Abstract
We have developed a technique called the Integrated Procedure for Gene Identification that modifies and integrates parts from several existing techniques to increase the efficiency for genome-wide gene identification. The procedure has the following features: (i) Only the 3′ portion of the expressed templates is used to ensure a match to 3′ expressed sequence tag (EST) sequences; (ii) the 3′ portion of the cDNA is poly dA/poly dT minus, which maintains complete representation of the expressed copies, particularly the rare copies, which otherwise would be lost heavily because of random poly dA/poly dT hybridization in the subtraction reaction; (iii) redundancy is decreased substantially by the subtraction reaction to reduce the effort for sequencing analysis; (iv) the nonsubtracted templates that largely contain the rare copies are amplified selectively with suppression PCR and are sequenced directly or through serial analysis of gene expression (SAGE); and (v) the identified sequences are matched to databases to determine whether they are cloned genes, ESTs, or novel sequences. Using this procedure in a model system, we showed that the redundant copies were largely removed, and the rates of EST matches and the novel sequence identification were significantly increased. Most of the plasmids containing the matched EST are readily available from the IMAGE consortium. This technique can be used to index genome-wide expressed genes and to identify differentially expressed genes in different cells. Compared with the existing techniques, this procedure is relatively efficient, simple, less expensive, and labor intensive. It is especially useful for standard molecular laboratories to perform genome-wide studies.
Genes expressed in a cell are regulated tightly in a temporal and tissue specific manner to maintain the normal growth and differentiation of the cell. Misregulation of genes in pathological situations changes the expression pattern, which alters the normal cell behavior and leads to various abnormalities, such as neoplasia. Analysis of gene expression in normal conditions can provide information to understand the basic cell physiology. Identification of abnormally expressed genes in pathological conditions can provide markers for early diagnosis, targets for drug design, indicators for treatment responsiveness, and prognosis.
The total number of expressed human genes has been estimated to be ≈100,000, with ≈11,000 genes expressed in any particular cell type (1). The level of expression of these genes varies, depending on a number of factors. Studies show that 85% genes from 49,000 genes identified were expressed at less than five transcripts per cell, constituting only 25% of the mass of total transcripts in a cell, whereas the remaining 15% genes were expressed at a much higher level and constituted 75% of the mass of total mRNA within a cell (2). Therefore, the majority of expressed genes belongs to the rare abundant class, and the processes for gene identification should focus on this category. More than 1,062,033 expressed sequence tags (ESTs) from the human genome are listed in the current EST database (National Center for Biotechnology Information dbEST database release, July 17, 1998). Ultimately, most of the expressed genes from the human genome should be indexed in the EST database. Maximal use of EST database information will accelerate greatly the identification of genes expressed in various conditions (3–4).
Because of the large size of the human genome, a critical issue for genome-wide gene identification is the availability of powerful techniques suitable to accomplish the task. Most of the methods currently used for genome-wide gene identification have inherent disadvantages as well as unique advantages. DNA microarray technology cannot be used to identify novel genes because the probes are based on known sequences (5–6). Large-scale sequencing of the expressed sequences such as in the cancer genome anatomy project (CGAP) is too costly for standard laboratories (7). The serial analysis of gene expression (SAGE) technique significantly decreases the overall work for genome-wide gene identification, but the scale is still very large (8). The differential display technique is relatively simple, but it does not provide sequence information directly, and it gives a high rate of false positives (9). More sophisticated techniques need to be developed to fulfil the special needs of the genome-wide studies. These techniques should be highly sensitive to identify the genes expressed at a low level, should require a relatively small amount of initial materials to conserve limited resources, should be largely based on routine molecular biology techniques to be applicable in standard laboratories, and, more importantly, they should use maximally the publically accessible databases for gene identification.
Through modification and integration of parts of several techniques, we developed a method called an Integrated Procedure for Gene Identification (IPGI). The procedure includes (i) collecting only the 3′ portion cDNA from all expressed genes to ensure that most of the sequences would be within the 3′ EST sequence range; (ii) removing poly dA/dT sequences from cDNA templates to avoid random hybridization between poly dA/poly dT sequences between templates during the subtraction process; (iii) performing subtraction hybridization to remove redundant templates and suppression PCR to selectively amplify the enriched copies; (iv) using multiplex quantitative PCR to verify the subtraction efficiency; and (v) identifying the resultant sequences through database match as cloned genes, EST sequences, or novel sequences. By using a model system, we demonstrate that these procedures allowed us to achieve our objectives; that is, the rare copies can be preserved largely for analysis, the EST database can be used maximally for gene identification, and the rate of identification of bona fide novel sequences can be increased significantly.
MATERIALS AND METHODS
Cell Culture.
