Abstract
The sorting of transmembrane proteins to endosomes and lysosomes is mediated by signals present in the cytosolic tails of the proteins. A subset of these signals conform to the [DE]XXXL[LI] consensus motif and mediate sorting via interactions with heterotetrameric adaptor protein (AP) complexes. However, the identity of the AP subunits that recognize these signals remains controversial. We have used a yeast three-hybrid assay to demonstrate that [DE]XXXL[LI]-type signals from the human immunodeficiency virus negative factor protein and the lysosomal integral membrane protein II interact with combinations of the γ and σ1 subunits of AP-1 and the δ and σ3 subunits of AP-3, but not the analogous combinations of AP-2 and AP-4 subunits. The sequence requirements for these interactions are similar to those for binding to the whole AP complexes in vitro and for function of the signals in vivo. These observations reveal a novel mode of recognition of sorting signals involving the γ/δ and σ subunits of AP-1 and AP-3.
Keywords: clathrin; adaptors; coats; endocytosis; endosomes
Introduction
The sorting of many transmembrane proteins at the plasma membrane, the TGN, and the endosomes is mediated by interactions of signals present in the cytosolic domains of the transmembrane proteins with adaptor proteins (APs) that are components of membrane coats (Mellman, 1996; Hirst and Robinson, 1998; Bonifacino and Traub, 2003). Most of these sorting signals consist of short, linear arrays of amino acid residues that fit one of several consensus motifs (Bonifacino and Traub, 2003).
“Tyrosine-based” sorting signals contain a critical tyrosine residue and conform to either NPXY or YXXØ motifs (Bonifacino and Traub, 2003) (N, asparagine; P, proline; X, any amino acid; Y, tyrosine; Ø, bulky hydrophobic amino acid). NPXY signals were originally discovered in the cytosolic tail of the low density lipoprotein receptor (Chen et al., 1990) and are now known to interact with the phosphotyrosine-binding domain of a family of monomeric clathrin adaptors that includes the autosomal recessive form of hypercholesterolemia protein (He et al., 2002; Mishra et al., 2002) and Disabled 2 (Morris and Cooper, 2001). YXXØ signals (Canfield et al., 1991; Collawn et al., 1991) are typically found in another subset of endocytic receptors such as the transferrin receptor, as well as in many transmembrane proteins targeted to endosomes and lysosomes (Bonifacino and Traub, 2003). Recognition of these signals is a function of the μ (medium) subunits of the heterotetrameric AP complexes AP-1, AP-2, AP-3, and AP-4 (Ohno et al., 1995, 1998; Owen and Evans, 1998; Fig. 1 A). AP-1 and AP-2 have long been known to be components of clathrin coats associated with the TGN/endosomes and the plasma membrane, respectively (Hirst and Robinson, 1998). AP-3 appears to exist as part of both clathrin and nonclathrin coats localized to endosomes, whereas AP-4 is most likely part of a nonclathrin coat associated with the TGN (Robinson and Bonifacino, 2001).
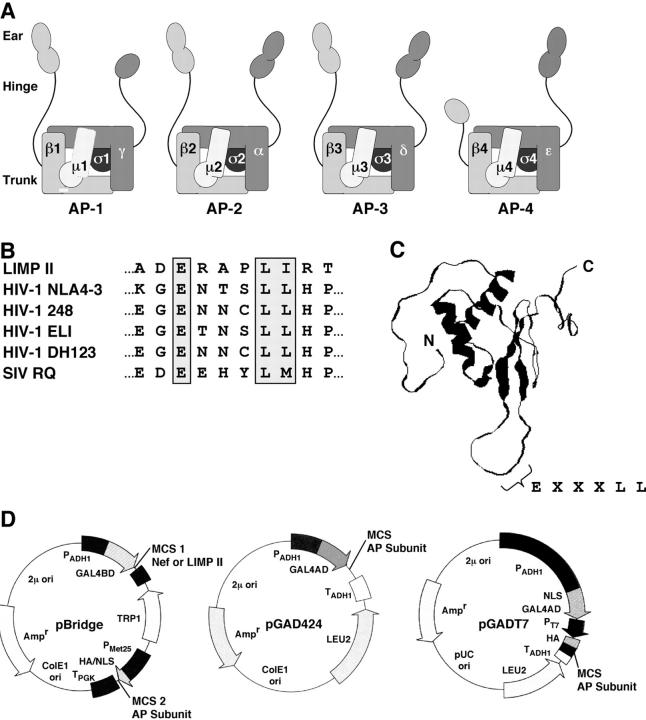
Figure 1.
Adaptors, signals, and plasmids used in this work. (A) Schematic representation of the heterotetrameric adaptor protein (AP) complexes. The designations of the generic subunits of each complex are indicated. The following subunits occur in various isoforms encoded by different genes, as indicated in parentheses: γ (γ1, γ2), μ1 (μ1A, μ1B), σ1 (σ1A, σ1B, σ1C), α (αA, αC), β3 (β3A, β3B), μ3 (μ3A, μ3B), and σ3 (σ3A, σ3B). The arrangement of the subunits was modeled after the crystal structure of the AP-2 core (Collins et al., 2002). The trunk, hinge, and ear domains of the large subunits are indicated. (B) Sequences of the dileucine-based sorting signals from LIMP-II and variants of HIV-1 and SIV Nef. The critical glutamate, leucine–leucine, leucine–isoleucine, and leucine–methionine residues are boxed. (C) NMR structure of HIV-1 Nef (BH10 isolate). The structure shown corresponds to the first of 40 structures determined by multidimensional heteronuclear NMR spectroscopy (Grzesiek et al., 1997). The locations of the NH2 and COOH termini, and of the EXXXLL sequence, are indicated. (D) Yeast expression plasmids used in two- and three-hybrid assays. The pBridge plasmid expresses two proteins, GAL4BD fused to Nef or the LIMP-II cytosolic tail (multiple cloning site 1, MCS 1) and an AP subunit (MCS 2). The pGAD424 and pGADT7 plasmids drive high-level expression of GAL4AD fused to another AP subunit.
Another major group of sorting signals is referred to as “dileucine-based” and contains critical leucine–leucine or leucine–isoleucine residues. It has recently become apparent that there are at least two distinct types of dileucine-based sorting signals defined by the consensus motifs DXXLL and [DE]XXXL[LI] (D, aspartate; E, glutamate; L, leucine; I, isoleucine) (Bonifacino and Traub, 2003). DXXLL signals occur within the cytosolic tails of intracellular sorting receptors such as the mannose 6-phosphate receptors (MPRs) and mediate sorting between the TGN and endosomes (Johnson and Kornfeld, 1992; Chen et al., 1997). It is now well established that DXXLL signals bind to the Vps27, Hrs, and Stam (VHS) domain of the Golgi-localized, γ-ear–containing, ARF-binding proteins (GGAs; Nielsen et al., 2001; Puertollano et al., 2001; Takatsu et al., 2001b; Zhu et al., 2001), which is another family of monomeric adaptors associated with clathrin coats at the TGN and endosomes.
[DE]XXXL[LI] signals (Letourneur and Klausner, 1992; Pond et al., 1995) are present in the cytosolic tails of numerous transmembrane proteins targeted to endosomes, lysosomes, and lysosome-related organelles (Bonifacino and Traub, 2003). A well-characterized example of proteins having [DE]XXXL[LI] signals (e.g., ERAPLI) is the lysosomal integral membrane protein II (LIMP-II; Barriocanal et al., 1986; Fig. 1 B). The leucine–leucine or leucine–isoleucine pairs are critical elements of the signals, whereas the aspartate or glutamate residue at position −4 from the first leucine is often important but not always essential for function. Interestingly, sequences resembling the canonical [DE]XXXL[LI] motif are also found in the negative factor (Nef) protein of all isolates of the human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV; Fig. 1 B). Unlike LIMP-II, Nef is not a membrane-spanning protein but an N-myristoylated protein tethered to the cytosolic leaflet of membranes (Geyer et al., 2001). A variable EXXXLL sequence is located in a solvent-exposed, unstructured loop near the COOH terminus of HIV-1 Nef (Lee et al., 1996; Grzesiek et al., 1997; Fig. 1 C). This sequence has proven critical for the ability of Nef to down-regulate the CD4 coreceptor from the surface of helper T cells, and has therefore been proposed to function in a manner similar to conventional [DE]XXXL[LI] signals (Bresnahan et al., 1998; Craig et al., 1998; Greenberg et al., 1998).
[DE]XXXL[LI] signals do not bind to the VHS domain of the GGAs (Puertollano et al., 2001), this is in part due to the unfavorable position of the acidic residue upstream of the dileucine pair (−4 vs. −3 relative to the first leucine; Misra et al., 2002; Shiba et al., 2002). Instead, some [DE]XXXL[LI] signals, including those of LIMP-II and HIV-1 Nef, bind to AP-1, AP-2, and AP-3 complexes in vitro (Dietrich et al., 1997; Bresnahan et al., 1998; Höning et al., 1998; Fujita et al., 1999; Hofmann et al., 1999; Peden et al., 2001; Kongsvik et al., 2002; Rodionov et al., 2002; Janvier et al., 2003). However, the identity of the subunits of the AP complexes that harbor the binding site for [DE]XXXL[LI] signals remains controversial. Using various in vitro binding and yeast two-hybrid assays, [DE]XXXL[LI] signals have been shown to interact with the μ subunits of AP-1, AP-2, and/or AP-3 in some studies (Bremnes et al., 1998; Rodionov and Bakke, 1998; Hofmann et al., 1999; Craig et al., 2000), and with the β subunits of AP-1 and AP-2 in other studies (Greenberg et al., 1998; Rapoport et al., 1998; Geyer et al., 2002).
