Abstract
Centromeres, telomeres, and ribosomal gene clusters consist of repetitive DNA sequences. To assess their contributions to the spatial organization of the interphase genome, their interactions with the nucleoskeleton were examined in quiescent and activated human lymphocytes. The nucleoskeletons were prepared using “physiological” conditions. The resulting structures were probed for specific DNA sequences of centromeres, telomeres, and ribosomal genes by in situ hybridization; the electroeluted DNA fractions were examined by blot hybridization. In both nonstimulated and stimulated lymphocytes, centromeric alpha-satellite repeats were almost exclusively found in the eluted fraction, while telomeric sequences remained attached to the nucleoskeleton. Ribosomal genes showed a transcription-dependent attachment pattern: in unstimulated lymphocytes, transcriptionally inactive ribosomal genes located outside the nucleolus were eluted completely. When comparing transcription unit and intergenic spacer, significantly more of the intergenic spacer was removed. In activated lymphocytes, considerable but similar amounts of both rDNA fragments were eluted. The results demonstrate that: (a) the various repetitive DNA sequences differ significantly in their intranuclear anchoring, (b) telomeric rather than centromeric DNA sequences form stable attachments to the nucleoskeleton, and (c) different attachment mechanisms might be responsible for the interaction of ribosomal genes with the nucleoskeleton.
Keywords: human lymphocytes, interphase nuclei, nucleoskeleton, nuclear matrix, fluorescence in situ hybridization
Repetitive DNA sequences in the human genome are numerous and diverse in their base composition. Several of them are clustered and possess clearly defined positions on chromosomes. The most prominent are the alpha-satellite repeats of centromeres, the telomeric repeats, and the clusters of ribosomal gene repeats. They have different functions as outlined in the following.
Alpha-satellite repeats are the main component of the centromeres and pericentromeric regions of all human chromosomes (Mitchell et al. 1992; Murphy and Karpen 1998). Centromeric DNA together with the associated proteins (CENPs; e.g., Haaf and Ward 1994; He et al. 1998) are the key structures where the mitotic spindle anchors. In most eucaryotes, the mitotic spindle is essential to accomplish an equal distribution of the genetic material into the two daughter cells (Rattner 1991; Wood et al. 1997). These DNA regions do not house active genes (Brown et al. 1997).
Telomeres generally consist of a highly repetitive hexanucleotide sequence of TTAGGG, although two other telomeric repeats have also been described (Allshire et al. 1989). The average length of these repeats are species and cell type specific (Lejnine et al. 1995; Lansdorp et al. 1996). Furthermore, since some telomeric DNA is lost in every round of DNA replication, the length of telomeres is indicative of the number of cell cycles a normal somatic cell has gone through. In cycling cells, it is therefore an indicator of cell age and may have an important role in the process of aging of individuals (Vaziri et al. 1993; Allsopp and Harley 1995; Allsopp et al. 1995; Henderson et al. 1996; Notaro et al. 1997; Weng et al. 1997; De Boer and Noest 1998). The telomeres are associated with a number of specific proteins (Zakian 1995; Gotta et al. 1996; Barinaga 1997; Cockell et al. 1998). A disturbance of the “telomeric complex” can result in end-to-end fusion of chromosomes (Hawley 1997; Mondello et al. 1997; Slijepcevic et al. 1997; van Steensel et al. 1998) and/or in malignant transformation of cells (Zakian 1995; Blasco et al. 1997). The telomeres are considered to be transcriptionally inactive.
Human ribosomal genes are organized in repeats at the secondary constriction of the five acrocentric chromosomes. In contrast to the centromeres and telomeres, this part of the genome can be transcriptionally highly active if cell proliferation and protein synthesis are high (Schwarzacher and Wachtler 1993; Raska 1995; Shaw and Jordan 1995; Sirri et al. 1995; Pederson 1998).
In addition to the above mentioned functions, it has been speculated that centromeres and telomeres have an organizing function in the spatial genome arrangement within the interphase nucleus (Haaf and Schmid 1991; He and Brinkley 1996). More than 100 yr ago, Rabl 1885 proposed a model of polarized orientation of interphase chromosomes with the centromeres at one side of the nucleus, and with the telomeres facing the other side. Since then, numerous studies on interphase chromosome arrangement have been performed and there is no doubt that the chromosomes in the interphase nucleus are not randomly distributed (e.g., Hilliker and Appels 1989; Cremer et al. 1993; Lamond and Earnshaw 1998). An ordered, nonrandom arrangement is especially evident for the centromeres, which tend to associate with the nuclear membrane and with the nucleolus (Manuelidis 1984; Billia and De Boni 1991; Ochs and Press 1992). The nonrandom distribution raises the question how this order is maintained and whether there exist structures that may serve as basic anchoring points that regulate the positioning of individual chromosomes and subchromosomal domains.
An underlying fibrillar structure such as the nucleoskeleton, or the nuclear matrix, is probably the best candidate for such a positioning structure (Berezney and Coffey 1975, Berezney and Coffey 1977; Mirkovitch et al. 1984; Hozák 1996; Nickerson et al. 1997). The fibrillar nature of the nucleoskeleton was clearly demonstrated, and some of the principal components of the nucleoskeleton have been identified in situ—such as heteronuclear RNPs, the nuclear mitotic apparatus protein, and intermediate filaments; e.g., lamins (Jackson and Cook 1988; He et al. 1990; Nakayasu and Berezney 1991; Hozák et al. 1995; Mancini et al. 1996; Mattern et al. 1997).
The question about the mechanisms of the interactions between the nucleoskeleton on one side, and the DNA or chromosomes on the other, has been addressed by many authors, sometimes with controversial results. Nevertheless, the postulated interactions can be divided into two principally different groups: first, interactions have been described between the nucleoskeleton and specific DNA sequences (MARs, SARs) that have rather permanent character and are responsible for chromatin loop formation (Getzenberg et al. 1991; Ivanchenko and Avramova 1992; Boulikas 1995; Gonzales and Sylvester 1995; Razin 1997). Second, interactions between functional DNA/protein complexes and the nucleoskeleton have been found, such as in transcription or replication foci. These attachments are thought to be rather transient than permanent (Jackson and Cook 1985b, Jackson and Cook 1986; Hozák et al. 1993; Hyrien et al. 1997; Jackson 1997; Stein et al. 1998).
