Abstract
Here, we show that efficient transport of membrane and secretory proteins from the ER of Saccharomyces cerevisiae requires concentrative and signal-mediated sorting. Three independent markers of bulk flow transport out of the ER indicate that in the absence of an ER export signal, molecules are inefficiently captured into coat protein complex II (COPII)-coated vesicles. A soluble secretory protein, glycosylated pro–α-factor (gpαf), was enriched ∼20 fold in these vesicles relative to bulk flow markers. In the absence of Erv29p, a membrane protein that facilitates gpαf transport (Belden and Barlowe, 2001), gpαf is packaged into COPII vesicles as inefficiently as soluble bulk flow markers. We also found that a plasma membrane protein, the general amino acid permease (Gap1p), is enriched approximately threefold in COPII vesicles relative to membrane phospholipids. Mutation of a diacidic sequence present in the COOH-terminal cytosolic domain of Gap1p eliminated concentrative sorting of this protein.
Keywords: intracellular membranes; endoplasmic reticulum; COPII-coated vesicle; protein transport; protein sorting signals
Introduction
Proteins of the secretory pathway are shuttled between organelles in membrane-bound transport vesicles that are generated by cytoplasmic coat proteins. Selective inclusion of proteins into these transport vesicles is thought to be the principle mechanism that restricts proteins to individual organelles (Schekman and Orci, 1996). However, it has been difficult to distinguish between active and passive processes of cargo protein capture. Measurements of passive (bulk flow) transport in mammalian cells using a glycosylated acyltripeptide led to the proposal that secretory proteins are nonselectively transported out of the ER (Wieland et al., 1987). ER resident proteins could then attain their steady-state localization by being retained in the ER or retrieved from the Golgi.
An alternative model postulates that secretory cargo proteins possess positively acting sorting signals that promote their enrichment in transport vesicles. The first direct evidence in support of this model came from immunoelectron microscopy (IEM)* of mammalian cells infected with vesicular stomatitis virus (VSV), which demonstrated enrichment of viral surface glycoprotein (VSV-G) at ER exit sites. The density of gold particles labeling VSV-G was almost 10-fold higher at ER exit sites and in ER-to-Golgi transport vesicles than it was elsewhere in the ER (Balch et al., 1994). Unfortunately, careful consideration of the immunological techniques used to obtain these data has cast doubt on their validity (Griffiths et al., 1995). Subsequent in vitro studies in both yeast and mammalian cells have supported the notion that membrane secretory proteins are concentrated as they exit the ER, but these studies have lacked markers of bulk flow transport for comparison (Rowe et al., 1996; Kuehn et al., 1998). Improved IEM techniques have been reapplied to this problem in an analysis of protein transport from the ER of pancreatic acinar cells. These experiments demonstrated that the SNARE protein rBet1 is enriched approximately sixfold in coat protein complex II (COPII) vesicles that bud from the ER. However, two soluble secretory proteins, amylase and chymotrypsinogen, were not enriched relative to membrane surface area as they exit the ER, and were instead concentrated in the tubular structures of the ER–Golgi intermediate compartment (ERGIC) (Martínez-Menárguez et al., 1999). It is not yet clear whether nonselective transport of soluble secretory proteins from the ER of pancreatic acinar cells represents a general phenomenon or one particular to this specialized cell type. In yeast, a reconstituted ER export assay using the glycosylated proform of the yeast mating pheromone (glycosylated pro–α-factor [gpαf]) has demonstrated that this soluble secretory protein is very efficiently packaged into transport vesicles, but enrichment of gpαf relative to markers of bulk flow transport has yet to be demonstrated biochemically (Yeung et al., 1995).
Identification of amino acid sequence motifs within secretory proteins that could promote their enrichment into transport vesicles has been complicated by the lack of a universal consensus sequence. However, directed mutagenesis of candidate domains within secretory proteins has led to the characterization of two types of ER export signal: dihydrophobic motifs, which are found at the extreme COOH terminus of several type I transmembrane proteins (ERGIC-53/Emp47p, p24 family); and diacidic motifs, which are found in the cytosolic COOH-terminal domain of a number of membrane proteins.
A diacidic ER export signal was first recognized in the COOH-terminal cytosolic domain of VSV-G (Nishimura and Balch, 1997). Although this transport signal is referred to as a diacidic sequence (DxE), neighboring amino acids have been shown to contribute to efficient ER export (Sevier et al., 2000). Strikingly, conservative substitutions of the DxE motif (e.g., D-to-E) result in delayed arrival of VSV-G at the Golgi (Nishimura and Balch, 1997). Diacidic ER export signals have also been characterized in the COOH-terminal domain of mammalian potassium channels (Kir1.1 and Kir2.1) (Ma et al., 2001), and in a single S. cerevisiae protein, Sys1p (Votsmeier and Gallwitz, 2001). Unlike VSV-G and the Kir proteins, the diacidic sequence in Sys1p is located at the extreme COOH terminus of the protein, and binds avidly to the COPII subunit Sec24p. Thus, the generality of diacidic ER export signals is not well established, nor is their mechanism of action. Here, we show that a diacidic signal conserved among yeast amino acid permeases is required for concentrative sorting of the general amino acid permease, Gap1p. Furthermore, we have used markers of bulk flow transport and biochemical assays to quantify the enrichment of membrane and soluble secretory proteins in ER-derived transport vesicles.