Our lab has made extensive use of the yeast two-hybrid system to characterize interactions of YXXØ signals with AP μ subunits (Ohno et al., 1995, 1998) and of DXXLL signals with the VHS domains of the GGAs (Puertollano et al., 2001; Kato et al., 2002). However, our attempts to demonstrate interactions of [DE]XXXL[LI] signals with single subunits of AP complexes using the yeast two-hybrid system have yielded only negative or marginally positive results. We reasoned that our failure to detect strong [DE]XXXL[LI]–AP subunit interactions using the yeast two-hybrid system could reflect a requirement for more than one subunit. This would be the case if the binding site involved residues on more than one subunit or if the isolated subunit containing the binding site did not fold properly in the absence of another subunit. To test this hypothesis, we resorted to a yeast three-hybrid system in which [DE]XXXL[LI] signals from HIV-1 Nef and LIMP-II were examined for interactions with combinations of two subunits from all four AP complexes.
Results
Yeast three-hybrid analysis of the interactions of HIV-1 Nef with combinations of two AP subunits
The yeast three-hybrid assay consisted of coexpressing the following: (a) the GAL4 DNA binding domain (GAL4BD) fused to HIV-1 Nef (NLA4-3 variant, herein referred to as Nef unless otherwise specified; cloned in multiple cloning site 1 of pBridge), (b) one of the AP subunits (cloned in multiple cloning site 2 of pBridge), and (c) the GAL4 transcription activation domain (GAL4AD) fused to another AP subunit (cloned in pGAD424 or pGADT7; Fig. 1 D). The NLA4-3 variant of Nef contains the dileucine-based signal ENTSLL (Fig. 1 B), which is required for induction of CD4 down-regulation in vivo (Bresnahan et al., 1998; Craig et al., 1998) and for interactions with the AP-1 and AP-3 complexes in vitro (Bresnahan et al., 1998; Janvier et al., 2003). Interactions in the three-hybrid system were detected by growth in histidine-deficient medium or expression of β-galactosidase activity. Using this assay, we were unable to detect interactions of Nef with different combinations of β with either μ (Fig. 2 A) or σ subunits (Fig. 2 B) from any of the four complexes. However, these assays revealed relatively strong interactions of Nef with γ1–σ1A and δ–σ3A, but not αC–σ2 and ɛ–σ4 (Fig. 2, B and D), pairs of subunits that are known or expected to interact with one another in the intact complexes (Page and Robinson, 1995; Takatsu et al., 2001a; Collins et al., 2002; Fig. 1 A). All of the homologous subunits were expressed at comparable levels (Fig. 2 C), indicating that the failure of the αC–σ2- and ɛ–σ4-transformed strains to grow was not due to the absence of these subunits. All other combinations of γ1/αC/δ/ɛ with σ1–4 tested negative in this assay (Fig. 2 D). Interactions of Nef with γ1–σ1A appeared stronger than those with δ–σ3A, as judged from the following: (a) the expression of β-galactosidase activity only by the γ1–σ1A-expressing strain (Fig. 2 D), (b) the faster growth in histidine-deficient liquid medium of the γ1–σ1A-expressing strain (Fig. 2 E), and (c) the higher concentrations of 3-aminotriazole (3AT) needed to inhibit growth of the γ1–σ1A-expressing strain (Fig. 2 F). These observations are entirely consistent with previous reports of interactions of HIV-1 Nef with the intact AP-1 and AP-3, but not AP-2, in vitro (Bresnahan et al., 1998; Janvier et al., 2003). Thus, the ability of Nef to interact with AP-1 and AP-3 appears to be a function of the γ1–σ1A and δ–σ3A hemicomplexes.
Figure 2.
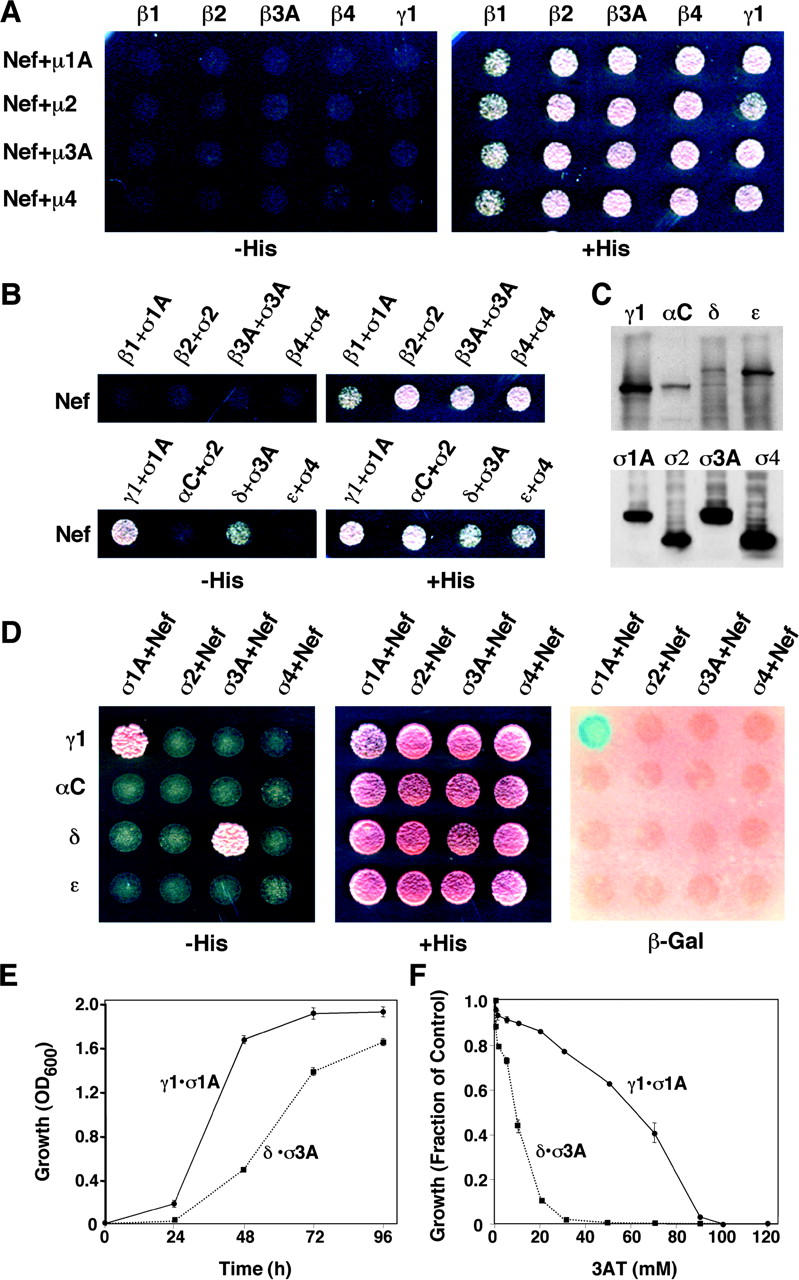
Three-hybrid analysis of the interaction of HIV-1 Nef (NLA4-3 variant) with different AP subunits. (A) GAL4BD-Nef and the μ subunits were expressed from pBridge, whereas the β subunits and γ1 were expressed as fusions with GAL4AD from pGADT7. (B) GAL4BD-Nef and the σ subunits were expressed from pBridge, whereas the β1, β2, β3A, β4, γ1, αC, δ, and ɛ were expressed as fusions with GAL4AD from pGADT7. (C) Immunoblot analysis (using anti-HA) of AP subunits in yeast cells cotransformed with γ1–σ1A, αC–σ2, δ–σ3A, or ɛ–σ4. The subunits were expressed as in B, and all subunits were tagged with the HA epitope. (D) GAL4BD-Nef and the σ subunits were expressed from pBridge, whereas γ1, αC, δ, and ɛ were expressed as fusions with GAL4AD from pGADT7. In A, B, and D, interactions were evidenced by growth on agar plates made with medium without histidine (−His) or by expression of β-galactosidase (β-Gal) activity. (E) Time course of growth of yeast strains coexpressing Nef with either γ1–σ1A (γ1•σ1A) or δ–σ3A (δ•σ3A) was measured by optical density at 600 nm (OD600). (F) Inhibition by 3AT of growth of yeast strains coexpressing Nef with either γ1–σ1A (γ1•σ1A) or δ–σ3A (δ•σ3A). Growth was measured by optical density at 600 nm after 48 h in culture. The value 1.0 on the y axis corresponds to the growth of each strain in the absence of 3AT. The expression of Nef and AP subunits from plasmids was as described in D. Values are the mean ± SD of triplicate determinations.
Interactions of Nef from different HIV-1 and SIV variants with γ1–σ1A and δ–σ3A
Both human and simian immunodeficiency viruses are highly variable genetically, owing to their error-prone reverse transcriptase (Roberts et al., 1988). However, the critical elements of the [DE]XXXL[LI]-type signal in Nef are conserved in all isolates of HIV-1, HIV-2, and SIV (Fig. 1 B; see supplemental material in Greenberg et al., 1998). The acidic residue is invariant, and the two bulky hydrophobic positions are occupied by leucine–leucine, leucine–valine, or leucine–methionine pairs in all Nefs. In contrast, the sequences flanking the [DE]XXXL[LI] signal and the X residues are variable (Fig. 1 B). To determine the generality of Nef–AP subunit interactions, we compared the binding of Nef gene products from four isolates of HIV-1 (NLA4-3, 248, ELI, and DH123) and one isolate of SIV (RQ). All five Nefs bound to γ1–σ1A, whereas only the NLA4-3 and 248 Nefs bound to δ–σ3A (Fig. 3). Thus, binding to γ1–σ1A appears to be the most general feature of all Nefs. Another general property revealed by these assays is the inability of all Nefs tested to bind to αC–σ2 and ɛ–σ4 (Fig. 3).