HL60 cells were cultured at 37°C in RPMI 1640 medium with 10% fetal calf serum. Cells were harvested at exponential phase for RNA isolation.
mRNA Isolation.
Total RNA was isolated with Trizol reagent (Life Technologies, Gaithersburg, MD) following the manufacture’s instruction. The isolated RNA then was treated with DNase I. mRNA was isolated from total RNA with Dynal dT25 (Dynal, Oslo) following the manufacture’s protocol.
cDNA Synthesis.
Double-stranded cDNAs were synthesized with a cDNA synthesis kit (Life Technologies) following the manufacture protocols, except that a mixture of three 3′ anchored and 5′ biotinylated primers 5′ biotin-TTTGCATGCTCGAG-T16-A/G/C was used for the reverse transcription reaction. The biotin was used for 3′ cDNA recovery. The anchored nucleotides were used to exclude poly dA sequences in cDNA templates. A SphI site was included in the primers for the addition of adapter.
3′ cDNA Recovery.
Double-stranded cDNAs were digested with NIaIII. 3′ cDNA was recovered with Dynal M280 avidin beads (Dynal) according to the manufacture’s protocol. After washing the unbound fragments, the bound 3′cDNA was released from the beads by mixing with phenol at 65°C for 30 min and vortexing at full speed for 10 min. Recovered 3′ cDNAs were precipitated, washed, and dissolved in TE buffer (10 mM Tris·Cl, pH 8.0/1 mM EDTA, pH 8.0). The purified 3′ cDNA was further digested with SphI to generate a CATG end within the reverse transcription primer sequence for adapter ligation.
Ligation of Adapters.
The 3′ cDNAs were divided into two groups. One was designated as tester, the other was designated as driver. The tester was divided further into two sets for ligation to adapter A or adapter B. The sequence of adapter A was sense, 5′ ATA CGA CTC ACT ATA GGG CTC GAG CGG CCG CAT ATG GGA CAT G 3′ and antisense, 5′ TCC CAT ATG C 3′. The sequence of adapter B was sense, 5′ ATA CGA CTC ACT ATA GGG CAG CTC GCC GGC GTA TAG GGA CAT G 3′ and antisense, 5′ TCC CTA TAC G 3′. The primers were developed further from the original adapter sequences (10) in such a way that the 5′ part of both adapters contained T7 promoter sequences, the 3′ part of both adapters contained GGGACATG, the BsmFI/NlaIII recognition sites used in SAGE adapters (8), and the sequences between 5′ and 3′ parts were counterpart between adapter A and adapter B for better suppressive PCR amplification of templates bearing heteroadapters. BsmFI is a type IIS that cuts at a distant site from its recognizing site and is used to obtain tag sequence in SAGE technique (8). As the adapter sequences are not phosphorylated, only the sense sequences can be ligated to the templates. The ligation reactions were carried out at 16°C overnight.
3′ cDNA Subtraction.
The subtraction reaction was performed following the protocol for suppression subtraction hybridization (11). Different ratios between tester and driver were set from 1:0 to 1:35. The first hybridization was carried out for 10 hr at 68°C. After mixing samples with adapter A and adapter B, the second hybridization was performed at 68°C for another 10 hours. Samples then were used for suppression PCR amplification.
Suppression PCR.
Suppression PCR was performed by using T7 primer. The reactions first were incubated at 74°C for 5 min to extend the 3′ end complementary to the adapter sequences to generate T7 primer binding sites for PCR. PCR was performed at 94°C for 10 sec, 66°C for 20 sec, and 72°C for 20 sec. NIaIII-digested pBR322 DNA ligated with adapter A, adapter B, and adapters A/B was set as the control to monitor the suppression effects of the reaction. The amplifications were stopped when clear signals were seen on the gel (A/B), but the noise signals represented on the control reactions (A or B) were not significantly amplified. The PCR products were then purified and adjusted to the same concentration.
Multiplex Quantitative PCR.