Results and discussion
Secretory and plasma membrane precursor proteins are enriched relative to bulk flow markers in COPII vesicles
Previous studies in yeast and mammalian cells have demonstrated that secretory and plasma membrane precursor proteins are enriched in relation to ER resident proteins in COPII-coated vesicles (Yeung et al., 1995; Rowe et al., 1996; Todorow et al., 2000). However, these experiments failed to distinguish between active capture of secretory cargo and active exclusion of ER residents. To address this question, we developed biochemical markers of bulk flow transport out of the ER.
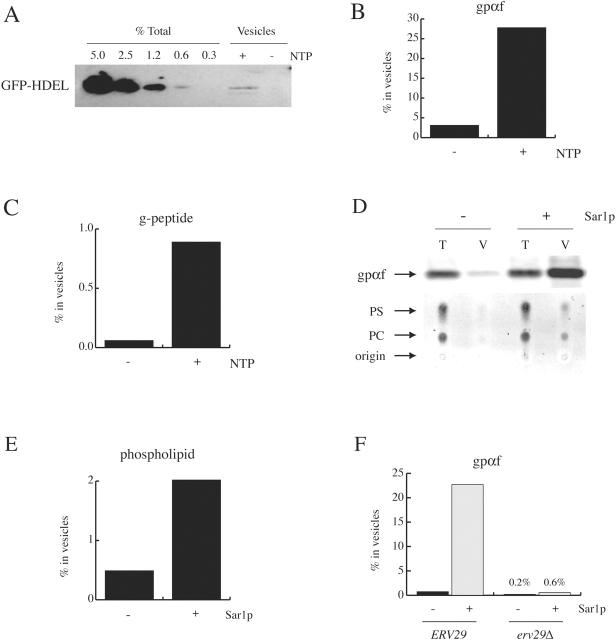
Markers of bulk flow transport should be neutral, lacking characteristics that either prevent or promote their transport. Retention of molecules in the ER is thought to occur via polymeric interactions between ER resident molecules (Ellgaard et al., 1999), whereas ER export is thought to be stimulated by the presence of transport signals. Therefore, we chose to divert a foreign cytosolic protein into the yeast secretory pathway to measure passive transport. GFP was targeted to the lumen of the yeast ER by appending the Kar2p signal sequence to the NH2 terminus of the GFP coding sequence (termed KGFP). To ensure that this hybrid protein would be located in the ER at steady-state, we also placed the ER retrieval sequence HDEL at the COOH terminus of GFP, yielding KGFP-HDEL. The GFP-HDEL hybrid protein was efficiently translocated into the ER lumen, was properly folded, as judged by its metabolic stability and intrinsic fluorescence, and was localized to the ER (data not shown; and Ng, D., personal communication). A microsomal membrane fraction enriched in ER membranes was prepared from cells expressing KGFP-HDEL and radiolabeled prepro–α-factor was then posttranslationally translocated into the lumen of these microsomes, so that the packaging of a soluble secretory protein could be measured alongside the packaging of GFP-HDEL. Membranes were incubated with purified COPII proteins in the presence and absence of guanine nucleotide, and a COPII-vesicle fraction was obtained by centrifugation. As shown in Fig. 1 A, ∼0.6% of the total GFP-HDEL present in the microsomal membranes was captured in vesicles in a nucleotide-dependent manner. In contrast, 28% of the gpαf was captured in COPII vesicles generated from reactions containing guanine nucleotide (Fig. 1 B).
Figure 1.
A soluble secretory protein (gpαf) is enriched in COPII vesicles relative to markers of bulk flow transport. (A) Microsomal membranes prepared from cells expressing GFP-HDEL (RSY255/pDN330) were combined with purified COPII proteins in the presence (+) or absence (−) of GMP-PNP (NTP). Serial dilutions of the total reaction (Total) and a vesicle-enriched (Vesicle) fraction were collected and analyzed by SDS-PAGE and immunoblotting using anti-GFP antibodies. (B) 35S-prepro–α-factor was translocated into microsomal membranes and these membranes were used in in vitro COPII-budding assays as described in A. The amount of gpαf in the vesicle fraction was quantified by PhosphorImager analysis. (C) Iodinated acyltripeptides were translocated into microsomal membranes prepared from RSY255. These membranes were used in in vitro COPII-budding assays, and the amount of membrane-associated glycosylated tripeptide (g-peptide) packaged into vesicles was quantified by scintillation counting (see Materials and methods). (D) Total (5%) and Vesicle (75%) fractions were collected from budding reactions containing all five purified COPII proteins (+) or lacking Sar1p (−). Fractions were analyzed for gpαf content by SDS-PAGE and for phospholipid content by organic extraction, followed by TLC and staining with a fluorescent dye (see Materials and methods). The percentage of phosphatidylcholine (PC) and phosphatidylserine (PS) (averaged) packaged into COPII-coated vesicles is shown in E. (F) 35S-prepro–α-factor was translocated into microsomal membranes prepared from ERV29 (BY4742) and erv29Δ (Y15936) cells. These membranes were used in in vitro COPII-budding assays as described in D and quantified as in B.
