C. Earnshaw, K.F. Sullivan, and B.R. Brinkley. 2001. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J. Cell Sci. 114:3529–3542. [DOI] [PubMed] [Google Scholar]
- Wigge, P.A., and J.V. Kilmartin. 2001. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J. Cell Biol. 152:349–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.