Multiplex quantitative PCR was performed to determine the subtraction efficiency. (β-actin (12), HSC70 (13), and HSP75 (14) were selected as the indicators.) The PCR primers for β-actin were sense, TGTTACAGGAAGTCCCTTGC and antisense, TAAGGTGTGCACTTTTATTC; for HSC70 were sense, CCAGGAGGAATGCCTGGG and antisense, TTAATCAACCTCTTCAATGG; and for HSP75 were sense, AGATAAAGGCACAAGACGTG and antisense, GCAGGTAATTGGTC CTTGAA. The locations of these primers are downstream of the 3′ last NIaIII site of these cDNAs. Control templates that were homologous to but shorter than the wild-type templates were generated through amplifying the corresponding cDNA with the antisense primer and a truncated sense primer. The truncated sense primers contained the same sense primer sequences but were connected further with downstream sequences resulting in a gap in between. The gap was 20 bp, 20 bp, and 10 bp for β-actin, HSP75, and HSC70, respectively. The templates generated by these primers had the same sequences as the wild-type but were shorter because of the deletion. The sequences of these truncated primers for β-actin, HSP75, and HSC70 were TGTTACAGGAAGTCCCTTGCTTCTCTCTAAGGAGAATGG C, CCAGGAGGAATGCCTGGGTGGTGGAGCTCCTCCT, and AGATAAAGGCACAAGACG TGTCTTCTGGTGGATTAAGCAA. The multiplex PCR reactions were performed by adding the DNA samples to the reaction mixtures containing the regular 5′ and 3′ primers from all of these genes, defined amounts of control templates, and α32P-dCTP. The PCR conditions were 94°C for 10 sec, 55°C for 20 sec, and 72°C for 20 sec for 38 cycles. The PCR products were fractionated on a 5% denaturing gel and were exposed on a PhosphorImager plate (Molecular Dynamics). The signal intensities were measured by ImageQuant (Molecular Dynamics). The ratio between wild-type and control templates was determined for each gene. The subtraction efficiencies were determined by comparison of these ratios among different samples.
cDNA Sequencing and Sequence Alignments.
The amplified DNAs from suppression PCR were cloned directly into pCR2.1 vector (Invitrogen). Sequencing reactions were performed by using M13 reverse primer and an Applied Biosystems cycle sequencing kit, and sequences were collected on an ABI377 autosequencer (Applied Biosystems). For database alignment, each sequence was matched first to GenBank databases by a blast search; if no match was found, the same sequence was used to match to the dbEST database. If no match was found in either database, the sequence was called a novel sequence.
Pre-SAGE Analysis.
The amplified DNA was digested with BsmFI to release the tag sequences from each template according to the SAGE protocol (8).
RESULTS
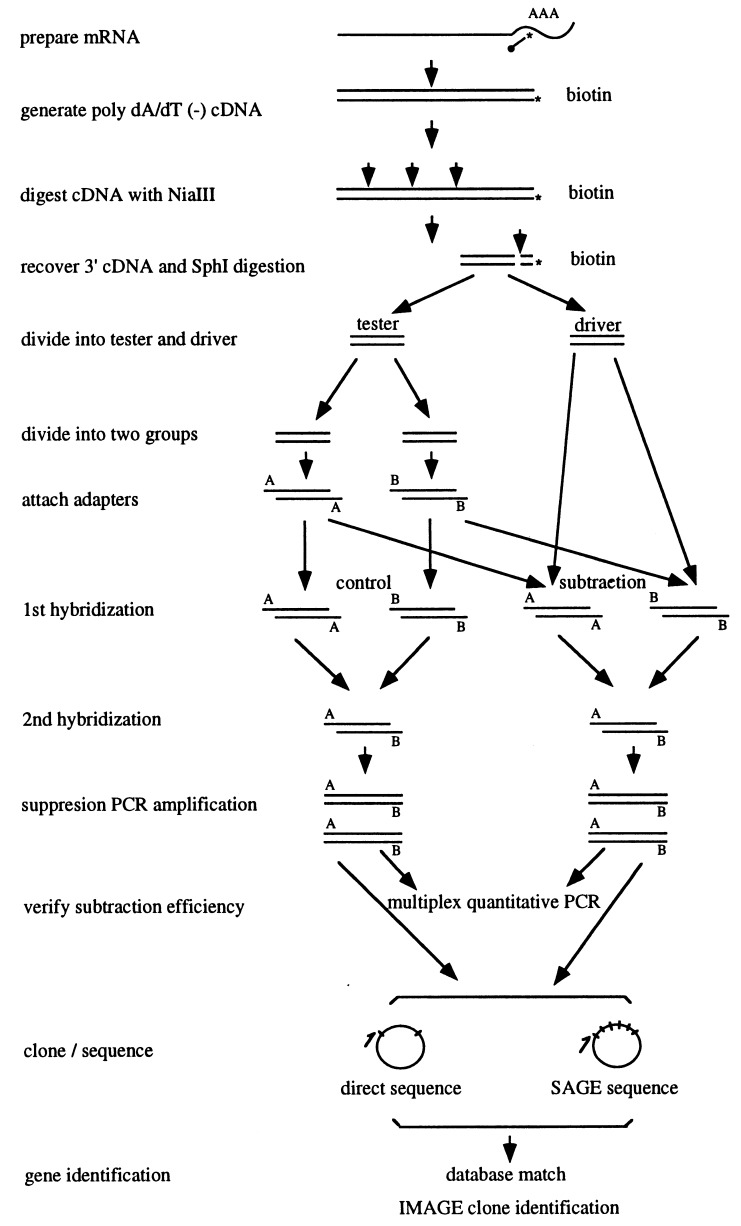
The strategy is illustrated in Table 1 and Fig. 1. Different parts of several techniques were integrated into a linear system for use in database matching and IMAGE consortium for clone identification. The same mRNA from HL60 cells was used for the analysis as in the normalization process. To validate the system, a control was set in which no driver was used for the subtraction reaction. The final results should indicate whether the system functions as expected; that is, the redundancy should decrease, the proportion of rare sequences should increase, and, among the identified sequences, many should match the EST database or lack a match and thus be classified as novel sequences.