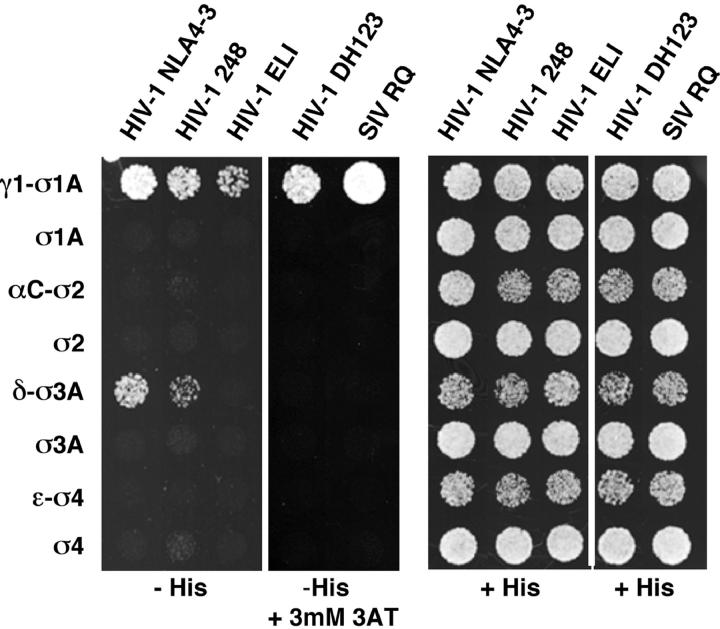
Figure 3.
Interaction of naturally occurring Nef variants with AP subunits. Three-hybrid analysis of the interaction of Nef from different isolates of HIV-1 or SIV fused to GAL4BD (pBridge) with different σ subunits (pBridge) and γ1, αC, δ, and ɛ subunits fused to GAL4AD (pGADT7). The sequences of the dileucine-based sorting signals from each of these constructs are indicated in Fig. 1 B. Interactions were evidenced by growth on agar plates made with medium without histidine (−His). The HIV-1 DH123 and SIV RQ Nef variants self-activated in this assay and had to be tested for growth in the presence of 3 mM 3AT.
Determinants of interactions of Nef with γ1–σ1A and δ–σ3A
The γ1 and δ subunits are organized into “trunk,” “hinge,” and “ear” domains (Fig. 1 A; Robinson and Bonifacino, 2001). Yeast three-hybrid analyses showed that interactions with Nef were mediated by the trunk, but not the hinge–ear domains, of γ1 and δ in combination with σ1A and σ3A, respectively (Fig. 4 A). To determine whether the interactions of Nef with AP subunits were mediated by the ENTSLL sequence (Fig. 4 B), and to delineate the sequence requirements for these interactions, we performed an alanine-scan mutagenesis. We observed that both leucine residues (L164 and L165) of the ENTSLL sequence were strictly required for interactions with γ1–σ1A and δ–σ3A (Fig. 4 C, asterisks). Mutation of the acidic residue (E160) of the signal diminished but did not completely abrogate the interaction with γ1–σ1A and abolished the interaction with δ–σ3A, as shown in both plate (Fig. 4 C, open triangle) and liquid medium growth assays (Fig. 4 D). These observations are in agreement with previous GST pull-downs assays, which showed that L164 and L165 are critical for binding to AP-1 and AP-3, and that E160 is only partially important for AP-1 but absolutely critical for AP-3 (Bresnahan et al., 1998; Janvier et al., 2003). Mutation of eleven residues other than E160, L164, and L165 had little or no effect on the interactions of Nef with both γ1–σ1A and δ–σ3A (Fig. 4 C).
Figure 4.
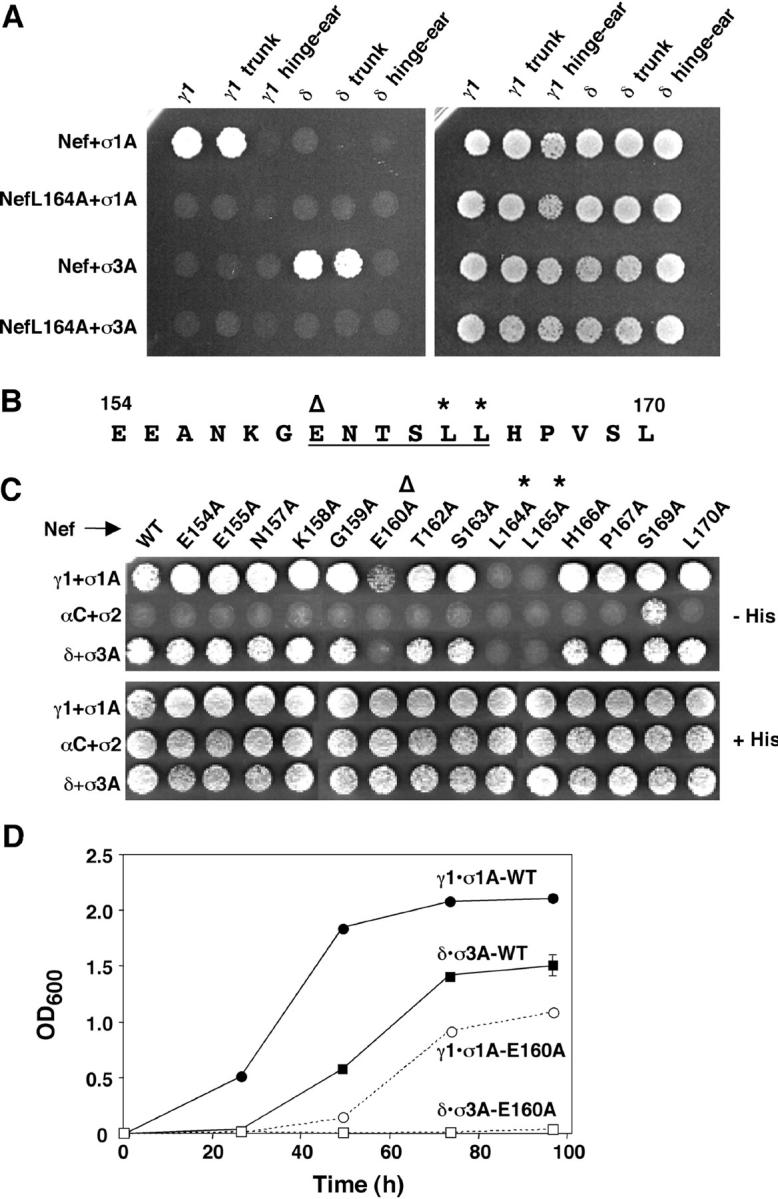
Mutational analyses of AP subunits and HIV-1 Nef. (A) Three-hybrid analysis of the interaction of Nef (NLA4-3 variant) or a NefL164A mutant GAL4BD (pBridge) with σ1A or σ3A (pBridge) and γ1, δ, or the trunk or hinge–ear fragments from these subunits fused to GAL4AD (pGADT7). (B) Sequence of the dileucine-based sorting signal (underlined) and flanking residues (154–170) in the NLA4-3 variant of HIV-1 Nef. In B and C, the critical glutamate and leucine residues are indicated by an open triangle and asterisks, respectively. (C) Three-hybrid analysis of the interaction of full-length, wild-type (WT) Nef (NLA4-3 variant) or alanine mutants of this Nef (pBridge) with σ1A, σ2, or σ3A (pBridge) and γ1, αC, or δ (pGADT7), as indicated in the figure. Interactions were detected by growth on agar plates made with medium without histidine (−His). (D) Time course of growth of yeast strains coexpressing wild-type (WT) or the E160A Nef mutant with either γ1–σ1A (γ1•σ1A) or δ–σ3A (δ•σ3A). Plasmid constructs were as described in C. Growth was measured by changes in the optical density at 600 nm (OD600). Values are the mean ± SD of triplicate determinations.
CD4 down-regulation induced by Nef mutants
To correlate the sequence requirements for Nef–AP subunit interactions and Nef function, we examined the ability of various Nef mutants to down-regulate expression of CD4. To this end, we used a variation of a well-established assay (Gratton et al., 1996) involving cotransfection of HeLa cells with plasmids encoding Nef, CD4, and a GFP-histone H2B chimera (this latter construct was included for the purpose of identifying the transfected cells and providing a landmark by staining of the nucleus). The localization of CD4 was determined by immunofluorescence microscopy (Fig. 5 A). As expected, CD4 was at the plasma membrane in mock-transfected cells, but intracellular in cells transfected with wild-type Nef (Fig. 5 A). Mutation of L164 and L165 abrogated the ability of Nef to redistribute CD4 from the plasma membrane to intracellular compartments, whereas mutation of other neighboring residues had no effect (Fig. 5 A). These results were confirmed using a flow cytofluorometric assay in which CD8 was substituted for GFP-histone H2B as a marker for the transfected cells. In agreement with the immunofluorescence microscopy assays, we found that the two leucines were the only essential residues among those tested (Fig. 5 B). Overall, these analyses demonstrated a very good correlation between the binding of Nef to AP-1 and AP-3 and the Nef-induced down-regulation of CD4 because Nef mutants that bound to γ1–σ1A and δ–σ3A down-regulated CD4, whereas those that did not bind to these subunits failed to down-regulate CD4. The only exception to this conclusion was the mutation of E160, which decreased the avidity for AP-1 and abrogated the interactions with AP-3 (Fig. 5; Janvier et al., 2003), but did not impair CD4 down-regulation (Fig. 5, A and B). Similar results were obtained with the 248 Nef variant (unpublished data). This behavior of the E160A Nef mutant could be explained if the activity of the NLA4-3 Nef is well above that required to effect full down-regulation of CD4, such that a weakening of the interaction of this particular Nef with AP-1 has no consequence for CD4 down-regulation.