To substantiate our observations with the GFP-HDEL molecule, we analyzed a second soluble bulk flow marker. An acyltripeptide containing the acceptor site for N-glycosylation (benzoyl-NYT) has been used to assess transport through the secretory pathway in several biological systems (Wieland et al., 1987; Geetha-Habib et al., 1990), but in yeast there is an ATP-dependent, nonvesicular transport route for this molecule out of the ER. We modified our in vitro packaging assay to suppress this nonvesicular process and to recover only vesicle-associated peptides (Römisch and Schekman, 1992). As shown in Fig. 1 C, glycosylated tripeptide was packaged into COPII vesicles very inefficiently (0.8% of total), albeit in a nucleotide-dependent manner.
To extend our analysis of bulk flow markers to membrane components, we modified the budding reaction to achieve the degree of sensitivity necessary to measure the phospholipid content of COPII vesicles. Membranes were washed extensively to remove contaminating phospholipids and large-scale incubations with COPII proteins were followed by fractionation. Biochemical analysis revealed that gpαf was efficiently incorporated into the vesicle fraction (∼22% of total) when all COPII proteins are provided, but not when Sar1p was omitted from the incubation (Fig. 1 D, top). In contrast, only 2.0% of the total phospholipid was released into the vesicle fraction when all COPII components were provided, whereas 0.5% was released in a control lacking Sar1p (Fig. 1, D and E). These observations indicate passive transport of membrane components may be more efficient than bulk flow of soluble molecules. Nonetheless, we found that the soluble secretory protein gpαf was enriched ∼10 fold in COPII vesicles relative to phospholipid. The concentrative sorting we observed for gpαf differs dramatically with the IEM analysis of amylase transport in pancreatic acinar cells, but is quantitatively similar to the enrichment of the membrane proteins rBet1 and VSV-G at ER exit sites (Balch et al., 1994; Martínez-Menárguez et al., 1999).
Belden et al. have demonstrated that efficient packaging of gpαf requires a transmembrane cargo-receptor, Erv29p, and that this membrane protein can be cross-linked to gpαf in COPII vesicles (Belden and Barlowe, 2001). In the absence of Erv29p, we presumed that gpαf would be packaged into COPII vesicles as inefficiently as the soluble bulk flow markers described above. We analyzed the efficiency of gpαf packaging from erv29Δ microsomes and found that it was quantitatively similar to GFP-HDEL and acyltripeptide budding (<1%; Fig. 1 F). This indicates that in the absence of a dedicated transport receptor, secretory proteins leave the ER solely by virtue of the nonspecific sampling of the ER lumen that occurs during vesicle biogenesis, as has been observed for amylase and chymotrypsinogen in pancreatic acinar cells (Martínez-Menárguez et al., 1999).
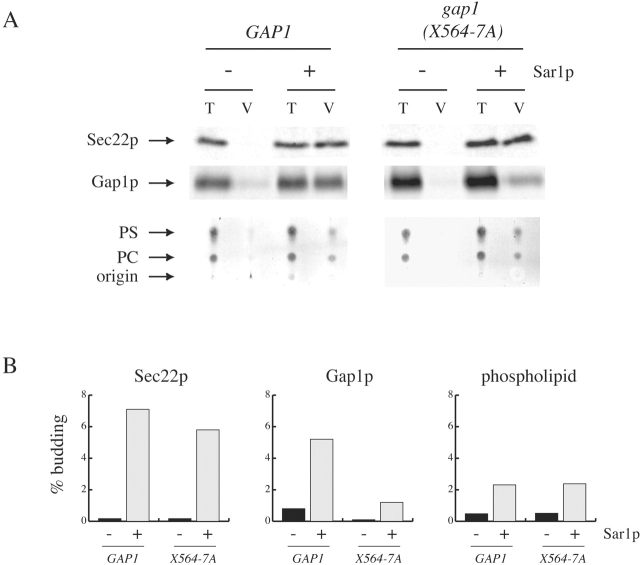
Two membrane proteins, Gap1p and Sec22p, were also analyzed using the assay described above, and the packaging of these molecules into COPII vesicles was determined by quantitative immunoblotting. We found that both Gap1p and Sec22p were also enriched in vesicles relative to membrane phospholipid (Fig. 2, A and B). The packaging efficiency of Gap1p and Sec22p (5% and 8% respectively) was low when compared with the packaging of gpαf (∼25%; Figs. 1 B and 2 B). This may be in part due to the extensive washing of membranes with 2.5 M urea that is required to obtain COPII-specific phospholipid packaging.
Figure 2.