Table 1.
The origin of integrated procedures for gene identification
| Original techniques | Integrated parts | Purpose |
|---|---|---|
| Differential display | Anchored oligo dT primers for reverse transcription | Generate poly dA/poly dT minus cDNAs to avoid the loss of low abundant copies in subtraction step |
| SAGE | NIaIII digestion Biotin labeling | Focus only on the 3′ sequences of any gene to maximally identify genes through matching EST |
| Suppression subtractive hybridization | Subtraction | Reduce mRNA requirement, and remove the abundant copies |
| Suppression subtractive hybridization | Suppressive PCR | Enrich the unsubtracted rare copies |
| Multiplex quantitative PCR | Relative quantification | Determine the subtraction efficiency |
| EST/CGAP | Large scale sequencing | Index the expressed genes |
| SAGE | Sequencing only 14 bases for each template | Index the expressed gene in a much smaller scale |
| Identify differentially expressed genes | ||
| GenBank database | National Center for Biotechnology Information blast search | Distinguish known genes, ESTs, and novel sequences |
| IMAGE consortium | EST plasmid stock | Obtain clones containing the matched EST sequences |
Figure 1.
Schematic of the integrated procedure for gene identification.
To isolate 3′ fragments from all of the expressed genes, the double stranded cDNAs were digested by the restriction enzyme NIaIII. NIaIII recognizes the CATG site that occurs on average every 256 bp (44) (8). The fragments generated by NlaIII digestion were distributed primarily between 300 to 500 bp (data not shown).
The 3′ cDNAs were isolated from the total NIaIII digested cDNA by using avidin beads. The recovered 3′ cDNA all included a CATG site at their 5′ end generated by NIaIII digestion. The 3′ cDNA was digested further by SphI to generate a CATG site at their 3′ ends. These sites are used for adapter ligation. The SphI site (GCATGC) was located in the reverse transcription primer. Even though it contained CATG, which is a NIaIII cleavage site, this site cannot be digested by NIaIII because of the shortness of the flanking sequences at the 3′ end.
A series of ratios between tester and driver was set for the normalization reactions to determine the subtraction patterns. The first round reaction was used to eliminate the redundant copies through the reassociation of the complementary templates between the two samples. The second round hybridization was continued to form the double strand templates, which were not subtracted in the first round of normalization. These represented the rare templates, and they were amplified selectively by suppression PCR. The annealing temperature in PCR was set at 66°C. At this temperature, the pan-structure by T7 sequences (19 bp) from the templates with heteroadapters would not form whereas the pan-structure from the whole adapter sequences (43 bp) from the templates with homoadapters would form. The T7 primer, therefore, can bind to the T7 site in the former templates but cannot bind to that in the latter templates. This leads to selective amplification of templates with heteroadapters but suppresses the amplification of templates with homoadapters (10).
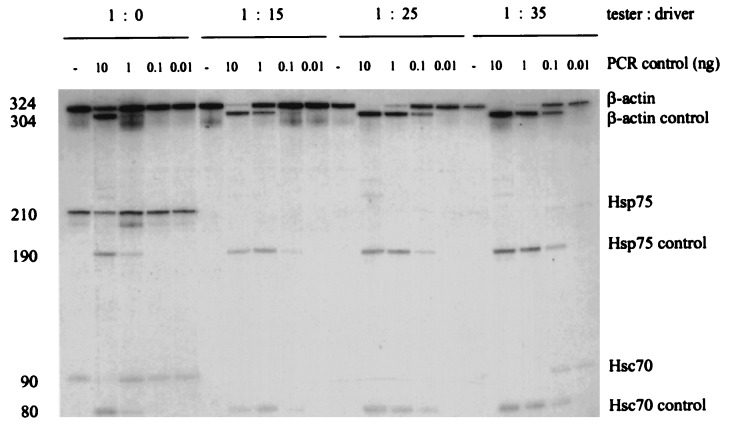
To determine the subtraction efficiency, the level of three genes was compared between the control and subtracted samples by using a multiplex quantitative PCR assay (Fig. 2). The β-actin gene is expressed at abundant level and has been used widely as a control in the analysis of gene expression. HSC70 and HSP75 were selected as representatives of intermediate abundant genes that are expressed at 100 copies per cell in human cell lines (S.M.W., K. Kaul, J. Khandekar, D. Winchester, and R. Morimoto, unpublished data). The level of β-actin copies decreased with the increasing amount of the driver DNA. The relative folds of subtraction were 4, 23, and 76 from the ratio (tester:driver) of 1:15, 1:25, and 1:35 respectively. HSC70 and HSP75 copies were hardly detectable after a ratio of 1:15.