Figure 5.
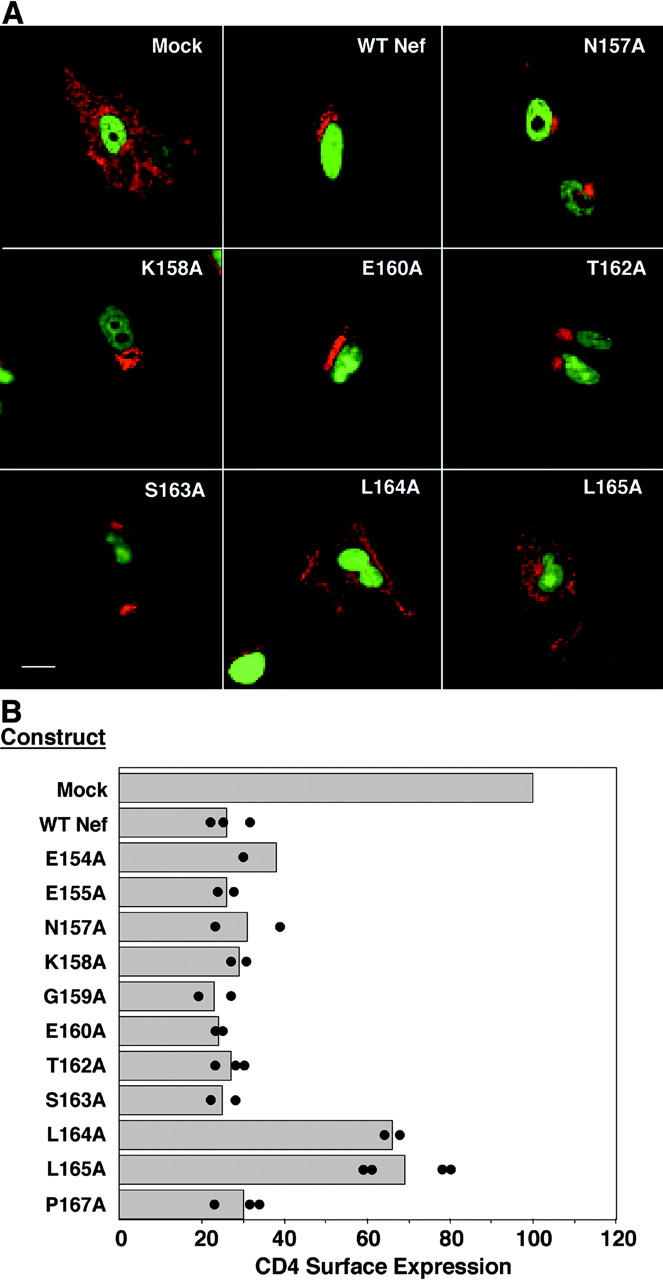
Down-regulation of CD4 by Nef or Nef mutants. (A) Analysis by immunofluorescence microscopy. HeLa cells were cotransfected with pCMV-CD4, pEGFP encoding GFP-histone H2B, and pCI-neo encoding either wild-type Nef (NLA4-3 variant) or Nef mutants. At 24 h after transfection, cells were fixed, permeabilized, and immunostained for CD4 (red). Cotransfected cells were identified by the expression of GFP-histone H2B in the nucleus (green). Images corresponding to selected Nef constructs are shown. Bar, 25 μm. (B) Analysis by FACS®. HeLa cells were cotransfected with pCMV-CD4, pCMV-CD8, and pCI-neo encoding either wild-type Nef (NLA4-3 variant) or Nef mutants. At 24 h after transfection, cells were coimmunostained with allophycocyanin-conjugated anti-CD4 and phycoerythrin-conjugated anti-CD8 antibodies. The percentage of CD8+ cells that also expressed CD4 on the surface was quantified by FACS® analysis, as described in Materials and methods. The results obtained in different determinations for each Nef construct are indicated by the individual dots. Bars represent the mean values for each Nef construct.
Interactions of the [DE]XXXL[LI] signal from LIMP-II with AP subunits
The results obtained with Nef beg the question of whether other [DE]XXXL[LI] signals also bind to the same combinations of AP subunits. To address this question, we examined the interaction of the four AP complexes with the cytosolic tail of LIMP-II, which has a well-characterized signal, ERAPLI (Ogata and Fukuda, 1994; Sandoval et al., 1994; Pond et al., 1995). We initially performed GST pull-downs using as sources of adaptors either HeLa cell cytosol or a mixture of AP complexes purified from bovine brain clathrin-coated vesicles. This latter preparation contained large quantities of AP-1 and AP-2, moderate quantities of AP-3, and traces of AP-4. Binding to the immobilized GST constructs was detected by immunoblotting with antibodies to specific AP subunits. We observed that GST–LIMP-II, like GST-Nef (Bresnahan et al., 1998; Janvier et al., 2003), bound to both AP-1 and AP-3, but not AP-2 and AP-4 (Fig. 6 A). The ability of the LIMP-II tail to bind AP-1 in this pull-down assay is at variance with previous findings made using surface plasmon resonance spectroscopy, which revealed binding only to AP-3 (Höning et al., 1998; Rodionov et al., 2002). As specificity controls, we showed that neither AP complex bound detectably to GST or to GST appended with the cation-independent MPR (CI-MPR) tail, which contains a DXXLL-type signal (Fig. 6 A). In addition, we found that mutations of the glutamate, leucine, or isoleucine residues to alanine residues completely abolished the interactions with both AP-1 and AP-3, thus, demonstrating that these interactions are mediated by the ERAPLI signal.
Figure 6.
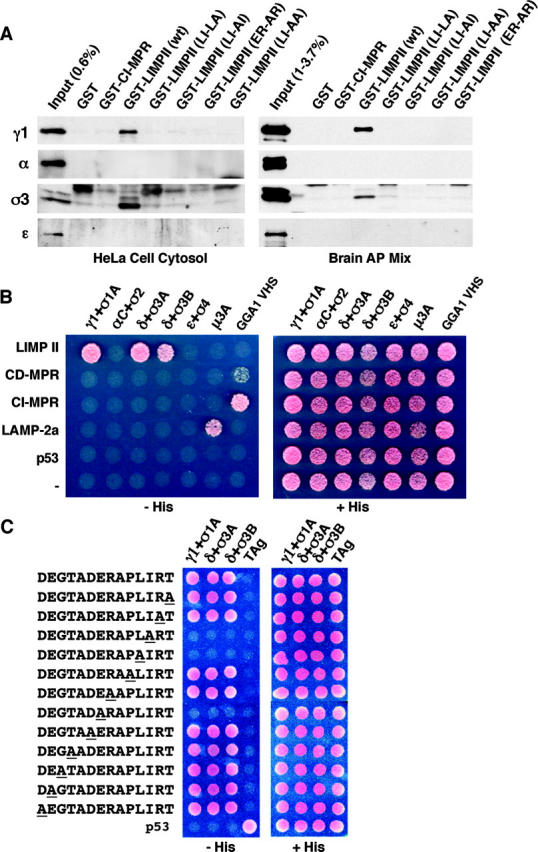
Interactions of the dileucine-based sorting signal from LIMP-II with AP complexes. (A) GST pull-down assays. GST fused to the cytosolic tail of the CI-MPR, LIMP-II, or mutants of this latter construct were incubated with either a cytosolic extract prepared from HeLa cells or a mixture of AP complexes prepared from bovine brain clathrin-coated vesicles. The LIMP-II tail mutants contain the amino acid substitutions indicated in the figure relative to the LIMP-II sequence shown in Fig. 1 B. Bound AP complexes were revealed by immunoblot analysis with antibodies specific for each complex. Some of the AP subunits (most notably σ3 and α in bovine brain) appeared as doublets. (B) Three-hybrid analysis of the interaction of the LIMP-II, CD-MPR, CI-MPR, and LAMP-2a cytosolic tails fused to GAL4BD (pBridge) with γ1, αC, δ, and ɛ (pBridge) and different σ subunits; μ3A and the VHS domain of GGA1 fused to GAL4AD (pGAD424). Interactions were detected by growth in the absence of histidine (−His). (C) Alanine-scan mutagenesis analysis of the interaction of LIMP-II cytosolic tail mutants with γ1–σ1A (γ1 + σ1A), δ–σ3A (δ + σ3A), or δ–σ3B (δ + σ3B) using the yeast-three hybrid system. Mutated residues are underlined. Plasmid constructs were as specified in B. The SV40 large T antigen (TAg) and p53 were used as controls. Interactions were detected by growth in the absence of histidine (−His).
In agreement with the GST pull-down assays, yeast three-hybrid analyses showed that the cytosolic tail of LIMP-II interacted with γ1–σ1A, δ–σ3A, and δ–σ3B (σ3A and σ3B are two isoforms of σ3), but not with αC–σ2, ɛ–σ4, μ3A, or the VHS domain of GGA1 (Fig. 6 B). In contrast, the cytosolic tails of the cation-dependent MPR (CD-MPR) and of the CI-MPR, which contain DXXLL-type signals, bound specifically to the VHS domain of GGA1 (Fig. 6 B; Puertollano et al., 2001; Takatsu et al., 2001b; Zhu et al., 2001). Finally, the cytosolic tail of LAMP-2a, which contains a YXXØ-type signal, interacted specifically with μ3A (Fig. 6 B; Gough et al., 1999). This experiment thus shows the specific recognition of three different types of sorting signals by their cognate recognition proteins.