A plasma membrane precursor protein requires an ER export signal for enrichment into COPII vesicles. (A) Microsomal membranes prepared from cells grown in SUD expressing HA-tagged Gap1p (Y17050/pPM11) or Gap1(X564–7A)p (Y17050/pPM12) were used in in vitro COPII-budding assays as described in the legend to Fig. 1 D. 5% of the total reaction (T), and 75% of the vesicle fraction (V) were analyzed by SDS-PAGE and immunoblotting (top two panels). A 35S-labeled secondary antibody and PhosphorImager analysis were used to quantify the packaging of Sec22p and Gap1p into COPII-coated vesicles (B). Phospholipids (A, bottom) were analyzed as in Fig. 1 D, and the percentage phospholipids packaged into COPII-coated vesicles was determined (B, right).
Amino acids in the cytosolic COOH terminus of Gap1p are required for enrichment into COPII vesicles
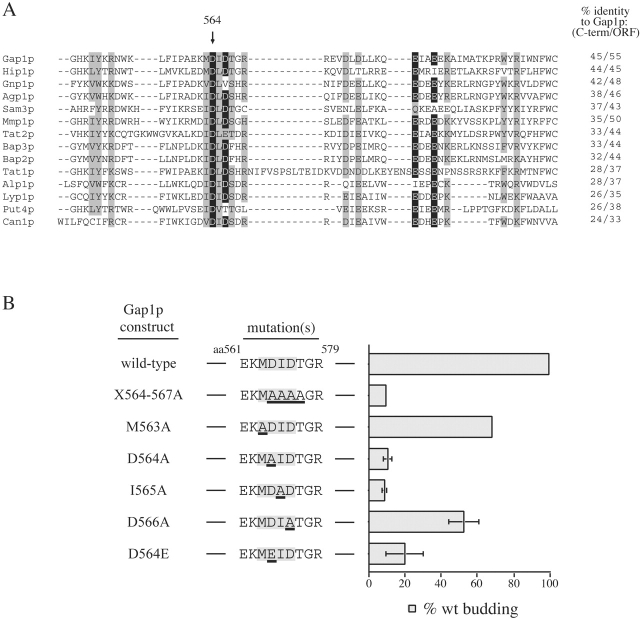
We sought to identify amino acids in Gap1p that are required for concentrative sorting of this protein into COPII vesicles. The work of Hein and André suggests that the COOH-terminal cytosolic domain of Gap1p (specifically residues 556–567) contains amino acids that are required for Gap1p to reach the plasma membrane, but that may not be required for enzymatic activity or regulation (Hein and André, 1997). Intriguingly, this region of Gap1p contains a diacidic sequence (DxD) that is highly conserved among yeast amino acid permeases (Fig. 3 A). We mutated this diacidic motif in Gap1p en bloc to alanine and assessed the packaging of this mutant protein into COPII vesicles. We found that only 1.2% of Gap1(X564–7A)p was packaged into COPII vesicles compared with 5.2% of wild-type Gap1 (Fig. 2, A and B). The packaging of Sec22p and phospholipid was not significantly affected by the presence of Gap1(X564–7A)p.
Figure 3.
Characterization of a diacidic ER export signal in the COOH-terminal domain of Gap1p. (A) The COOH- terminal domain of Gap1p (amino acids 546–602) was used in a BLAST search of the S. cerevisiae proteome to find related sequences (see Materials and methods). Gap1p and the 13 amino acid permeases identified in the BLAST search were aligned using ClustalW. Residues shaded black are identical in >80% of the aligned sequences and those shaded gray are similar in >80% of the aligned sequences. (B) Cells expressing HA-tagged Gap1p constructs were metabolically labeled for 3 min. Semi-intact cells (SICs) prepared from labeled cells were used in in vitro COPII-budding assays and analyzed by immunoprecipitation, SDS-PAGE, and PhosphorImager analysis. The packaging of Gap1p diacidic mutant proteins was normalized to the packaging of wild-type Gap1p.
If the diacidic sequence at positions 564–6 of Gap1p is truly an ER export signal, we expected that subtle perturbations in this amino acid sequence would cause packaging defects, as has been observed for VSV-G and Kir2.1 (Nishimura and Balch, 1997; Ma et al., 2001). Gap1p mutants were examined in an in vitro COPII-budding assay using gently-washed membranes prepared from metabolically-labeled cells which permits one to assess the packaging of newly-synthesized membrane proteins. Approximately 16% of radiolabeled Gap1p was captured into COPII vesicles in this assay. To allow comparison between experiments, the packaging of mutant proteins into COPII vesicles is expressed as a percentage of wild-type Gap1p (Fig. 3 B). Strikingly, even the conservative mutation of D564E in Gap1p caused a severe defect in ER export. The data in Fig. 3 B suggest that D564 and I565 are the two residues most sensitive to perturbation. This pair of residues is also highly conserved in the COOH-terminal domain of the yeast amino acid permeases (Fig. 3 A). Mutations in the diacidic sequence of Gap1p did not abrogate ER export by causing misfolding, as indicated by the stability and functionality of Gap1p diacidic mutants (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200208074/DC1).