As outlined above, the centromeres, telomeres, and ribosomal gene repeats have very different functions; in addition, they all might contribute to or influence the spatial organization of the genome in different ways. These contributions are poorly understood, and the mechanisms of the interactions have been identified only in a few cases.
Telomeric repeats have been shown to be matrix-associated in cultured cells (de Lange 1992; Ludérus et al. 1996). Conflicting results exist concerning the presence of attachment sites within the centromeric alpha-satellite repeats (Jackson et al. 1996; Strissel et al. 1996; Craig et al. 1997). For the ribosomal gene repeat, diverging observations were reported with respect to intranuclear attachment. Several studies describe an interaction with the nuclear matrix only for the intergenic spacer (Smith and Rothblum 1987; Stephanova et al. 1993). Others, however, have observed rDNA/nuclear matrix attachments throughout the entire ribosomal gene repeat (Keppel 1986; Maric and Hyrien 1998). Using gentle techniques for nucleoskeleton preparation (Jackson et al. 1988), associations of the transcription unit with the nucleoskeleton were observed in HeLa cells, whereas the intergenic spacer was attached to the nucleoskeleton only rarely (Dickinson et al. 1990; Weipoltshammer et al. 1996b).
The terminology used above illustrates the methodological difficulties of mapping the attachment sites: “nuclear matrix” protocols involve high-salt extraction, “nuclear scaffold” preparations involve the use of lithium diiodosalicylate, whereas “nucleoskeleton” preparations are generated with a more gentle, “physiological” method of chromatin removal. The many different approaches and cell models used make it very difficult to compare data on chromatin attachment. In addition, it is generally accepted that changes in the expression level of a gene can change its intranuclear attachment; however, only a few systematic studies exist on that issue (Craig et al. 1997; for a review see Stein et al. 1998). Throughout this paper, we use the terminology matching the extraction protocols applied.
In this paper, we compare the patterns of attachment of three functionally different repetitive genome elements, centromeres, telomeres, and ribosomal genes, to the nucleoskeleton, using a gentle and controlled method of chromatin extraction. Human lymphocytes before and after growth stimulation were used to compare nuclear attachment in cells with low and high levels of nuclear transcription. By using in situ evaluation, we took a different approach than in previous studies. This in situ method allowed us to investigate qualitative and quantitative signal characteristics at the single-cell level. We combined the in situ analysis of the DNA sequences remaining in the nucleus after chromatin removal with an analysis of the extracted DNA fraction by Southern blot hybridization. Double in situ hybridization experiments were performed to investigate attachment characteristics of the repetitive genome elements in single cells: telomeres plus ribosomal genes or centromeres plus ribosomal genes were detected simultaneously in control and extracted cells. The amount of DNA remaining in the nucleus after electroelution was quantified by densitometrically evaluating the signals after in situ hybridization. This method is complementary to procedures using biochemical fractionation of isolated DNA before and after chromatin depletion (e.g., Jackson et al. 1996). The method of quantification we used enables us to evaluate the degree of nucleoskeleton anchoring and at the same time to visualize attached DNA fragments at the single cell level.
The aim of this study is to investigate whether: (a) the various repetitive DNA sequences differ in their intranuclear anchoring, (b) transcriptional activation of cells results in a change of attachment characteristics of the three repetitive DNA sequences studied, and (c) the activation of nucleolar transcription is connected with a spatial rearrangement of specific rDNA elements relative to the nucleoskeleton.
Materials and Methods
Cells
Human lymphocytes were separated according to standard procedures. The separated lymphocytes were washed twice in PBS and embedded in low temperature-melting agarose (type VII; Sigma Chemical Co.) according to the protocol of Weipoltshammer et al. 1996b. One part of the embedded cells was subjected to the nucleoskeleton preparation procedure. The other part was incubated in RPMI medium (supplemented with 20% fetal calf serum and phytohemagglutinin; Abbott Laboratories) at 37°C for 72 h. Stimulated lymphocytes were then washed twice in PBS and subsequently nucleoskeletons were produced.
Preparation of the Nucleoskeletons
All solutions were prepared with diethylpyrocarbonate (DEPC)-treated distilled water (0.1%, inactivated) to minimize RNase content. Agarose-embedded cells were transferred into 0.1 M Soerensen buffer (SB; 70 mM Na2HPO4, 30 mM KH2PO4, pH 7.4), and then incubated in 0.05 M SB for 5 min at 37°C to disperse the granular component of nucleoli (Hozák et al. 1990, Hozák et al. 1992). Afterwards, cells were permeabilized in two changes of lysis buffer (70% PB diluted with DEPC-water, supplemented with 0.2% Triton X-100, 0.1 mM PMSF, and 2.5 U/ml RNase inhibitor; Amersham International), 5 min each on ice, and washed in “physiological buffer” (PB; 70 mM K-acetate, 30 mM KCl, 10 mM Na2HPO4, 1 mM MgCl2, 1 mM Na2ATP, 1 mM DTT, 0.1 mM PMSF, pH 7.4 adjusted with KH2PO4, 2.5 U/ml RNase inhibitor; Amersham International) 4 × 10 min on ice. Chromatin in permeabilized cells was cut using 340 U/ml HaeIII (Roche Laboratories) and 1,650 U/ml EcoRI (Roche Laboratories) in the presence of 25 U/ml RNase inhibitor (Amersham International); the reaction was carried out in a 3-ml vol bead suspension in PB for 20 min at 33°C. For the telomeric repeat, we cross-checked our results by cutting one sample of stimulated lymphocytes with RsaI (1,000 U/ml; Sigma Chemical Co.) and HinfI (1,000 U/ml; Roche Laboratories) that cut within the subtelomeric DNA sequences (Vaziri et al. 1993). No difference between the two samples was found. Subsequently, the bead suspension was introduced into the slots of an agarose gel (0.8% in tetraethylammonium [TAE] buffer, supplemented with 2.5 U/ml RNase inhibitor). Electrophoresis was run for 30 min at 20 V, and then for 2 h and 30 min at 35 V. For electrophoresis, PB diluted with DEPC-water to 70% and containing 0.1 mM PMSF and 0.5 U/ml RNase inhibitor was used as a running buffer. Cells were retrieved from the slots and fixed in 4% formaldehyde in PBS for 30 min.