To corroborate the specificity of the three-hybrid interactions, we performed an alanine-scan mutagenesis of the LIMP-II cytosolic tail. Of 12 residues that were mutated, only three were essential for interactions with γ1–σ1A, δ–σ3A, and δ–σ3B, the leucine-isoleucine pair and the glutamate residue at position −4 (Fig. 6 C). Mutation of other residues had no effect on the interactions. Together, these experiments demonstrate that the LIMP-II cytosolic tail binds to the AP-1 and AP-3 complexes through γ1–σ1A and δ–σ3 (A or B isoforms). Binding to both complexes is mediated by the ERAPLI signal, with only the glutamate, leucine, and isoleucine residues being absolutely required for interactions. These sequence requirements are in agreement with the requirements for function of the ERAPLI signal in sorting to lysosomes (Ogata and Fukuda, 1994; Sandoval et al., 1994; Pond et al., 1995), which provides strong support for the physiological significance of these interactions.
Discussion
Identity of the AP subunits that bind [DE]XXXL[LI] signals
The use of the yeast three-hybrid system has allowed us to detect relatively strong interactions of the [DE]XXXL[LI] signals from HIV-1 Nef and LIMP-II with specific combinations of two subunits from the AP-1 and AP-3 complexes. Strikingly, the subunits that interact with these signals when expressed together, γ1–σ1 and δ–σ3, are precisely the two subunits from each complex that had not been previously implicated in the recognition of [DE]XXXL[LI] signals. A previous yeast two-hybrid study had indicated a role for the μ subunits of AP-1, AP-2, and AP-3 in the recognition of the EXXXLL sequence from HIV-1 Nef (Craig et al., 2000). Although we were able to confirm the interaction of Nef with μ1A, this was much weaker than the interaction of Nef with γ1–σ1 and δ–σ3 (unpublished data). Other studies involving competition of labeling with photoactivatable peptides (Greenberg et al., 1998), yeast two-hybrid assays (Geyer et al., 2002), and binding of in vitro translated proteins to GST fusions (Geyer et al., 2002) had indicated that the EXXXLL sequence from Nef interacts with the β1 and β2 subunits of AP-1 and AP-2, respectively. These studies are representative of others in which [DE]XXXL[LI] signals from several proteins were shown to interact with either μ or β subunits (Bremnes et al., 1998; Rapoport et al., 1998; Rodionov and Bakke, 1998; Hofmann et al., 1999).
In view of the aforementioned discrepancies, what gives us confidence that the interactions with γ1–σ1 and δ–σ3 reported herein reflect the actual mode of recognition of [DE]XXXL[LI] signals by AP complexes? We think that the weight of our evidence comes from the extensive correlative analyses performed in our study. The fact that the [DE]XXXL[LI] signals from both Nef and LIMP-II interact with γ1–σ1 and δ–σ3 but not αC–σ2 and ɛ–σ4 correlates well with the interaction of those signals with the intact AP-1 and AP-3, but not AP-2, in GST pull-down assays (Bresnahan et al., 1998; Janvier et al., 2003; this paper). This correlation holds true even in the one instance in which the results of our GST pull-down assays are at variance with previously published results, as is the case for LIMP-II, which was previously reported not to bind AP-1 (Höning et al., 1998). A second type of correlation concerns the residues of the signals required for interactions in the yeast three-hybrid system and in GST pull-downs. Again, there is excellent correspondence between these two sets of data: the key glutamate and leucine–leucine or leucine–isoleucine residues are required for interactions in both assays. Even the fact that the key glutamate is less important for Nef than for LIMP-II interactions is reflected quantitatively in both assays (compare the results in Fig. 4 C in this paper with those in Fig. 7 B of Janvier et al. [2003], and in Fig. 6, A and C, in this paper). Thus, binding to γ1–σ1 and δ–σ3 recapitulates the binding to the intact complexes, implying that both are manifestations of the same molecular recognition event.
Structural implications for the recognition of [DE]XXXL[LI] signals
Although two subunits are necessary to detect binding of [DE]XXXL[LI] signals in the yeast three-hybrid assays, at present, we cannot ascertain whether the binding site involves residues on the surface of both subunits or is located exclusively on one subunit, with the other subunit being required for proper folding or stability of the hemicomplex. The structures of the γ1–σ1 and δ–σ3 hemicomplexes can be modeled after that of the α–σ2 hemicomplex in the context of the AP-2 core, which was recently solved by X-ray crystallography (Collins et al., 2002). The NH2-terminal trunk domain of each large subunit (γ1 or δ) would be expected to consist of a long α-helical superhelix with an “elbow-like” bend near the middle of the domain. The small chain (σ1 or σ3) would nestle at the bend, forming a tight heterodimer (Collins et al., 2002). The trunk domains of γ1 or δ seem particularly suited to harbor peptide-binding sites because of the many grooves predicted to occur on the surface of their superhelical folds. Indeed, the spatial arrangement of the NH2-terminal α-helices of the large subunits (Collins et al., 2002) resembles that of the eight α-helices that make up the VHS domain of the GGAs (Misra et al., 2002; Shiba et al., 2002), as previously proposed (Geyer et al., 2002). A groove between helices 6 and 8 of the GGA VHS domain constitutes the binding site for DXXLL signals (Misra et al., 2002; Shiba et al., 2002), and it is possible that an analogous groove might be present in the trunk domain of the large AP subunits. We would expect this binding site to occur in a membrane-proximal region of the γ1 or δ trunks because some [DE]XXXL[LI] signals (e.g., that of LIMP-II) are close to the transmembrane domain. Although it might be possible to map the peptide-binding site on the γ1–σ1 and δ–σ3 hemicomplexes by mutational analyses, its definitive identification will require X-ray crystallographic analyses.
Our observations demonstrate that [DE]XXXL[LI] and YXXØ signals (Ohno et al., 1995, 1998) bind to different subunits of the AP complexes (γ1–σ1 and δ–σ3 vs. μ subunits). This conclusion agrees with previous observations from in vivo overexpression/saturation experiments, which showed that [DE]XXXL[LI] signals compete with other [DE]XXXL[LI] signals but not with YXXØ signals for engagement of the protein trafficking machinery; the same is the case for YXXØ signals (Marks et al., 1996; Craig et al., 1998). The binding of both types of signals to the AP complexes may explain why these signals have similar functions in sorting to endosomes, lysosomes, and lysosome-related organelles. In contrast, the spatial separation of the two binding sites may afford differential regulation of signal recognition.
Possible role of AP-1 in Nef-induced CD4 down-regulation
We have shown that the ability of Nef to down-regulate CD4 correlates best with binding to AP-1, pointing to a likely role for this complex in CD4 down-regulation. AP-3 could also play a role, at least for some Nef variants. The localization of these two AP complexes to the TGN and endosomes makes it likely that the Nef-induced down-regulation of CD4 results mainly from impairment of its transport from intracellular compartments to the plasma membrane. The fact that AP-2 does not interact with Nef suggests that this adaptor is not a direct target for regulation by HIV-1. Rather, another protein such as the regulatory V1H subunit of the vacuolar ATPase may be responsible for the Nef-induced removal of CD4 from the cell surface (Lu et al., 1998). Alternatively, CD4 may be internalized constitutively in HeLa cells or upon dissociation of the tyrosine kinase lck in T cells (Pelchen-Matthews et al., 1991, 1998; Gratton et al., 1996). This internalization may be mediated by direct interaction of the phosphorylated CD4 tail with AP-2, which occurs even in the absence of Nef (Pitcher et al., 1999). Whichever the mechanism of endocytosis, the internalized CD4 could be captured by AP-1 and AP-3 in the presence of Nef, preventing its return to the cell surface and possibly routing it to lysosomes. An alternative scenario would involve interaction of newly-synthesized CD4 with Nef and AP-1 or AP-3, resulting in reduced transport to the cell surface and enhanced delivery to the endosomal–lysosomal system. Both scenarios would be in line with the observation that intracellular retention plays a critical role in the down-regulation of CD4 by Nef (Mangasarian et al., 1997).
The molecular mechanism of down-regulation likely involves the formation of a tripartite complex between Nef, CD4, and AP-1. The cytosolic tail of CD4 contains a determinant encompassing a dileucine pair at positions 413 and 414 that does not conform to the [DE]XXXL[LI] consensus (Pitcher et al., 1999). This determinant is required for Nef-induced down-regulation (Aiken et al., 1994; Hua et al., 1997; Pitcher et al., 1999) via interactions with a hydrophobic patch on Nef that includes the side chains of W57, L58, and L110 (Grzesiek et al., 1996). The [DE]XXXL[LI] signal in Nef (residues 160–165) in turn binds to the AP-1 γ1–σ1 and AP-3 δ–σ3 subunits. Thus, Nef can be thought of as a “connector” (Mangasarian et al., 1997) that bridges the tail of CD4 to AP-1. Alternatively, the interaction of Nef with AP-1 and AP-3 could modify its properties, enabling it to bind directly to the CD4 tail. In this regard, expression of Nef has been shown to stabilize the association of AP-1 and AP-3 with membranes by an Arf-independent mechanism (Janvier et al., 2003). This stabilization could enhance an otherwise weak binding of CD4 to AP-1 and AP-3.