The Gap1p ER export signal is transferable
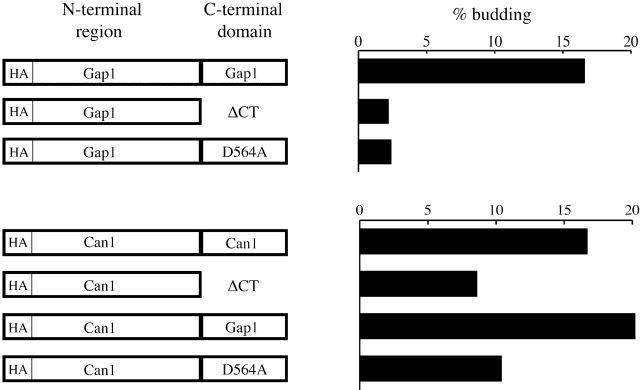
The potassium channel proteins Kir2.1 and Kir1.1 contain COOH-terminal diacidic signals required for their ER export (Ma et al., 2001). Ma et al. showed that the ER export signal of Kir2.1 was sufficient to promote the transport of a channel protein (Kir3.1, 1–373) that normally accumulates in the ER. We took an analogous approach to determine whether the ER export signal we identified in the COOH-terminal domain of Gap1p could mediate ER export of another protein.
We constructed HA-tagged chimeras of Gap1p and the distantly related arginine permease (Can1p) to determine whether the COOH-terminal domain of Gap1p could substitute for that of Can1p. Radiolabeled membranes were prepared from cells expressing HA-tagged Gap1p and Can1p proteins and used in in vitro COPII-budding assays. As shown in Fig. 4, full-length Can1p and Gap1p were packaged into vesicles with similar efficiency (∼17% of total radiolabeled protein). Removal of the Gap1p COOH-terminal domain (Gap1ΔCT) caused a severe defect in ER export, as did mutation of the diacidic signal in Gap1p (D564A), reducing packaging to ∼2–3%. The truncated version of Can1p (Can1ΔCT) was also packaged into COPII vesicles less efficiently than the full-length protein (∼9%), but the effect of truncation was not as dramatic as it was for Gap1p. This suggests that Can1p may also rely on ER export determinants present outside the COOH-terminal domain. Appending the COOH terminus of Gap1p to truncated Can1p resulted in a chimeric molecule that was packaged into COPII vesicles even more efficiently that the full length Can1p protein (∼20%). Appending the diacidic mutant Gap1p COOH terminus (D564A) to truncated Can1p did not promote incorporation into COPII vesicles. This observation indicates that the Gap1p ER export signal is transferable to Can1p and that it may act additively with other export determinants present in Can1p.
Figure 4.
Packaging of Gap1/Can1p chimeras into COPII vesicles. The first 535 residues of Gap1p and 540 residues of Can1p (NH2-terminal region) were fused to the cytosolic COOH-terminal domain of Gap1p, Gap1(D564A)p, or to a truncation possessing the first seven residues of the Gap1p COOH-terminal domain followed by two serines and a stop codon (ΔCT). HA-tagged chimeras were expressed from multicopy plasmids in BY4742 cells. Radiolabeled SICs prepared from these cells were used in in vitro COPII-budding assays (as described in the legend to Fig. 3 B). The percentage of radiolabeled chimera packaged into COPII vesicles is plotted on the right.
Mechanism of Gap1p packaging into COPII vesicles
Two biochemical intermediates in COPII vesicle formation that contain Gap1p have been identified. One is a complex between Gap1p and an ER-localized accessory protein, Shr3p. Shr3p is required for ER export of Gap1p, and a mutant shr3–23 allele has been shown to encode a protein that is defective in binding Gap1p (Gilstring et al., 1999); therefore, the defects we observed in packaging of Gap1p diacidic mutants into COPII vesicles could be due to their inability to bind Shr3p. We found, however, that our Gap1p diacidic mutants interacted with GST-Shr3p as efficiently as wild-type Gap1p (Gilstring, F., and P. Ljungdahl, personal communication).
A second biochemical intermediate in transport vesicle formation, termed a prebudding complex, contains the COPII subunits Sar1p and Sec23/24p, as well as secretory cargo (e.g., Gap1p and gpαf), but not ER resident molecules (e.g., Shr3p and Sec61p) (Kuehn et al., 1998). We observed that Gap1p diacidic mutants were not captured in prebudding complexes as efficiently as wild-type Gap1p (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200208074/DC1). Therefore, the diacidic ER export signal in Gap1p may be recognized by COPII subunits (Sar1p and/or Sec23/24p) after dissociating from Shr3p.