Control samples were fixed with 4% formaldehyde in PBS for 30 min without any enzymatic digestion of DNA. All samples were washed twice in PBS, dehydrated in graded series of methanol, and stored in 100% methanol at −20°C for further analysis.
After recovering agarose beads from the gel slots, the gel was treated with RNAse A (0.5 μg/ml, 1 h, 30°C), proteinase K (5 μg/ml, 1 h, 20°C), and stained with ethidium bromide as described (Jackson and Cook 1986). The gel was photographed and blotted onto a Nylon membrane (Roche Laboratories) for hybridization.
As an internal control, DNA was isolated from three fractions of cells by phenol extraction: (a) before cutting with restriction enzymes, (b) after cutting with EcoRI and HaeIII before electrophoresis, and (c) after electrophoresis. With these three samples of isolated DNA, gel electrophoresis (1.7% agarose in TAE buffer) was performed.
DNA In Situ Hybridization
The agarose-embedded cells were spread over aminoalkylsilane-coated slides (Polysciences Inc.) and immobilized by incubating the slides at 65°C for 3 d. Fluorescence DNA in situ hybridization on nucleoskeleton preparations and control cells was performed according to Wachtler et al. 1991. Instead of proteinase K, 0.1% pepsin in 0.01 N HCl (pH < 2) was used (Dirks et al. 1989). To optimize hybridization conditions for the embedded cells, the formamide content of the hybridization mix was reduced to 30%. Sample and probe were denatured simultaneously on the slide for 10 min at 95°C. The hybridization took place at 37°C overnight in a moist chamber. Stringency washes were carried out at 42°C with 0.6× standard sodium citrate containing 20% formamide (three changes, 10 min each).
For the detection of telomeres and centromeres, commercially available probes were used: the digoxigenin-labeled “all human telomere DNA-probe” consisting of the TTAGGG repeat (Vysis), and the biotin-labeled “all human centromere probe” containing centromeric alpha-satellite sequences (Vysis). Alternatively, we used a Cy3-labeled PNA-telomere probe (DAKO SA) that has a higher sensitivity but a lower staining intensity. The probes for the detection of the ribosomal gene repeat were the EcoRI A fragment (part of the transcribed unit; stretching from the 3′ end of 18S to the 3′ end of 28S subunit, thereby including both internal transcribed spacers and the 5.8S subunit of the ribosomal gene; the A fragment measures 7.1 kb), and the HindIII DHH fragment (located within the noncoding intergenic spacer; it is part of the EcoRI-defined D fragment; the DHH fragment measures 9 kb). These probes were kindly donated by Prof. James Sylvester. They were labeled by nick-translation (nick-translation kit; Roche Laboratories) with either digoxigenin or biotin. For simultaneous detection of telomeres and rDNA, the digoxigenin-labeled “all human telomere probe” in combination with either A or DHH fragment, both biotin-labeled, was used. Simultaneous detection of centromeres and rDNA was carried out with the biotin-labeled “all human centromere” probe plus the digoxigenin-labeled rDNA fragments. Detection of DNA probes was performed with commercially available antibodies against digoxigenin (rhodamine conjugated), or using FITC-conjugated avidin in the biotin/avidin detection system (Pinkel et al. 1986) as in Weipoltshammer et al. 1996a.