Figure 2.
Determination of subtraction efficiency by multiplex quantitative PCR. The level of β-actin, Hsc70, and Hsp75 templates in different subtracted samples were quantified with the same set of control DNA and primers. The ratio between each set of wild-type and control templates reflects their relative content in each sample.
A total of 93 clones were sequenced, including 41 clones from the control sample and 52 clones from the subtracted sample at a ratio of tester to driver equaling 1:35. (Table 2 and Table 3). Analysis of these sequences showed that, of the 41 sequences from control clones, 78% (32 clones) matched to well defined sequences in the GenBank, European Molecular Biology Laboratory and DNA Data Base in Japan databases, with most of them being housekeeping or abundant genes; for example, 10 of the 32 clones were ribosomal protein sequences. Seventeen percent (seven clones) matched to EST sequences, of which two clones were ribosomal protein genes. Five percent (two clones) had no match. In contrast, of the 52 sequences from the subtracted sample, 40% (21 clones) matched to the GenBank sequences, with only 6 being ribosomal protein sequences; most of the others were functional genes. Forty percent (21 clones) matched EST sequences, and 20% (10 clones) had no match to any sequences. These two categories constitute 60% of the total clones. Through UniGene database searching, three of the EST sequences were identified as cloned genes, three had partial similarities to some genes, and two were ribosomal protein genes. Twenty plasmids containing the matched EST sequences are available from the IMAGE consortium. Therefore, through the IPGI process, we have achieved a substantial enrichment of sequences that matched to the EST database as well as bona fide novel sequences.
Table 2.
Sequence alignment of unsubtracted cDNA clones
| Clone | GenBank + EMBL + DDBJ. | dbEST | UniGene | Plasmid ID |
|---|---|---|---|---|
| 1 | ribosomal L5 | |||
| 2 | ribosomal S28 | |||
| 3 | elongation factor 1-alpha | |||
| 4 | — | AA374089 | — | 178491 |
| 5 | mitochondrial trnas and partial proteins 4 and 5 | |||
| 6 | ribosomal S3 | |||
| 7 | ribosomal protein L23a | |||
| 8 | BN51 | |||
| 9 | KIAA0002 | |||
| 10 | DNA polymerase delta small subunit | |||
| 11 | prothymosin alpha | |||
| 12 | elongation factor 1-alpha | |||
| 13 | elongation factor 1-alpha | |||
| 14 | — | D45527 | bleomycin hydrolase | lg1240 |
| 15 | — | AA526048 | hypothetical 75.2-kDa protein | 982571 |
| 16 | cytochrome c oxidase SII | |||
| 17 | ribosomal protein S28 | |||
| 18 | leucine-rich protein | |||
| 19 | leucine-rich protein | |||
| 20 | — | F20343 | ribosomal protein S12 | 036-X3-02 |
| 21 | T-cell cyclophilin | |||
| 22 | elongation factor 1-alpha | |||
| 23 | elongation factor 1-alpha | |||
| 24 | — | T23659 | — | b4HB3ma |
| 25 | elongation factor 1-alpha | |||
| 26 | ribosomal protein S8 | |||
| 27 | ubiquitin activating enzyme E1 | |||
| 28 | — | W07352 | elongation factor 1-alpha | 300651 |
| 29 | cytochrome c-1 | |||
| 30 | ribosomal protein S28 | |||
| 31 | ribosomal protein S24 | |||
| 32 | — | — | ||
| 33 | ribosomal protein L37a | |||
| 34 | — | — | ||
| 35 | mitochondrial genome X62996 | |||
| 36 | — | N75815 | ribosomal protein S23 | 300310 |
| 37 | ribosomal protein L37a | |||
| 38 | vacuolar H(+)-ATPase subunit | |||
| 39 | elongation factor 1-alpha | |||
| 40 | 23-kDa highly basic protein | |||
| 41 | 23-kDa highly basic protein |
Table 3.