The binding interaction between COPII subunits and Gap1p is weak and can only be detected by Western blotting or by immunoprecipitation of metabolically labeled protein. In contrast, Sec23/24p binds avidly to the COOH-terminal domain of Sys1p. Unlike the complexes formed with Gap1p, activated Sar1p(GTP) seems not to be required for binding of Sec23/24p to Sys1p. Instead, Sec23/24p binds directly to GST-Sys1(CT) or to Sys1p COOH-terminal peptides (Votsmeier and Gallwitz, 2001; data not shown). Thus, the two known diacidic ER export signals in yeast differ in precise sequence, in position relative to transmembrane domains and the COOH terminus, and in the manner they bind to COPII. This raises the possibility that diacidic signals may mediate secretory protein transport via different pathways. Although the COOH-terminal domain of VSV-G, Kir2.1, and Sys1p have all been shown to promote ER export of poorly transported reporter proteins, no experiments have addressed whether these COOH-terminal domains are interchangeable, or whether they compete for common transport factors (Nishimura and Balch, 1997; Ma et al., 2001; Votsmeier and Gallwitz, 2001).
Structural analysis of the COPII coat and directed mutagenesis of its subunits may help refine our understanding of the mechanism of cargo protein capture. Low-resolution electron microscopy images of the Sec23/24p and Sec13/31p complexes have revealed the general shape of these coat subunits (Lederkremer et al., 2001; Matsuoka et al., 2001). Combined with atomic resolution crystallographic studies, the binding interfaces between subunits and the location of membrane proximal domains will be identified (Bi et al., 2002). With this information in hand, mutagenesis of surface residues in coat subunits may be used to create COPII coats that are capable of forming vesicles, but that are defective in packaging of certain cargo molecules. The sequence similarity of these cargo molecules may then reveal new ER export motifs, as well as confirming the generality of old ones. Combined with the available biochemical tools for analysis of COPII vesicle formation, such mutants may provide valuable insights into the mechanism underlying the selective capture of secretory proteins.
Materials and methods
Yeast strains, plasmids, growth conditions, and reagents
Yeast strains and plasmids used in this study are listed in Table SI (available at http://www.jcb.org/cgi/content/full/jcb.200208074/DC1). Plasmid pPM11 was generated by cutting pPL257 with SpeI and SacII, treating with mung bean nuclease, and ligating with T4 DNA Ligase (New England Biolabs, Inc.). Plasmids pPM12 through pPM18 were made using standard PCR mutagenesis and subcloning techniques as described (Ausubel et al., 1987). Plasmids pPM60 and pPM61 were made by digestion of pAS55 with XbaI and EcoRI, and insertion of a PCR product corresponding to nucleotides 1639–2667 of GAP1 or gap1(D564A) (obtained using pPM11 or pPM16 as templates). The NH2-terminal regions of GAP1 (4–1638) and CAN1 (4–1632) were obtained by PCR using a genomic DNA template and were introduced into pPM60 and pPM61. Subsequently, the COOH-terminal domain of CAN1 (1645–2746) was similarly cloned to generate plasmids pPM63 through pPM79. Plasmids pPM115 and pPM117 were made by inserting annealed oligonucleotides encoding the amino acid sequence YKRSS** into Bgl2-digested pPM63 and pPM73, respectively. Primers were purchased from Sigma-Aldrich, and nucleotide sequences were confirmed by sequencing (Elim Biopharmaceuticals).
Yeast were transformed using a standard lithium acetate procedure (Schiestl and Gietz, 1989). Yeast cells were grown in minimal (SD) or rich (YPD) media at 30°C. SCD contained 0.67 g/liter yeast nitrogen base without amino acids and ammonium sulfate, 0.05% citrulline, and 2% glucose. SUD medium contained 0.67 g/liter yeast nitrogen base without amino acids and ammonium sulfate, 1 g/liter urea, and 2% glucose, supplemented with required amino acids.
Immunoblots were conducted using the following primary antibodies: monoclonal anti-GFP (Covance Inc.), affinity-purified polyclonal anti-Sec61p, and polyclonal anti-Sec22p. Secondary antibodies conjugated to HRP were visualized using a SuperSignal West Pico chemiluminescent substrate (Pierce Chemical Co.). Alternatively, 35S-labeled secondary antibodies (Amersham Biosciences) were used for quantitative immunoblotting. Immunoprecipitations were conducted essentially as described (Kuehn et al., 1996), using monoclonal anti-HA (HA.11; BAbCo), polyclonal anti-Sec23p, and polyclonal anti-Gap1p.