Southern Blots
After electroelution, the agarose gel containing the eluted fraction of chromatin was blotted onto a Nylon membrane using 0.4 N NaOH overnight. DNA was fixed on the membrane by baking for 30 min at 110°C. The total extracted DNA was detected by hybridization with human genomic DNA as a probe (Sigma Chemical Co.). Specific sequences (telomeres, centromeres, and the ribosomal gene segments A and DHH) were detected with the same probes used for in situ hybridization. All DNA probes were digoxigenin-labeled, and then detected with antidigoxigenin antibodies conjugated with alkaline phosphatase (Roche Laboratories); NBT/X-phosphate solution (Roche Laboratories) was used as a substrate.
Imaging and Evaluation In Situ
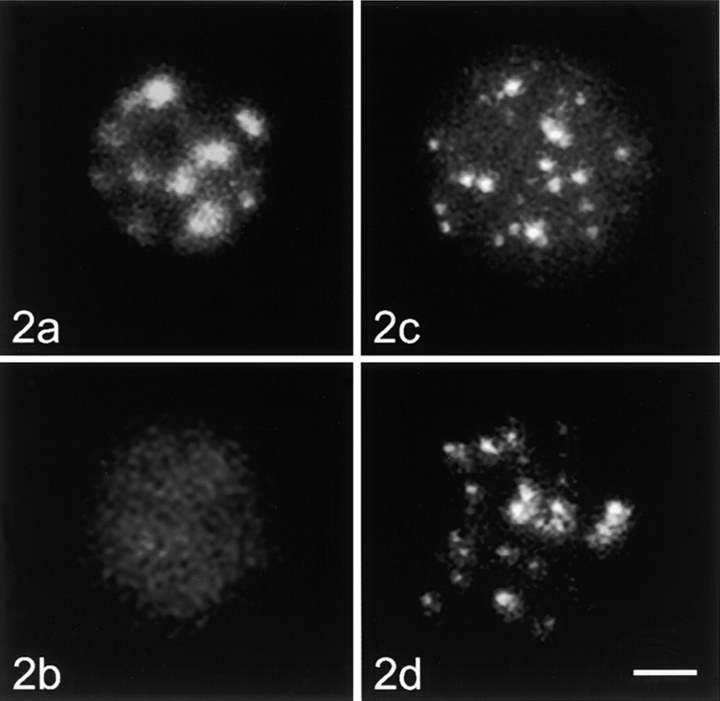
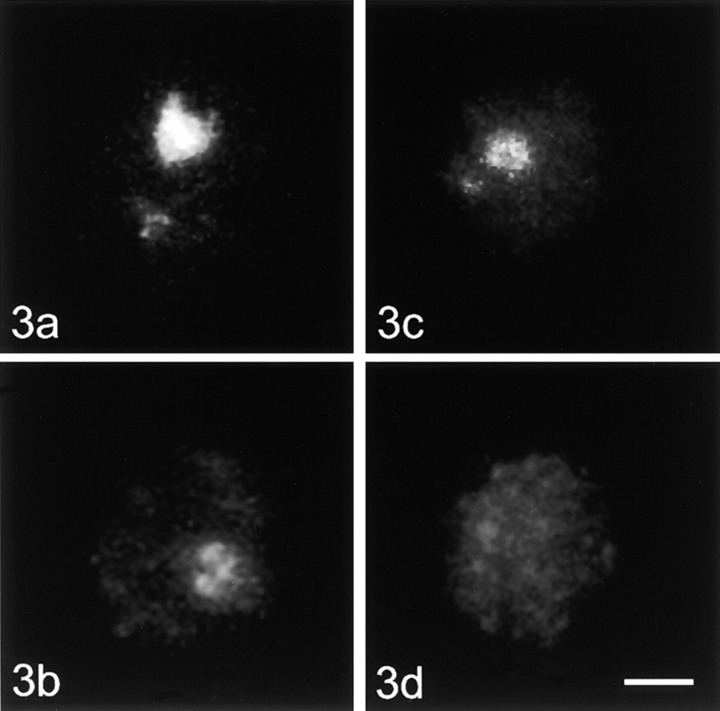
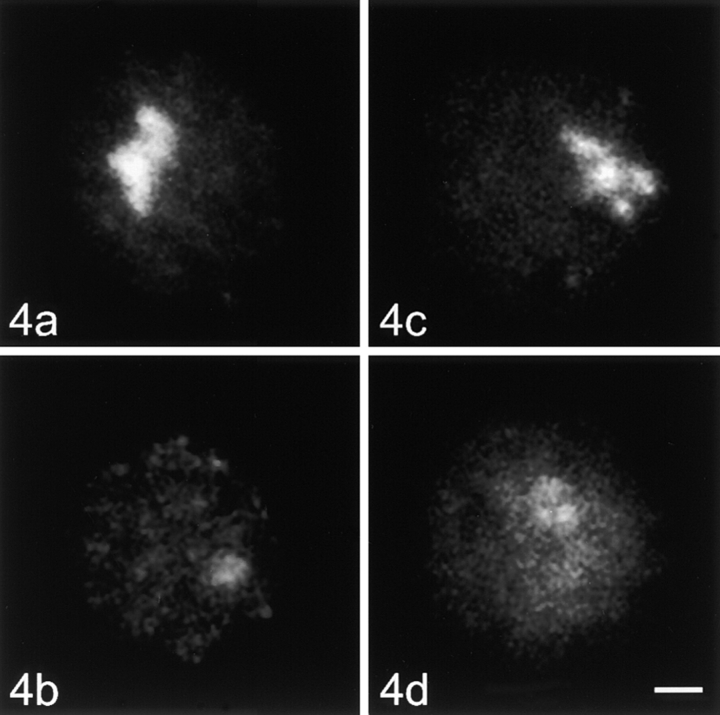
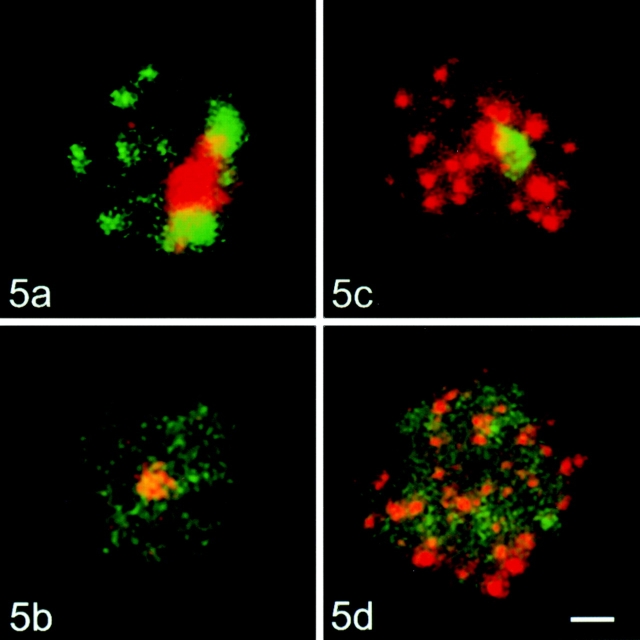
Optical sections of nuclei were recorded with a confocal laser scanning microscope (LSM 410; Carl Zeiss, Inc.). The nuclei displayed in Fig. 2 Fig. 3 Fig. 4 Fig. 5 are projections of stacks of optical sections.
Figure 2.
Unstimulated human lymphocytes, control cells, and nucleoskeleton preparations hybridized with the centromeric alpha-satellite probe and the telomeric repeat probe. Nucleoskeleton preparations were produced by permeabilizing the agarose-encapsulated cells with 0.2% Triton X-100 and cutting of DNA with the restriction enzymes EcoRI and HaeIII. Afterwards, the cells were placed into the slots of an agarose gel, electroeluted, and used for in situ hybridization. All figures are projections of stacks of optical sections. (a) Control cell, centromeric probe; (b) nucleoskeleton preparation, centromeric probe; (c), control cell, telomeric probe; (d) nucleoskeleton preparation, telomeric probe. Bar, 2 μm.
Figure 3.
Unstimulated human lymphocytes, control cells, and nucleoskeleton preparations hybridized with the A (part of the transcribed unit) and DHH (part of the intergenic spacer) fragments of the ribosomal DNA. (a) Control cell, A fragment; (b) nucleoskeleton preparation, A fragment; (c) control cell, DHH fragment; (d) nucleoskeleton preparation, DHH fragment. Bar, 2 μm.
Figure 4.
Stimulated human lymphocytes, control cells, and nucleoskeleton preparations hybridized with the A (part of the transcribed unit) and DHH (part of the intergenic spacer) fragments of the ribosomal DNA. (a) Control cell, A fragment; (b) nucleoskeleton preparation, A fragment; (c) control cell, DHH fragment; (d) nucleoskeleton preparation, DHH fragment. Bar, 2 μm.
Figure 5.