Conclusion
In summary, our studies have uncovered a novel mode of recognition of [DE]XXXL[LI] signals from HIV-1 Nef and LIMP-II involving two subunits of the AP-1 and AP-3 complexes, γ1–σ1 and δ–σ3, respectively. The assays developed to study these interactions are robust and should be applicable to the analysis of the interaction of other [DE]XXXL[LI] signals with AP complexes. In particular, it will be of interest to determine whether other [DE]XXXL[LI] signals are recognized by α–σ2 or ɛ–σ4, which tested negative in our assays despite having structures similar to γ1–σ1 and δ–σ3. Finally, our studies support a prominent role for the AP-1 complex in the Nef-induced down-regulation of CD4, implying that this down-regulation occurs mainly from endosomes or the TGN.
Materials and methods
Recombinant DNA constructs
GAL4AD fusion constructs were made by PCR amplification and in-frame cloning into yeast two-hybrid vectors (CLONTECH Laboratories, Inc.; Fig. 1 D). γ1, αC, δ, and the trunk and hinge–ear fragments of γ1 and δ (BamHI–XhoI) and β1 and β3A (SmaI–SalI) were cloned into pGADT7. μ3A and σ1A (EcoRI–SalI), and σ2, σ3A, σ3B, and σ4 (BamHI–XhoI) were cloned into pGAD424. ɛ and β4 in pGADT7 (Boehm et al., 2001), as well as the GGA1 VHS domain and the SV40 T-antigen (TAg) in pGAD424 (Kato et al., 2002), have been described previously. β2 in pGAD424 was a gift of M.S. Robinson (University of Cambridge, Cambridge, UK).
GAL4BD fusion constructs were also made by PCR amplification and in-frame cloning into multiple cloning site 1 of pBridge (CLONTECH Laboratories, Inc.; Fig. 1 D, MCS1). Nef from different strains of HIV-1 or SIV, the cytosolic tails of LIMP-II (20 COOH-terminal residues), CI-MPR (114 COOH-terminal residues), CD-MPR (41 COOH-terminal residues), and p53 were cloned into the EcoRI–SalI sites of pBridge. The LAMP-2a cytosolic tail was cloned into the EcoRI–PstI sites of pBridge. Mutants of GAL4BD-Nef and GAL4BD–LIMP-II were obtained by PCR-directed mutagenesis.
For three-hybrid analyses, the AP subunit cDNAs were amplified by PCR and subcloned into the multiple cloning site 2 (Fig. 1 D, MCS2) of the recombinant pBridge constructs expressing the GAL4BD fusion proteins as described in the previous paragraph. The σ and μ subunits of the AP complexes were cloned into the NotI–BglII sites of the pBridge-Nef constructs. γ1, αC, and δ were cloned into the NotI site of pBridge constructs expressing the GAL4BD–LIMP-II, –CI-MPR, –CD-MPR, and -p53 fusion proteins described in the previous paragraph. ɛ was cloned into the BglII site of the same pBridge recombinant constructs.
For expression in mammalian cells, wild-type and mutant Nef cDNA were PCR amplified and cloned into the EcoRI–SalI sites of the pCI-neo vector (Promega). The GFP-tagged histone H2B construct was made as described previously (Dey et al., 2000). The pCMV-CD4 and pCMV-CD8 expression vectors were obtained from the AIDS Reference and Reagent Program, National Institute of Allergy and Infectious Diseases.
The GST–LIMP-II and GST–CI-MPR constructs were obtained by in frame cloning of sequences encoding the 20 COOH-terminal residues of LIMP-II and the 114 COOH-terminal residues of the CI-MPR into the EcoRI–SalI sites of pGEX-5X-1 (Amersham Biosciences). The mutant pGEX–LIMP-II constructs were obtained by PCR-directed mutagenesis. The GST fusion proteins were produced by transformation of the Escherichia coli strain BL21 (DE3) pLysS (Novagen) and purified according to the manufacturer's instructions.
Antibodies
The following mouse mAbs were used: 100/3 to γ1-adaptin and 100/2 to α-adaptin (Sigma-Aldrich), anti–ɛ-adaptin (Transduction Laboratories), S3.5 to human CD4, anti-CD4, allophycocyanin-conjugated anti-CD4, and phycoerythrin-conjugated anti-CD8 antibodies (Caltag Laboratories), and HA.11 to the HA tag (Covance). The following polyclonal antibodies were also used: rabbit antibody to σ3 (Dell'Angelica et al., 1997), Alexa 594–conjugated anti–mouse IgG (Molecular Probes), and HRP-conjugated anti–mouse and anti–rabbit IgG (Amersham Biosciences).
Yeast culture, transformation, and two- and three-hybrid assays
The Saccharomyces cerevisiae strain HF7c (Clontech), was maintained on dropout agar plates lacking methionine. Transformation was performed by the lithium acetate procedure as described in the instructions for the MATCHMAKER two-hybrid kit (CLONTECH Laboratories, Inc.). HF7c transformants were selected by spreading on plates lacking leucine, tryptophan, and methionine. For colony growth assays, HF7c transformants were dotted on plates lacking leucine, tryptophan, methionine, and histidine, and allowed to grow at 30°C for 3–5 d. Quantitative growth assays were performed as described previously (Aguilar et al., 1997). Filter-based β-galactosidase assays were performed according to the instructions for the MATCHMAKER two-hybrid kit (CLONTECH Laboratories, Inc.).
Preparation of yeast lysates
Yeast were grown in synthetic medium at 30°C to an optical density of 0.6. 3 OD units of yeast cells were resuspended in 200 μl of ice-cold 10% trichloroacetic acid, and then transferred to Eppendorf tubes containing 200 μl of acid-washed glass beads. The cells were broken by vigorous vortexing for 1 min, followed by chilling on ice for another 1 min. This cycle was repeated 10 times. Proteins contained in the supernatant were precipitated by centrifugation at top speed, followed by a wash in ice-cold acetone. The protein extract was resuspended in 100 μl of Laemmli loading buffer, and then analyzed by immunoblotting using an anti-HA antibody, followed by chemiluminescent detection (Amersham Biosciences).
Transfection
HeLa cells (American Type Culture Collection) were transfected using the lipofectamine reagent (Invitrogen) according to the manufacturer's instructions. For immunofluorescent staining, cells were cotransfected with mammalian expression vectors encoding CD4 and GFP-histone H2B along with expression vectors encoding Nef or Nef mutants. For FACS® analysis, the GFP-histone H2B construct was substituted by a vector encoding CD8.
Immunofluorescence microscopy
24 h after transfection, HeLa cells grown on glass coverslips were fixed in 4% PFA in PBS, quenched for 10 min with 50 mM NH4Cl in PBS, and permeabilized for 10 min with 0.1% (wt/vol) Triton X-100 in PBS. After permeabilization, the cells were blocked for 30 min with 10% (vol/vol) normal goat serum in PBS and incubated for 1 h at RT with the primary antibody, washed with PBS, and incubated for 1 h with the secondary antibody. The coverslips were washed and mounted on slides. Images were acquired on a confocal microscope (model LSM 410; Carl Zeiss MicroImaging, Inc.).
Flow cytofluorometry
Cotransfections of HeLa cells with plasmids encoding CD4 and CD8 were optimized ahead of time to obtain equivalent mean fluorescence value ratios, which were typically achieved with 0.5 μg CD8 and 0.8 μg CD4 plasmids. For determination of cell surface antibody binding, 106 cells transfected with CD4, CD8, and Nef (or Nef mutants) were collected by centrifugation and washed with PBS. Cells were incubated for 10 min at RT with allophycocyanin-conjugated anti-CD4 and phycoerythrin-conjugated anti-CD8 antibodies. The cells were washed three times with ice-cold PBS containing BSA or FBS, and fixed in PBS containing 4% PFA. Flow cytofluorometric data were acquired by using a two-laser, four-color FACSCalibur flow cytometer (Becton Dickinson). Data analysis was done using CELLQuest v3.3 (Becton Dickinson) and FlowJo v3.3.4 software (Treestar Corp.).
GST pull-downs
GST pull-downs were performed using either a cytosolic extract of HeLa cells prepared in 25 mM Hepes, pH 7.4, 150 mM NaCl, 1 mM EGTA, 0.5 mM MgCl2, and 0.5% (wt/vol) Triton X-100 (lysis buffer) or a purified fraction of AP complexes prepared from bovine brain clathrin-coated vesicles as described previously (Jiang et al., 2000; provided by L. Greene, National Heart, Lung, and Blood Institute, NIH). Purified GST, GST-CI-MPR, or GST-LIMP-II (wild-type or mutated) proteins were immobilized on glutathione-agarose beads (Amersham Biosciences), and then incubated overnight with 1 ml HeLa cell lysate (corresponding to 107 cells) or for 5 h with 0.2 ml of the purified AP complexes (in extraction buffer containing 0.5 M Tris-HCl, pH 7.0, 2 mM DTT, and 1 mM EDTA) diluted to 1 ml in lysis buffer. The beads were washed three times with lysis buffer and once with lysis buffer without Triton X-100. Bound AP complexes were resolved by SDS-PAGE and revealed by immunoblotting using antibodies to γ1-, α-, σ3-, or ɛ-adaptin followed by chemiluminescent detection (Amersham Biosciences).
Acknowledgments
We thank X. Zhu and A. San Miguel for excellent technical assistance, M.S. Robinson and L. Greene for kind gifts of reagents, and R. Mattera and R. Puertollano for critical review of the manuscript.
Y. Kato's present address is Faculty of Pharmaceutical Sciences, Kanazawa University, 13-1 Takara-machi, Kanazawa, 920-0934, Japan.