In vitro COPII-budding assays
A microsomal membrane fraction enriched in ER membranes was prepared as described (Wuestehube and Schekman, 1992) from cells expressing KGFP-HDEL (RSY255/pDN330), Gap1p (Y17050/pPM11), or Gap1(X564–7A)p (Y17050/pPM12), or from untransformed cells (RSY255). 35S-labeled prepro–α-factor, generated by in vitro transcription/translation, was translocated into the ER (Baker et al., 1988). Acyltripeptides were iodinated and translocated into ER membranes (Römisch and Schekman, 1992). 1 mg of microsomal membranes was washed 4× with 1 ml of 2.5 M urea in B88, 2× with 1 ml of 0.1 μM GMP-PNP in B88, and 3× with 1 ml of B88 (all at 4°C). Washing was done by centrifuging membranes at 14,000 g in a fixed-angle rotor (TOMY, TMA-11) for 2 min at 4°C and resuspending gently with a micropipet. These membranes were then incubated with 0.1 μM GMP-PNP, and COPII proteins (±20 μg/ml Sar1, 40 μg/ml Sec23/24p, 40 μg/ml Sec13/31p) in 1 ml of B88 at 25°C for 20 min. Reactions were then placed on ice for 5 min. An aliquot (5%) of the reaction was set aside (Total fraction), and the remainder was centrifuged at 19,000 g in a swing-out rotor (TOMY, TMS-4) for 7 min at 4°C to yield a medium-speed supernatant (MSS). To isolate vesicle-associated glycopeptides, Total and MSS fractions were loaded on top of a 15–30–50% sucrose step gradient and centrifuged for 40 h. Fractions were collected from the top, and the vesicle peak was quantified by scintillation counting. In other experiments, membranes were recovered from Total and MSS fractions by centrifugation in a TL-100 ultracentrifuge (Beckman Coulter) at 70,000 rpm (TLA-100.3 rotor) for 20 min at 4°C, and were then resuspended in a small volume of B88. Aliquots of the Total and MSS were analyzed by SDS-PAGE and PhoshorImaging (gpαf) or immunoblotting (Gap1p). Phospholipids were extracted from aliquots of the Total and MSS fractions using the Bligh/Dyer method in mildly acidic conditions. Lipids were resolved on silica gel TLC plates in chloroform/methanol/acetate (65:35:5), stained with 0.005% primulin (Sigma-Aldrich), and visualized by blue fluorescence on a Typhoon PhosphoImager (Molecular Dynamics; Amersham Biosciences). Quantitation was performed using Molecular Dynamics ImageQuant software.
Alternatively, the packaging of newly synthesized membrane proteins was analyzed as described (Shimoni and Schekman, 2002). 35S-ProMix was purchased from Amersham Biosciences.
BLAST searches and sequence alignments
The amino acid sequence of the COOH-terminal domain of Gap1p was compared with Saccharomyces cerevisiae ORFs using four iterations of PSI-BLASTp (NCBI). The COOH-terminal domain (from the boundary of the last transmembrane domain to the COOH terminus) of the 14 S. cerevisiae permeases was aligned using ClustalW (v1.74, EBMnet server). The output of the ClustalW alignment was converted using BOXSHADE (v3.21). Boxed residues were identical (black) or similar (gray) in >80% of the aligned sequences.
Online supplemental material
Strains and plasmids used in this study are listed in Table S1. Fig. S1 shows the stability and functionality of Gap1p diacidic mutants. Fig. S2 shows that Gap1p diacidic mutants were not captured in prebudding complexes as efficiently as wild-type Gap1p. All supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200208074/DC1.
Acknowledgments
We would like to thank B. André (Free University of Brussels, Gosselies, Belgium), M. Hall (University of Basel, Basel, Switzerland), and D. Ng (Penn State University, University Park, PA) for providing strains and plasmids. Special thanks to F. Gilstring and P. Ljungdahl for collaborating with us in GST-Shr3p binding studies, and to B. Lesch and C. Chan (Howard Hughes Medical Institute) for providing reagents. Thanks also to E. Miller and M. Lee for comments on the manuscript and to members of the Schekman laboratory for both critical and supportive discussions.
This work was supported by the Howard Hughes Medical Institute.
The online version of this article includes supplemental material.
F. Jiang's present address is MedAmerica, Inc., 2101 Webster Street, Oakland, CA 94612.
Footnotes
Abbreviations used in this paper: COPII, coat protein complex II; ERGIC, ER–Golgi intermediate compartment; gpαf, glycosylated pro–α-factor; IEM, immunoelectron microscopy; VSV, vesicular stomatitis virus; VSV-G, viral surface glycoprotein.
References
- Ausubel, F.M., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl. 1987. Current Protocols in Molecular Biology. V.B. Chanda, editor. John Wiley & Sons, Inc., New York.