Stimulated human lymphocytes, control cells, and nucleoskeleton preparations hybridized simultaneously with centromeric alpha-satellite probe and ribosomal DNA probe, or with telomeric repeat probe and ribosomal DNA probe. (a) Control cell, centromeric probe detected with FITC-labeled avidin and A fragment detected with rhodamine-labeled antibodies; (b) nucleoskeleton preparation, hybridization protocol as in a; (c) control cell, DHH fragment detected with FITC-labeled avidin, telomeric probe detected with rhodamine-labeled antibodies; (d) nucleoskeleton preparation, hybridization protocol as in c. Bar, 2 μm.
Control and eluted cells were densitometrically evaluated for several criteria. For all in situ evaluations, confocal series of nuclei were gained under identical conditions (pinhole, resolution, z distance, gain, contrast, etc.).
First of all, the elution efficiency for DNA was checked. Control and eluted cells of unstimulated and stimulated lymphocytes were stained with quinacrin mustard. Samples were recorded and densitometric quantification of the sections was carried out with KS400 software (Kontron). The samples were segmented with a fixed threshold. The total signal density per nucleus was calculated from the optical sections and the mean signal density per nucleus was determined for each sample. Elution efficiency was expressed as the relation of mean signal density per nucleus of eluted cells to the control cells.
The same method was used to compare hybridization signals between control and eluted cells. For all sequences hybridized (telomeric repeat, alpha-satellite repeat, and A and DHH fragments) the relations of mean signal density per nucleus of eluted cells to the corresponding control cells were calculated.
Imaging and Evaluation of Blot Hybridizations
The hybridized membranes were digitized with a flat-bed scanner. The signal was densitometrically evaluated and normalized against the membrane background with KS400 software. Linearity of the detection system was checked with a dilution series of the alkaline phosphatase-labeled antibody. For comparison of A and DHH fragments, the densities of the hybridization signals were measured and given as relative value. To correct for differences in probe characteristics, a dilution series of total human DNA was blotted and hybridized with the A as well as the DHH fragment. The relation of the signal intensity of A and DHH fragments was the same for all DNA concentrations. The obtained ratio of signal intensity of A versus DHH fragment was used to normalize the hybridization signal of the electroeluted DNA fraction. The amount of extracted A and DHH fragments was compared within the sample of unstimulated lymphocytes and within the sample of stimulated lymphocytes densitometrically. The extracted total DNA was visualized on blots of the same elution experiments after hybridization with a total genomic probe.
Results
Nuclear Extraction
Nuclear morphology after the electroelution was well preserved as checked at the electron microscopic level (data not shown). In both unstimulated and stimulated lymphocytes, the DNA was generally extracted in equal amounts (>80% of DNA) as assessed with densitometric evaluation after quinacrin mustard staining (Table ). The amount of extracted DNA is in line with data from the literature (compare Dickinson et al. 1990). The permeabilization conditions used in our experiments represent a good balance between preservation of nuclear morphology and a high yield of DNA extracted.
Table 1.
In Situ Quantification of Chromatin Depletion
| Unstimulated lymphocytes | Stimulated lymphocytes | |||
|---|---|---|---|---|
| Control | Eluted | Control | Eluted | |
| % | % | % | % | |
| Total DNA | 100 ± 19.9 | 18.3 ± 5.3 | 100 ± 38.7 | 19.9 ± 9.4 |
| Centromeric DNA | 100 ± 20.7 | 1.7 ± 0.5 | 100 ± 33.1 | 1.8 ± 0.9 |
| Telomeric DNA | 100 ± 20.6 | 92.4 ± 1.7 | 100 ± 30.6 | 87.7 ± 13.8 |
| rDNA | ||||
| A fragment | 100 ± 60.4 | 11.1 ± 6.8 | 100 ± 42.7 | 9.5 ± 5.4 |
| DHH fragment | 100 ± 44.9 | 6.1 ± 6.3* | 100 ± 39.3 | 9.8 ± 3.8 |
Summary of the densitometric evaluation of control and electroeluted cells in situ. Total DNA was stained with quinacrin mustard, centromeric and telomeric repeats and rDNA fragments were labeled by fluorescence in situ hybridization. Cells were recorded and mean values of signal intensities in control and corresponding electroeluted samples were evaluated as described in Materials and Methods. The mean values are displayed as a percentage of signal intensities in electroeluted and control cells (set to 100%). Standard deviations are given as a percentage of the mean value of the control sample. Student's t test was performed for all experiments. Differences in signal intensity between control and eluted samples were highly significant (P < 0.001), with the exception of samples probed for telomeres (NS). When comparing unstimulated and stimulated lymphocytes, no significant differences in elution efficiency were found except for the DHH fragment of rDNA (*P < 0.05). In addition, the DHH fragment is extracted to a greater amount (*P < 0.05) than the A fragment in unstimulated lymphocytes, whereas no significant difference can be found in stimulated lymphocytes. For each sample, at least 20 randomly chosen cells were evaluated.
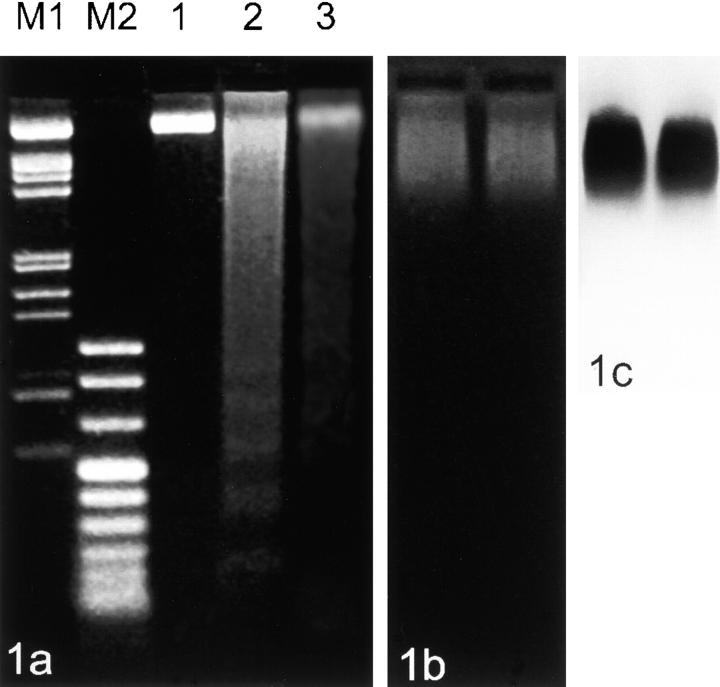
The reliability of the electroelution procedure was checked on the DNA level at several points. DNA was isolated from the agarose-embedded cells after restriction enzyme cutting (EcoRI, HaeIII) and directly after electroelution. Both samples were subjected to electrophoresis (Fig. 1 a). Total DNA after restriction enzyme cutting showed a DNA smear plus nucleosomal bands in the lower weight range (lane 2). The DNA fraction remaining in the nucleus after electroelution also shows a smear, the nucleosomal bands are no longer found (lane 3). This pattern is well in line with published pictures of extracted DNA from nucleoskeleton preparations (e.g., Jackson et al. 1996).