M. Boehm's present address is ALTANA Pharma AG, Byk-Gulden-Strasse 2, 78467 Konstanz, Germany.
Abbreviations used in this paper: 3AT, 3-aminotriazole; AP, adaptor protein; CD-MPR, cation-dependent MPR; CI-MPR, cation-independent MPR; GAL4AD, GAL4, transcription activation domain; GAL4BD, GAL4 DNA binding domain; GGAs, Golgi-localized, γ-ear–containing, Arf-binding proteins; HIV, human immunodeficiency virus; LIMP-II, lysosomal integral membrane protein II; MPR, mannose 6-phosphate receptor; Nef, negative factor; SIV, simian immunodeficiency virus; VHS, Vps27, Hrs, and Stam.
References
- Aguilar, R.C., H. Ohno, K.C. Roche, and J.S. Bonifacino. 1997. Functional domain mapping of the clathrin-associated adaptor medium chains mu1 and mu2. J. Biol. Chem. 272:2760–2766. [DOI] [PubMed] [Google Scholar]
- Aiken, C., J. Konner, N.R. Landau, M.E. Lenburg, and D. Trono. 1994. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell. 76:853–864. [DOI] [PubMed] [Google Scholar]
- Barriocanal, J.G., J.S. Bonifacino, L. Yuan, and I.V. Sandoval. 1986. Biosynthesis, glycosylation, movement through the Golgi system, and transport to lysosomes by an N-linked carbohydrate-independent mechanism of three lysosomal integral membrane proteins. J. Biol. Chem. 261:16755–16763. [PubMed] [Google Scholar]
- Boehm, M., R.C. Aguilar, and J.S. Bonifacino. 2001. Functional and physical interactions of the adaptor protein complex AP-4 with ADP-ribosylation factors (ARFs). EMBO J. 20:6265–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino, J.S., and L.M. Traub. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72:395–447. [DOI] [PubMed] [Google Scholar]
- Bremnes, T., V. Lauvrak, B. Lindqvist, and O. Bakke. 1998. Selection of phage displayed peptides from a random 10-mer library recognising a peptide target. Immunotechnology. 4:21–28. [DOI] [PubMed] [Google Scholar]
- Bresnahan, P.A., W. Yonemoto, S. Ferrell, D. Williams-Herman, R. Geleziunas, and W.C. Greene. 1998. A dileucine motif in HIV-1 Nef acts as an internalization signal for CD4 downregulation and binds the AP-1 clathrin adaptor. Curr. Biol. 8:1235–1238. [DOI] [PubMed] [Google Scholar]
- Canfield, W.M., K.F. Johnson, R.D. Ye, W. Gregory, and S. Kornfeld. 1991. Localization of the signal for rapid internalization of the bovine cation-independent mannose 6-phosphate/insulin-like growth factor-II receptor to amino acids 24-29 of the cytoplasmic tail. J. Biol. Chem. 266:5682–5688. [PubMed] [Google Scholar]
- Chen, H.J., J. Yuan, and P. Lobel. 1997. Systematic analysis of the cation-independent mannose 6-phosphate receptor/insulin-like growth II factor receptor cytoplasmic domain. J. Biol. Chem. 272:7003–7012. [DOI] [PubMed] [Google Scholar]
- Chen, J.-J., J.L. Goldstein, and M.S. Brown. 1990. NPXY, a sequence often found in cytoplasmic tails, is required for coated pit-mediated internalization of the low density lipoprotein receptor. J. Biol. Chem. 265:3116–3123. [PubMed] [Google Scholar]
- Collawn, J.F., L.A. Kuhn, L.-F.S. Liu, J.A. Tainer, and I.S. Trowbridge. 1991. Transplanted LDL and mannose-6-phosphate receptor internalization signals promote high-efficiency endocytosis of the transferrin receptor. EMBO J. 10:3247–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, B.M., A.J. McCoy, H.M. Kent, P.R. Evans, and D.J. Owen. 2002. Molecular architecture and functional model of the endocytic AP2 complex. Cell. 109:523–535. [DOI] [PubMed] [Google Scholar]
- Craig, H.M., M.W. Pandori, and J.C. Guatelli. 1998. Interaction of HIV-1 Nef with the cellular dileucine-based sorting pathway is required for CD4 down-regulation and optimal viral infectivity. Proc. Natl. Acad. Sci. USA. 95:11229–11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig, H.M., T.R. Reddy, N.L. Riggs, P.P. Dao, and J.C. Guatelli. 2000. Interactions of HIV-1 nef with the mu subunits of adaptor protein complexes 1, 2, and 3: role of the dileucine-based sorting motif. Virology. 271:9–17. [DOI] [PubMed] [Google Scholar]
- Dell'Angelica, E.C., H. Ohno, C.E. Ooi, E. Rabinovich, K.W. Roche, and J.S. Bonifacino. 1997. AP-3: an adaptor-like protein complex with ubiquitous expression. EMBO J. 15:917–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey, A., J. Ellenberg, A. Farina, A.E. Coleman, T. Maruyama, S. Sciortino, J. Lippincott-Schwartz, and K. Ozato. 2000. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G(2)-to-M transition. Mol. Cell. Biol. 20:6537–6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, J., J. Kastrup, B.L. Nielsen, N. Odum, and C. Geisler. 1997. Regulation and function of CD3γ DxxxLL motif: a binding site for adaptor protein-1 and adaptor protein-2 in vitro. J. Cell Biol. 138:271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita, H., M. Saeki, K. Yasunaga, T. Ueda, T. Imoto, and M. Himeno. 1999. In vitro binding study of adaptor protein complex (AP-1) to lysosomal targeting motif (LI-motif). Biochem. Biophys. Res. Commun. 255:54–58. [DOI] [PubMed] [Google Scholar]
- Geyer, M., O.T. Fackler, and B.M. Peterlin. 2001. Structure-function relationships in HIV-1 Nef. EMBO Rep. 2:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer, M., O.T. Fackler, and B.M. Peterlin. 2002. Subunit H of the V-ATPase involved in endocytosis shows homology to beta-adaptins. Mol. Biol. Cell. 13:2045–2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough, N.R., M.E. Zweifel, O. Martinez-Augustin, R.C. Aguilar, J.S. Bonifacino, and D.M. Fambrough. 1999. Utilization of the indirect lysosome targeting pathway by lysosome-associated membrane proteins (LAMPs) is influenced largely by the C-terminal residue of their GYXXphi targeting signals. J. Cell Sci. 112:4257–4269. [DOI] [PubMed] [Google Scholar]
- Gratton, S., X.J. Yao, S. Venkatesan, E.A. Cohen, and R.P. Sekaly. 1996. Molecular analysis of the cytoplasmic domain of CD4: overlapping but noncompetitive requirement for lck association and down-regulation by Nef. J. Immunol. 157:3305–3311. [PubMed] [Google Scholar]
- Greenberg, M., L. DeTulleo, I. Rapoport, J. Skowronski, and T. Kirchhausen. 1998. A dileucine motif in HIV-1 Nef is essential for sorting into clathrin-coated pits and for downregulation of CD4. Curr. Biol. 8:1239–1242. [DOI] [PubMed] [Google Scholar]
- Grzesiek, S., S.J. Stahl, P.T. Wingfield, and A. Bax. 1996. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry. 35:10256–10261. [DOI] [PubMed] [Google Scholar]
- Grzesiek, S., A. Bax, J.S. Hu, J. Kaufman, I. Palmer, S.J. Stahl, N. Tjandra, and P.T. Wingfield. 1997. Refined solution structure and backbone dynamics of HIV-1 Nef. Protein Sci. 6:1248–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, G., S. Gupta, M. Yi, P. Michaely, H.H. Hobbs, and J.C. Cohen. 2002. ARH is a modular adaptor protein that interacts with the LDL receptor, clathrin, and AP-2. J. Biol. Chem. 277:44044–44049. [DOI] [PubMed] [Google Scholar]
- Hirst, J., and M.S. Robinson. 1998. Clathrin and adaptors. Biochim. Biophys. Acta. 1404:173–193. [DOI] [PubMed] [Google Scholar]
- Hofmann, M.W., S. Honing, D. Rodionov, B. Dobberstein, K. von Figura, and O. Bakke. 1999. The leucine-based sorting motifs in the cytoplasmic domain of the invariant chain are recognized by the clathrin adaptors AP1 and AP2 and their medium chains. J. Biol. Chem. 274:36153–36158. [DOI] [PubMed] [Google Scholar]
- Höning, S., I.V. Sandoval, and K. von Figura. 1998. A di-leucine-based motif in the cytoplasmic tail of LIMP-II and tyrosinase mediates selective binding of AP-3. EMBO J. 17:1304–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua, J., W. Blair, R. Truant, and B.R. Cullen. 1997. Identification of regions in HIV-1 Nef required for efficient downregulation of cell surface CD4. Virology. 231:231–238. [DOI] [PubMed] [Google Scholar]
- Janvier, K., H. Craig, D. Hitchin, R. Madrid, N. Sol-Foulon, L. Renault, J. Cherfils, D. Cassel, S. Benichou, and J. Guatelli. 2003. HIV-1 Nef stabilizes the association of adaptor protein complexes with membranes. J. Biol. Chem. 278:8725–8732. [DOI] [PubMed] [Google Scholar]
- Jiang, R., B. Gao, K. Prasad, L.E. Greene, and E. Eisenberg. 2000. Hsc70 chaperones clathrin and primes it to interact with vesicle membranes. J. Biol. Chem. 275:8439–8447. [DOI] [PubMed] [Google Scholar]