- Baker, D., L. Hicke, M. Rexach, M. Schleyer, and R. Schekman. 1988. Reconstitution of SEC gene product-dependent intercompartmental protein transport. Cell. 54:335–344. [DOI] [PubMed] [Google Scholar]
- Balch, W.E., J.M. McCaffery, H. Plutner, and M.G. Farquhar. 1994. Vesicular stomatitis virus glycoprotein is sorted and concentrated during export from the endoplasmic reticulum. Cell. 76:841–852. [DOI] [PubMed] [Google Scholar]
- Belden, W.J., and C. Barlowe. 2001. Role of Erv29p in collecting soluble secretory proteins into ER-derived transport vesicles. Science. 294:1528–1531. [DOI] [PubMed] [Google Scholar]
- Bi, X., R.A. Corpina, and J. Goldberg. 2002. Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature. 419:271–277. [DOI] [PubMed] [Google Scholar]
- Ellgaard, L., M. Molinari, and A. Helenius. 1999. Setting the standards: quality control in the secretory pathway. Science. 286:1882–1888. [DOI] [PubMed] [Google Scholar]
- Geetha-Habib, M., H.R. Park, and W.J. Lennarz. 1990. In vivo N-glycosylation and fate of Asn-X-Ser/Thr tripeptides. J. Biol. Chem. 265:13655–13660. [PubMed] [Google Scholar]
- Gilstring, C.F., M. Melin-Larsson, and P.O. Ljungdahl. 1999. Shr3p mediates specific COPII coatomer-cargo interactions required for the packaging of amino acid permeases into ER-derived transport vesicles. Mol. Biol. Cell. 10:3549–3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths, G., R.W. Doms, T. Mayhew, and J. Lucocq. 1995. The bulk-flow hypothesis: not quite the end. Trends Cell Biol. 5:9–13. [DOI] [PubMed] [Google Scholar]
- Hein, C., and B. André. 1997. A C-terminal di-leucine motif and nearby sequences are required for NH4(+)-induced inactivation and degradation of the general amino acid permease, Gap1p, of Saccharomyces cerevisiae. Mol. Microbiol. 24:607–616. [DOI] [PubMed] [Google Scholar]
- Kuehn, M.J., J.M. Herrmann, and R. Schekman. 1998. COPII-cargo interactions direct protein sorting into ER-derived transport vesicles. Nature. 391:187–190. [DOI] [PubMed] [Google Scholar]
- Kuehn, M.J., R. Schekman, and P.O. Ljungdahl. 1996. Amino acid permeases require COPII components and the ER resident membrane protein Shr3p for packaging into transport vesicles in vitro. J. Cell Biol. 135:585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederkremer, G.Z., Y. Cheng, B.M. Petre, E. Vogan, S. Springer, R. Schekman, T. Walz, and T. Kirchhausen. 2001. Structure of the Sec23p/24p and Sec13p/31p complexes of COPII. Proc. Natl. Acad. Sci. USA. 98:10704–10709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, D., N. Zerangue, Y.F. Lin, A. Collins, M. Yu, Y.N. Jan, and L.Y. Jan. 2001. Role of ER export signals in controlling surface potassium channel numbers. Science. 291:316–319. [DOI] [PubMed] [Google Scholar]
- Martínez-Menárguez, J.A., H.J. Geuze, J.W. Slot, and J. Klumperman. 1999. Vesicular tubular clusters between the ER and Golgi mediate concentration of soluble secretory proteins by exclusion from COPI-coated vesicles. Cell. 98:81–90. [DOI] [PubMed] [Google Scholar]
- Matsuoka, K., R. Schekman, L. Orci, and J.E. Heuser. 2001. Surface structure of the COPII-coated vesicle. Proc. Natl. Acad. Sci. USA. 98:13705–13709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura, N., and W.E. Balch. 1997. A di-acidic signal required for selective export from the endoplasmic reticulum. Science. 277:556–558. [DOI] [PubMed] [Google Scholar]
- Römisch, K., and R. Schekman. 1992. Distinct processes mediate glycoprotein and glycopeptide export from the endoplasmic reticulum in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 89:7227–7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe, T., M. Aridor, J.M. McCaffery, H. Plutner, C. Nuoffer, and W.E. Balch. 1996. COPII vesicles derived from mammalian endoplasmic reticulum microsomes recruit COPI. J. Cell Biol. 135:895–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman, R., and L. Orci. 1996. Coat proteins and vesicle budding. Science. 271:1526–1533. [DOI] [PubMed] [Google Scholar]
- Schiestl, R.H., and R.D. Gietz. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16:339–346. [DOI] [PubMed] [Google Scholar]
- Sevier, C.S., O.A. Weisz, M. Davis, and C.E. Macham. 2000. Efficient export of the vesicular stomatitis virus G protein from the endoplasmic reticulum requires a signal in the cytoplasmic tail that includes both tyrosine-based and di-acidic motifs. Mol. Biol. Cell. 11:13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoni, Y., and R. Schekman. 2002. Vesicle budding from endoplasmic reticulum. Methods Enzymol. 351:258–278. [DOI] [PubMed] [Google Scholar]
- Todorow, Z., A. Spang, E. Carmack, J. Yates, and R. Schekman. 2000. Active recycling of yeast Golgi mannosyltransferase complexes through the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA. 97:13643–13648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votsmeier, C., and D. Gallwitz. 2001. An acidic sequence of a putative yeast Golgi membrane protein binds COPII and facilitates ER export. EMBO J. 20:6742–6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland, F.T., M.L. Gleason, T.A. Serafini, and J.E. Rothman. 1987. The rate of bulk flow from the endoplasmic reticulum to the cell surface. Cell. 50:289–300. [DOI] [PubMed] [Google Scholar]
- Wuestehube, L.J., and R.W. Schekman. 1992. Reconstitution of transport from endoplasmic reticulum to Golgi complex using endoplasmic reticulum-enriched membrane fraction from yeast. Methods Enzymol. 219:124–136. [DOI] [PubMed] [Google Scholar]
- Yeung, T., C. Barlowe, and R. Schekman. 1995. Uncoupled packaging of targeting and cargo molecules during transport vesicle budding from the endoplasmic reticulum. J. Biol. Chem. 270:30567–30570. [DOI] [PubMed] [Google Scholar]