Figure 1.
Gel electrophoresis from different steps of nucleoskeleton preparations from stimulated human lymphocytes. (a) DNA was purified from agarose-embedded cells without cutting with enzymes (controls, 1), after cutting with EcoRI and HaeIII (2), and after cutting and removal of chromatin (3). Note the presence of nucleosomal bands in lane 2. Samples of DNA (1 μg in 1 and 2 and 0.5 μg in 3) were subjected to electrophoresis on a 1.7% agarose gel (M1: λ/EcoRI, HindIII; M2: pUCBM21/HpaII, DraI, HindIII). (b) Extracted fraction of chromatin after gel electrophoresis of cells cut with EcoRI and HaeIII. The two lanes displayed were loaded with roughly equal numbers of beads. Electrophoresis was performed on a 0.8% agarose gel that was subsequently treated with RNAse A and proteinase K (see Materials and Methods) before staining with ethidium bromide. (c) DNA hybridization of the blotted extracted fraction of chromatin using a human total genomic probe.
The extraction of chromatin from nuclei results in a smear in the agarose gel (Fig. 1 b). Nucleosomal bands are not clearly visible and are likely to be obscured by other extracted charged molecules (compare Jackson and Cook 1986). The corresponding blot hybridization with a total genomic probe is shown in Fig. 1 c. A prominent portion of DNA from eluted chromatin is present in the high–molecular weight range. In this respect, it should be added that it has been shown that chromatin containing DNA fragments larger than 125 kb can be removed from nuclei during preparation of nucleoskeletons (Jackson and Cook 1985a).
Centromeres
In metaphase cells, the “all human centromere DNA-probe” hybridized to all centromeres, resulting in bright foci, including the pericentromeric regions (data not shown). In the agarose-embedded control interphase nuclei, the hybridization signal was highly clustered (Fig. 2 a) and located preferentially at the nuclear periphery and around the nucleolus in unstimulated as well as stimulated lymphocytes (see Fig. 5 a). In nucleoskeleton preparations, the hybridization signal patterns were dramatically changed. The overall signal intensity was reduced and distinct signal clusters were no longer clearly visible in both unstimulated and stimulated lymphocytes (see Fig. 2 b and 5 b). Quantification revealed that <2% of signal specific for centromeric DNA remained in the nucleus after electroelution (Table ).
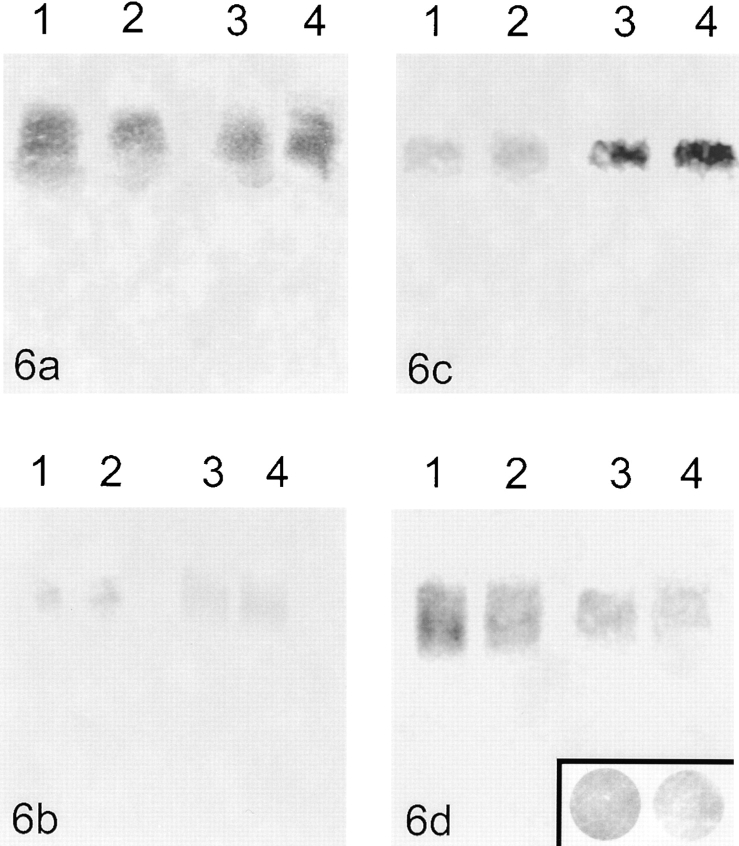
The blot hybridization of the extracted chromatin fraction (i.e., the unattached chromatin fragments) proved that a considerable amount of centromeric alpha-satellite sequences was removed during the extraction procedure regardless of the overall transcriptional activity of the cells used for elution (see Fig. 6 a).
Figure 6.
Blot hybridization of chromatin electroeluted from beads. (a) Hybridization with the centromeric alpha-satellite probe; (1 and 2) unstimulated lymphocytes, (3 and 4) stimulated lymphocytes. (b) Hybridization with the telomeric probe; (1 and 2) unstimulated lymphocytes, (3 and 4) stimulated lymphocytes. The very faint signal represents most likely nonattached extratelomeric TTAGGG repeats. (c) Unstimulated lymphocytes; (1 and 2) hybridization with the A fragment (part of the transcribed unit of rDNA), (3 and 4) hybridization with the DHH fragment (part of the intergenic spacer of rDNA). (d) Stimulated lymphocytes; (1 and 2) hybridization with the A fragment, (3 and 4) hybridization with the DHH fragment. The insert shows dot blots of equal amounts of human placenta-DNA hybridized with the A (left) and DHH (right) fragments. To be able to compare the amount of eluted A and DHH fragment in unstimulated as well as stimulated lymphocytes, the signal intensities in the blots c and d were normalized against these dot blots. This results in relative values for signal intensities A:DHH = 1.62:1 in unstimulated lymphocytes and A:DHH = 0.97:1 in stimulated lymphocytes.