Sequence alignment of subtracted cDNA clones (Tester:Driver = T:35)
| Clone | GenBank + EMBL + DDBJ | dbEST | UniGene | Plasmid ID |
|---|---|---|---|---|
| 1 | — | N75293 | Cbf5p homolog | 298739 |
| 2 | ribosomal protein L17 | |||
| 3 | — | T23947 | — | HB3MA-4 |
| 4 | — | AA142952 | similar to EGF-R substrate 15 | 504699 |
| 5 | — | — | ||
| 6 | — | AA256523 | — | 682558 |
| 7 | — | AA570755 | similar to MDR-1 | 914430 |
| 8 | — | AA427866 | — | 771016 |
| 9 | — | AA218963 | — | 650193 |
| 10 | — | W69264 | — | 343643 |
| 11 | — | — | ||
| 12 | Human Hlark mRNA | |||
| 13 | mitochondrial ubiquinone binding protein | |||
| 14 | eosinophil granule major basic protein | |||
| 15 | — | — | ||
| 16 | LLRep3 | |||
| 17 | TI-227H | |||
| 18 | — | H07593 | — | — |
| 19 | ribosomal protein L37a | |||
| 20 | LLRep3 | |||
| 21 | — | AA159050 | glucose phosphate isomerase | 591011 |
| 22 | — | AA310374 | — | 180113 |
| 23 | — | — | ||
| 24 | SnRNP core protein SmD3 | |||
| 25 | transferrin receptor | |||
| 26 | — | R48155 | — | 153719 |
| 27 | — | AA524203 | — | 936933 |
| 28 | KIAA0174 gene | |||
| 29 | — | C18312 | Mitochondrial NADH Fe-S protein | 18312 |
| 30 | acidic ribosomal phosphoprotein P2 | |||
| 31 | actin-related protein Arp3 | |||
| 32 | — | — | ||
| 33 | — | T24112 | — | Cot274 |
| 34 | ribosomal protein L5 | |||
| 35 | — | AA583472 | — | 1086940 |
| 36 | — | AA583472 | — | 1086940 |
| 37 | human protective protein | |||
| 38 | — | AA306845 | Highly similar to AUP46 precursor | 160936 |
| 39 | ribosomal protein S17 | |||
| 40 | ribosomal protein S28 | |||
| 41 | — | — | ||
| 42 | mitochondrial ubiquinone binding protein | |||
| 43 | — | H05063 | — | 43295 |
| 44 | ferritin light subunit | |||
| 45 | elongation factor 1-alpha | |||
| 46 | NADH-ubiquinone | |||
| 47 | — | AA484025 | ribosomal protein L19 | 910266 |
| 48 | — | — | ||
| 49 | — | AA156023 | ribosomal protein S8 | 590124 |
| 50 | — | — | ||
| 51 | ribosomal protein L27a | |||
| 52 | — | — |
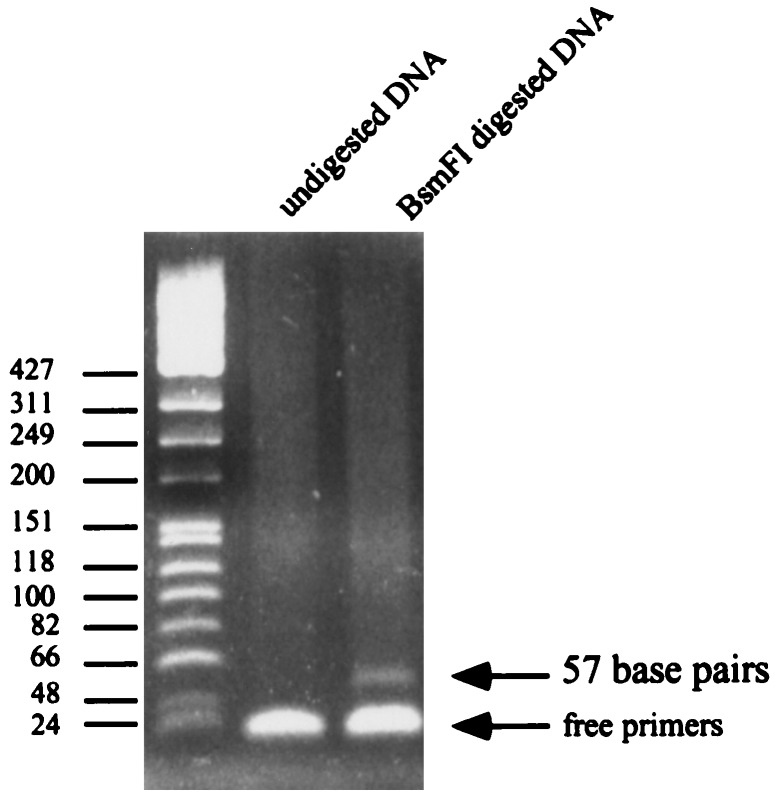
Direct sequencing of the amplified DNA demonstrated the presence of a BsmFI/NlaIII site in the adapter sequence. The amplified DNAs were digested with BsmFI to see whether tags can be released from each template for SAGE analysis. As shown in Fig. 3, a 57bp band containing the tag (14 bp) and the adapter sequence (41 bp) was identified clearly after the digestion. The tags released from the 3′ portion were removed by SphI digestion to maintain only one unique 5′ tag from each template for SAGE analysis.