- Johnson, K.F., and S. Kornfeld. 1992. A His-Leu-Leu sequence near the carboxyl terminus of the cytoplasmic domain of the cation-dependent mannose 6-phosphate receptor is necessary for the lysosomal enzyme sorting function. J. Biol. Chem. 267:17110–17115. [PubMed] [Google Scholar]
- Kato, Y., S. Misra, R. Puertollano, J.H. Hurley, and J.S. Bonifacino. 2002. Phosphoregulation of sorting signal VHS domain interactions by a direct electrostatic mechanism. Nat. Struct. Biol. 9:532–536. [DOI] [PubMed] [Google Scholar]
- Kongsvik, T.L., S. Honing, O. Bakke, and D.G. Rodionov. 2002. Mechanism of interaction between leucine-based sorting signals from the invariant chain and clathrin-associated adaptor protein complexes AP1 and AP2. J. Biol. Chem. 277:16484–16488. [DOI] [PubMed] [Google Scholar]
- Lee, C.H., K. Saksela, U.A. Mirza, B.T. Chait, and J. Kuriyan. 1996. Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain. Cell. 85:931–942. [DOI] [PubMed] [Google Scholar]
- Letourneur, F., and R.D. Klausner. 1992. A novel di-leucine motif and a tyrosine-based motif independently mediate lysosomal targeting and endocytosis of CD3 chains. Cell. 69:1143–1157. [DOI] [PubMed] [Google Scholar]
- Lu, X., H. Yu, S.H. Liu, F.M. Brodsky, and B.M. Peterlin. 1998. Interactions between HIV1 Nef and vacuolar ATPase facilitate the internalization of CD4. Immunity. 8:647–656. [DOI] [PubMed] [Google Scholar]
- Mangasarian, A., M. Foti, C. Aiken, D. Chin, J.L. Carpentier, and D. Trono. 1997. The HIV-1 Nef protein acts as a connector with sorting pathways in the Golgi and at the plasma membrane. Immunity. 6:67–77. [DOI] [PubMed] [Google Scholar]
- Marks, M.S., L. Woodruff, H. Ohno, and J.S. Bonifacino. 1996. Protein targeting by tyrosine- and di-leucine–based signals: evidence for distinct saturable components. J. Cell Biol. 135:341–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman, I. 1996. Endocytosis and molecular sorting. Annu. Rev. Cell Dev. Biol. 12:575–625. [DOI] [PubMed] [Google Scholar]
- Mishra, S.K., S.C. Watkins, and L.M. Traub. 2002. The autosomal recessive hypercholesterolemia (ARH) protein interfaces directly with the clathrin-coat machinery. Proc. Natl. Acad. Sci. USA. 99:16099–16104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra, S., R. Puertollano, Y. Kato, J.S. Bonifacino, and J.H. Hurley. 2002. Structural basis for acidic-cluster-dileucine sorting-signal recognition by VHS domains. Nature. 415:933–937. [DOI] [PubMed] [Google Scholar]
- Morris, S.M., and J.A. Cooper. 2001. Disabled-2 colocalizes with the LDLR in clathrin-coated pits and interacts with AP-2. Traffic. 2:111–123. [DOI] [PubMed] [Google Scholar]
- Nielsen, M.S., P. Madsen, E.I. Christensen, A. Nykjaer, J. Gliemann, D. Kasper, R. Pohlmann, and C.M. Petersen. 2001. The sortilin cytoplasmic tail conveys Golgi-endosome transport and binds the VHS domain of the GGA2 sorting protein. EMBO J. 20:2180–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata, S., and M. Fukuda. 1994. Lysosomal targeting of Limp II membrane glycoprotein requires a novel Leu-Ile motif at a particular position in its cytoplasmic tail. J. Biol. Chem. 269:5210–5217. [PubMed] [Google Scholar]
- Ohno, H., J. Stewart, M.C. Fournier, H. Bosshart, I. Rhee, S. Miyatake, T. Saito, A. Gallusser, T. Kirchhausen, and J.S. Bonifacino. 1995. Interaction of tyrosine-based sorting signals with clathrin-associated proteins. Science. 269:1872–1875. [DOI] [PubMed] [Google Scholar]
- Ohno, H., R.C. Aguilar, D. Yeh, D. Taura, T. Saito, and J.S. Bonifacino. 1998. The medium subunits of adaptor complexes recognize distinct but overlapping sets of tyrosine-based sorting signals. J. Biol. Chem. 273:25915–25921. [DOI] [PubMed] [Google Scholar]
- Owen, D.J., and P.R. Evans. 1998. A structural explanation for the recognition of tyrosine-based endocytotic signals. Science. 282:1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page, L.J., and M.S. Robinson. 1995. Targeting signals and subunit interactions in coated vesicle adaptor complexes. J. Cell Biol. 131:619–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden, A.A., G.Y. Park, and R.H. Scheller. 2001. The Di-leucine motif of vesicle-associated membrane protein 4 is required for its localization and AP-1 binding. J. Biol. Chem. 276:49183–49187. [DOI] [PubMed] [Google Scholar]
- Pelchen-Matthews, A., J.E. Armes, G. Griffiths, and M. Marsh. 1991. Differential endocytosis of CD4 in lymphocytic and nonlymphocytic cells. J. Exp. Med. 173:575–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelchen-Matthews, A., R.P. da Silva, M.J. Bijlmakers, N. Signoret, S. Gordon, and M. Marsh. 1998. Lack of p56lck expression correlates with CD4 endocytosis in primary lymphoid and myeloid cells. Eur. J. Immunol. 28:3639–3647. [DOI] [PubMed] [Google Scholar]
- Pitcher, C., S. Honing, A. Fingerhut, K. Bowers, and M. Marsh. 1999. Cluster of differentiation antigen 4 (CD4) endocytosis and adaptor complex binding require activation of the CD4 endocytosis signal by serine phosphorylation. Mol. Biol. Cell. 10:677–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pond, L., L.A. Kuhn, L. Teyton, M.P. Schutze, J.A. Tainer, M.R. Jackson, and P.A. Peterson. 1995. A role for acidic residues in di-leucine motif-based targeting to the endocytic pathway. J. Biol. Chem. 270:19989–19997. [DOI] [PubMed] [Google Scholar]
- Puertollano, R., R.C. Aguilar, I. Gorshkova, R.J. Crouch, and J.S. Bonifacino. 2001. Sorting of mannose 6-phosphate receptors mediated by the GGAs. Science. 292:1712–1716. [DOI] [PubMed] [Google Scholar]
- Rapoport, I., Y.C. Chen, P. Cupers, S.E. Shoelson, and T. Kirchhausen. 1998. Dileucine-based sorting signals bind to the beta chain of AP-1 at a site distinct and regulated differently from the tyrosine-based motif-binding site. EMBO J. 17:2148–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts, J.D., K. Bebenek, and T.A. Kunkel. 1988. The accuracy of reverse transcriptase from HIV-1. Science. 242:1171–1173. [DOI] [PubMed] [Google Scholar]
- Robinson, M.S., and J.S. Bonifacino. 2001. Adaptor-related proteins. Curr. Opin. Cell Biol. 13:444–453. [DOI] [PubMed] [Google Scholar]
- Rodionov, D.G., and O. Bakke. 1998. Medium chains of adaptor complexes AP-1 and AP-2 recognize leucine-based sorting signals from the invariant chain. J. Biol. Chem. 273:6005–6008. [DOI] [PubMed] [Google Scholar]
- Rodionov, D.G., S. Honing, A. Silye, T.L. Kongsvik, K. von Figura, and O. Bakke. 2002. Structural requirements for interactions between leucine-sorting signals and clathrin-associated adaptor protein complex AP3. J. Biol. Chem. 277:47436–47443. [DOI] [PubMed] [Google Scholar]
- Sandoval, I.V., J.J. Arredondo, J. Alcalde, A. Gonzalez Noriega, J. Vandekerckhove, M.A. Jimenez, and M. Rico. 1994. The residues Leu(Ile)475-Ile(Leu, Val, Ala)476, contained in the extended carboxyl cytoplasmic tail, are critical for targeting of the resident lysosomal membrane protein LIMP II to lysosomes. J. Biol. Chem. 269:6622–6631. [PubMed] [Google Scholar]
- Shiba, T., H. Takatsu, T. Nogi, N. Matsugaki, M. Kawasaki, N. Igarashi, M. Suzuki, R. Kato, T. Earnest, K. Nakayama, and S. Wakatsuki. 2002. Structural basis for recognition of acidic-cluster dileucine sequence by GGA1. Nature. 415:937–941. [DOI] [PubMed] [Google Scholar]
- Takatsu, H., M. Futatsumori, K. Yoshino, Y. Yoshida, H.W. Shin, and K. Nakayama. 2001. a. Similar subunit interactions contribute to assembly of clathrin adaptor complexes and COPI complex: analysis using yeast three-hybrid system. Biochem. Biophys. Res. Commun. 284:1083–1089. [DOI] [PubMed] [Google Scholar]
- Takatsu, H., Y. Katoh, Y. Shiba, and K. Nakayama. 2001. b. Golgi-localizing, gamma-adaptin ear homology domain, ADP-ribosylation factor-binding (GGA) proteins interact with acidic dileucine sequences within the cytoplasmic domains of sorting receptors through their Vps27p/Hrs/STAM (VHS) domains. J. Biol. Chem. 276:28541–28545. [DOI] [PubMed] [Google Scholar]
- Zhu, Y., B. Doray, A. Poussu, V.P. Lehto, and S. Kornfeld. 2001. Binding of GGA2 to the lysosomal enzyme sorting motif of the mannose 6-phosphate receptor. Science. 292:1716–1718. [DOI] [PubMed] [Google Scholar]