Telomeres
In chromosome spreads, the all human telomere DNA-probe produced bright dots at the chromosomal ends (data not shown). In interphase control nuclei, the hybridization signal was visible as bright foci distributed throughout the entire volume of the nucleus, in both unstimulated (Fig. 2 c) and stimulated (see Fig. 5 c) cells. Hybridization with the PNA-telomere probe generated identical results (data not shown). In nucleoskeleton preparations of unstimulated and stimulated cells, no apparent differences in signal intensity and distribution were found when compared with the control cells (see Fig. 2 d and 5 d). Densitometric evaluation showed that ∼90% of telomeric DNA remained in the nucleus after nuclear extraction (Table ).
Southern blot hybridization of extracted chromatin fragments with the all human telomere probe produced only a very faint signal (see Fig. 6 b). This faint signal is most likely attributed to extra-telomeric TTAGGG repeats (de Lange 1992; Azzalin et al. 1997).
Ribosomal Genes
In metaphase spreads, both rDNA probes marked the nucleolus organizer regions on all five acrocentric chromosome pairs (data not shown). In interphase control nuclei, both probes hybridized to the nucleolus. In unstimulated cells, this signal was usually roundish or ring-shaped (Fig. 3, a and c). In addition to the typical nucleolar signal, one to several small dot-like signals were observed in ∼50% of the unstimulated lymphocytes. These dots correspond to the silent ribosomal gene clusters located outside of the active nucleolus (Wachtler et al. 1986). In stimulated lymphocytes, the nucleolar signal was much more prominent and extended; the extra-nucleolar signals were absent (Fig. 4, a and c).
When in situ hybridization was performed on chromatin-depleted nuclei of unstimulated lymphocytes, the remaining nucleolar signal consisted of one or occasionally a cluster of several dots for both rDNA fragments. The small extranucleolar signals representing silent gene repeats were completely removed from nuclei. An overall loss of signal intensity was seen for the A as well as the DHH fragment (Fig. 3b and Fig. d). However, densitometric quantification revealed that significantly more (factor 1.8) of the A than the DHH fragment remained in the nucleus after electroelution (Table ). Some of the cells had no hybridization signals at all; the percentage of such cells was different for the two rDNA fragments (34.6% for the DHH fragment and 14.6% for the A fragment).
In the case of in situ hybridizations to stimulated lymphocytes after chromatin depletion, the signal consisted of one to several dots for both probes (Fig. 4b and Fig. d). The signal intensity was also considerably lower than in control cells. In this case, however, densitometry showed that equal amounts of both fragments were removed from nuclei (Table ). The percentage of cells lacking any signal was practically identical for both probes hybridized (22.1% for A fragment and 23.4% for DHH fragment). The results of double in situ hybridization with telomeric and rDNA probes (or with centromeric and rDNA probes) proved that the differences in attachments of the various repetitive genome elements were reproducible on a single cell level (Fig. 5, a–d); thus, these differences cannot result from cell-to-cell variations in the efficiency of chromatin depletion.
The eluted chromatin fragments were blotted and hybridized with A and DHH probes. The hybridization intensities were semiquantitatively compared under normalized conditions. In unstimulated lymphocytes, significantly more of the DHH than the A fragment was removed from the cells by the extraction procedure (relative gray values: A:DHH = 1.62:1; a lower value means higher signal intensity; Fig. 6 c). In stimulated lymphocytes, however, no difference between the amount of eluted A and DHH fragments was detected (relative gray values: A:DHH = 0.97:1; Fig. 6 d). The fraction of stimulated lymphocytes contained ∼2% of mitotic cells with potentially different parameters for detaching chromatin fragments, as indicated by Gerdes et al. 1994 and Craig et al. 1997. The low abundance of mitotic cells allows us to neglect the possible effect on the quantified hybridization values.
Discussion
The Approach
The principal aim of this paper was to compare the attachment of three functionally different repetitive genome elements: centromeres, telomeres, and ribosomal genes to the nucleoskeleton. The experimental model, human lymphocytes during their metabolic activation, was chosen for two reasons. Firstly, the model provides an excellent system to study possible changes in chromatin/nucleoskeleton attachment during transcriptional activation. Secondly, the model allows us to use native cells without the possible effects of long-term cultivation. Our approach is based on a gentle and controlled removal of unattached chromatin fragments from permeabilized cells (Jackson et al. 1988), followed by a combination of detection of the DNA sequences remaining in the nucleus by in situ hybridization, and that of the extracted DNA fraction by Southern blot hybridization. The extraction method used is known to preserve not only the basic morphological characteristics of the nucleoskeleton, but also the synthetic activities of cell nuclei (Jackson and Cook 1985b, Jackson and Cook 1986; Hozák et al. 1993). The in situ approach allowed us to investigate signal intensity as well as signal distribution at the single cell level. Double in situ hybridization experiments served as a reliable control to describe attachment characteristics of the repetitive genome elements simultaneously in a single cell, thus eliminating possible errors when comparing parallel experiments.
Centromeric and Telomeric DNA Sequences Differ in Their Attachments
We found that centromeric and telomeric DNA sequences show great differences in their interactions with the nucleoskeleton. The majority of centromeric alpha-satellite repeats are removed during the extraction procedure; i.e., they cannot be anchored by a massive number of attachment sites to the nucleoskeleton (see also Jackson et al. 1996; Craig et al. 1997). The general attachment pattern observed was not connected with the activity of cell metabolism as the results were identical in both unstimulated and stimulated lymphocytes. However, it is possible that a minority of the centromeric sequences is still attached as Strissel et al. 1996 mapped SARs on chromosomes 1 and 16, and were able to hybridize the SAR fraction to the centromeres of the other mitotic chromosomes.