Figure 3.
Tag fragments released from DNA templates. DNA amplified by suppressive PCR was digested by BsmFI to obtain the fragments containing tag from each template for SAGE analysis.
DISCUSSION
Analysis of gene expression has moved rapidly from classical studies on single or a few genes toward genome-wide studies on multiple genes. As most of the traditional techniques have very limited capacity for analysis on such a scale, and current techniques for genome-wide studies of gene expression have various limitations, we developed this procedure by integrating different parts of several techniques into a linear system to make maximal use of databases. The data we obtained show that we largely achieved our objective. The unique features of this system include the following: The cDNA templates do not contain poly dA/poly dT sequences to prevent random polydA/polydT hybridization between the templates in the subtraction process. This feature largely will conserve the rare copies after subtraction for gene identification. The templates are located at the 3′ end and contain mostly 300 to 500 bp. This feature guarantees the maximal use of EST information. The redundancy can be decreased largely through subtraction reaction. By using quantitative multiplex PCR to verify the subtraction efficiency, the mRNA requirement can be decreased significantly and the subtraction efficiency can be determined precisely. The inclusion of the SAGE technique further decreases the scale of analysis for genome-wide scanning.
Our data show that these features provide a cDNA population in which (i) much of the sequence redundancy can be reduced significantly and the rare templates can be enriched, (ii) the sequences identified can be used directly to match the EST database for gene identification, (iii) any sequences unmatched to the EST database are likely to be bona fide novel sequences not yet existing in the databases. In the report of Diatchenko et al. (11), 55 sequences of 62 sequences identified through subtraction suppressive PCR were considered “novel” sequences because they were not matched to databases. However, as these sequences could have came from anywhere in the cDNA template, these novel sequences could represent genes whose 3′ or 5′ sequences were already in the EST databases. To analyze these “novel” genes further, one would need to use traditional methods of screening cDNA libraries to clone these “novel” cDNAs. Our approach has a very high likelihood of distinguishing EST sequences from real novel sequences.
Compared with the 5′ EST sequences, the 3′ EST sequences are the most reliable ones because the cDNAs generated in reverse transcription are frequently unable to reach the 5′ end of the templates because of the existence of mRNA secondary structure (4). Several approaches have been developed in recent years for using the 3′ portion of cDNA for gene identification (8, 15–17). The advantage of focusing on 3′ sequences is well demonstrated by our system. First, it provides a better representation of genes because the 3′ part is the highly heterogeneous portion of the gene. Second, because sequences are relatively short for each gene, the probability of false hybridization among different genes will be decreased, which further increases the specificity of the subtraction reaction. Third, each expressed gene in the analysis has only one marker represented by its 3′ sequence, thus avoiding the uncertainty that multiple sequences may be generated for the same genes because of the analysis of the 5′ and 3′ portions when regular cDNA libraries are used. Fourth, and of most importance, it guarantees that the sequences identified will have the highest likelihood of matching to any existing EST sequences. The size of 300–500 bp parallels with the length of EST sequences and is sufficient as a specific marker for each gene. For these novel sequences, techniques such as 5′ rapid amplification of cDNA ends can be used to obtain more 5′ sequences if necessary (18).
A potential application of the IPGI technique would be for the EST project. The number of EST sequences currently is increasing rapidly, but the number of unique genes identified from these sequencing efforts is diminishing (http://www.ncbi.nlm.nih.gov/ncicgap/gene_discovery.html). This could suggest that most of the expressed genes might have been identified through the current EST project. However, when comparing data from SAGE analysis and the EST project, it is interesting to note that nearly half of the sequences identified by SAGE have no match in databases, including the EST database (2), indicating the potential of more sequences waiting to be identified. Many of these unidentified sequences may express at a very low level. The libraries used for the EST project are generated by oligo dT priming in reverse transcription, which generates cDNAs all containing poly dA/poly dT sequences at their 3′ ends, and undergo normalization/subtraction before being used for sequencing reaction (19). The random hybridization between poly dA and poly dT sequences in the normalization/subtraction process may lead to heavy loss of the rare copies by the abundant copies. This can be one of the major reasons why the current EST project has difficulty identifying more genes, particularly the genes expressed at a rare level. With the approaches described here, it should be possible to generate the libraries with better coverage of the rare copies. This may significantly increase the rate for novel sequence identification.
The IPGI procedures also may be applied to CGAP. The priority in the current CGAP is to index all genes expressed in primary tumors (7). Because of the large size of the human genome and the redundancy of the expressed transcripts, it is difficult, if not impossible, to identify all of the expressed genes by direct sequencing of the primary cDNA library from each tumor. The normalization/subtraction strategy would be a necessary step to decrease the redundancy for the analysis. On the other hand, it is very likely that, in many tumor cells, the abnormally expressed genes account for only a small portion of the total expressed genes and the majority of expressed genes would be the same as these expressed in normal cells (2). The EST project provides a large number of sequences expressed in normal cells. Maximal use of EST information will decrease significantly the cost for indexing genes expressed in tumor cells. The features of IPGI make it a good choice for CGAP: The sequences generated through the IPGI technique provide a high degree of completeness in covering most of the expressed templates, particularly the rare copies; only one unique 3′ marker for each gene will be generated, which will increase the specificity for gene identification and cut the cost in half if regular libraries are used for 5′ and 3′ sequencing; the overall work can be decreased significantly through the normalization process; and the EST information can be used maximally.
In summary, the establishment of IPGI procedures provides a tool for genome-wide gene analysis. It should find wide application in functional genomic studies. Of equal importance is the fact that it can be used in standard molecular biology laboratories to answer genome-wide questions that were not heretofore available.
Acknowledgments
We thank Nancy Zeleznik-le, Scott Fears, and Peter Domer for helpful discussion. This project is supported by an National Institutes of Health Grant CA425570 (to J.D.R.) and a grant from the G. Harold and Lelia Y. Mathers Foundation (to J.D.R.).
ABBREVIATIONS
- CGAP
cancer genome anatomy project
- EST
expressed sequence tag
- SAGE
serial analysis of gene expression
- IPGI
integrated procedure for gene identification
Footnotes
References
- 1. Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J D. In: Molecular Biology of the Cell. Robertson M, editor. New York: Garland; 1994. p. 369. [Google Scholar]
- 2.Zhang L, Zhou W, Velculescu V E, Kern S E, Hruban R H, Hamilton S R, Vogelstein B, Kinzler K W. Science. 1997;276:1268–1272. doi: 10.1126/science.276.5316.1268. [DOI] [PubMed] [Google Scholar]
- 3.Boguski M S. Trends Biochem Sci. 1995;20:295–296. doi: 10.1016/s0968-0004(00)89051-9. [DOI] [PubMed] [Google Scholar]
- 4.Gerhold D, Caskey C T. BioEssays. 1996;18:973–981. doi: 10.1002/bies.950181207. [DOI] [PubMed] [Google Scholar]
- 5.Lockhart D J, Dong H, Byrne M C, Follettie M T, Gallo M V, Chee M S, Mittmann M, Wang C, Kobayashi M, Horton H, Brown E L. Nat Biotechnol. 1996;14:1675–1680. doi: 10.1038/nbt1296-1675. [DOI] [PubMed] [Google Scholar]
- 6.DeRisi J, Penland L, Brown P O, Bittner M L, Meltzer P S, Ray M, Chen Y, Su Y A, Trent J M. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 7.Strausberg R L, Dahl C A, Klausner R D. Nat Genet. 1997;15:415–416. doi: 10.1038/ng0497supp-415. [DOI] [PubMed] [Google Scholar]
- 8.Velculescu V E, Zhang L, Vogelstein B, Kinzler K W. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 9.Liang P, Pardee A B. Science. 1992;257:967–970. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 10.Siebert P D, Chenchik A, Kellogg D E, Lukyanov K L, Lukyanov S A. Nucleic Acid Res. 1995;23:1087–1088. doi: 10.1093/nar/23.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diatchenko L, Lau Y C, Campbell A P, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov E D, Siebert P D. Proc Natl Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ponte P, Ng S Y, Engel J, Gunning P, Kedes L. Nucleic Acids Res. 1984;12:1687–1696. doi: 10.1093/nar/12.3.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dworniczak B, Milault M E. Nucleic Acids Res. 1987;15:5181–5197. doi: 10.1093/nar/15.13.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhattacharyya T, Karnezis A N, Murphy S P, Hoang T, Freeman B, Phillips B, Morimoto R I. J Biol Chem. 1995;270:1705–1710. doi: 10.1074/jbc.270.4.1705. [DOI] [PubMed] [Google Scholar]
- 15.Ivanova N B, Belyavsky A V. Nucleic Acids Res. 1995;23:2954–2958. doi: 10.1093/nar/23.15.2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato K. Nucleic Acids Res. 1995;23:3685–3690. doi: 10.1093/nar/23.18.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prashar Y, Weissman S M. Proc Natl Acad Sci USA. 1996;93:659–663. doi: 10.1073/pnas.93.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertling W M, Beier F, Reichenberger E. PCR Methods Appl. 1993;3:95–99. doi: 10.1101/gr.3.2.95. [DOI] [PubMed] [Google Scholar]
- 19.Bonaldo M F, Lennon G, Soares M B. Genome Res. 1996;6:791–806. doi: 10.1101/gr.6.9.791. [DOI] [PubMed] [Google Scholar]