In contrast, telomeric chromatin is tightly attached to the nucleoskeleton in unstimulated as well as stimulated human lymphocytes. This observation is in agreement with the outcome of studies on nuclear matrix preparations of several established cell lines (de Lange 1992). In two studies on further characterization of the sequences retained in nuclei after nucleoskeleton preparation, telomeres or telomeric repeats were not described (Jackson et al. 1996; Craig et al. 1997). The reason for this difference to the findings presented here might be found in different methods of evaluation used after the electroelution step.
Our results on nucleoskeleton attachment of alpha-satellite repeats and telomeric repeats prove that two genomic elements can vary dramatically in their attachment properties despite of the fact that they are both transcriptionally silent.
Moreover, it seems that the attachment of telomeric and alpha-satellite repeats is similar throughout interphase. It is known that unstimulated lymphocytes are in G0 phase of the cell cycle (Wachtler et al. 1982). In our samples of stimulated lymphocytes, we counted that ∼58% of the cells were in G1 and 40% were in S and G2 phase (∼2% were mitotic figures). Nevertheless, the amount of electroeluted alpha-satellite and telomeric sequences was similar in both samples of lymphocytes.
It has been reported that the different chromatin-depletion protocols preferentially retain either structural (nuclear matrix, nuclear scaffold) or functional (nucleoskeleton) sequences (Jackson et al. 1996; Craig et al. 1997). This may explain discrepancies of our findings to reports about nuclear scaffold attachment of centromeres (Strissel et al. 1996). However, in the case of telomeres, our results are comparable to the study of Ludérus et al. 1996, although different preparation techniques were used (nucleoskeleton, nuclear scaffold).
The nature of the attachments of centromeres and telomeres to the nucleoskeleton is not yet fully understood. As both centromeres and telomeres are transcriptionally inactive, the attachment sites cannot be attributed to anchoring via transcriptional complexes. de Lange 1992 suggested that a nucleoprotein complex containing TTAGGG repeats could be the element responsible for nuclear scaffold attachments as TTAGGG repeats introduced by DNA transfection did not behave as matrix-attached loci. The exact nature of these interactions still remains to be defined. However, we can conclude that telomeres rather than centromeres contribute to the formation of intranuclear order by being anchored to the nucleoskeleton.
Activation Is Correlated to Spatial Rearrangement of rDNA Elements
The interaction of ribosomal genes with the nucleoskeleton is of a more complex nature. (a) Clusters of nontranscribed ribosomal genes, which are found outside the nucleolus in unstimulated human lymphocytes (Wachtler et al. 1986), are completely removed during the extraction procedure. (b) When comparing the attachment pattern of the transcription unit (A fragment) and the intergenic, nontranscribed spacer (DHH fragment), considerable differences between unstimulated and activated lymphocytes can be observed. In unstimulated lymphocytes, the amount of DHH fragment removed during extraction is significantly higher than the amount of A fragment. Blot hybridization confirmed the results obtained by in situ analysis. A fraction of cells exists with weaker signal than seen in control cells, or with no signal at all for both the A and DHH fragments. The cell fraction without hybridization signal is larger for the DHH fragments. In activated lymphocytes, the amount of A and DHH fragments eluted is roughly the same.
The results on unstimulated lymphocytes can be partly explained by an attachment via the transcription complexes (Dickinson et al. 1990; Hozák et al. 1994; Weipoltshammer et al. 1996b). Within the extranucleolar ribosomal gene clusters, no transcription takes place (Wachtler et al. 1986). As expected, the transcription unit (A) and the intergenic spacer (DHH) are extracted completely. Within the nucleolus, a preferential extraction of the DHH fragment is observed.
Nevertheless, a certain amount of DHH fragment remains within the nucleolus. Furthermore, in stimulated lymphocytes, the amount of A and DHH fragment removed during the extraction procedure is roughly the same.
These results imply that additional mechanisms of attachment of rDNA to the nucleoskeleton must be of importance. It could, for instance, be assumed that the permanent attachment sites (matrix attachment regions, MARs) that organize the DNA into loops are responsible for at least part of the attachments. MARs have been found preferentially within the intergenic spacers by several authors (Smith and Rothblum 1987; Stephanova et al. 1993; Gonzales and Sylvester 1995). One also has to bear in mind that unstimulated lymphocytes are in G0 phase of the cell cycle, whereas stimulated lymphocytes consist of cells in various phases of cell cycle. This could explain the somewhat unexpected finding that the rDNA fragments of stimulated lymphocytes are not significantly more retained in nuclei than those of unstimulated cells. Therefore, in contrast to telomeric and alpha-satellite repeats, a cell-cycle dependence of the rDNA attachment to the nucleoskeleton cannot be ruled out at this point.
In addition, differences might exist in nucleoskeleton attachment of ribosomal genes between different cell types studied. As we have shown in Weipoltshammer et al. 1996b, in nucleoskeleton preparations of HeLa cells, the A fragment remains in the nucleus, whereas the DHH fragment is almost completely removed. All these nucleoskeleton preparations were performed several times with each cell type (unstimulated/stimulated lymphocytes, HeLa cells growing in suspension), and the results were highly reproducible. The reasons for these differences are unknown. However, one can speculate that the differences are due to the cell types studied. In contrast to lymphocytes of the peripheral blood, HeLa cells are malignant, long-term cultured cells. They show alterations in genome (more rDNA gene repeats present and presumably different transcriptional activity of ribosomal genes).
Thus, the nucleoskeleton attachment characteristics of ribosomal genes are dependent on the level of transcriptional activation of ribosomal genes. The results obtained by our study cannot be explained only by one model of nucleoskeleton attachment. Concerning the nature of the interaction of ribosomal genes with the nucleoskeleton, it is most probable that the attachment characteristics we observe result from a combination of sequence-specific DNA/nucleoskeleton attachments (attached either directly or via a mediating protein complex), and of functional attachments, mediated above all by RNA polymerase I transcriptional complexes.
Conclusions
The results demonstrate that: (a) the various repetitive DNA sequences differ significantly in their intranuclear anchoring, (b) telomeric rather than centromeric DNA sequences form stable attachments with the nucleoskeleton, and (c) the activation of nucleolar transcription is connected with a spatial rearrangement of specific rDNA elements relative to the nucleoskeleton. In conclusion, we can observe that very stable DNA/nucleoskeleton attachment sites exist that seem to be independent of cell type and activation state of the cell such as the telomeres (Ludérus et al. 1996). It can be speculated that telomeres play an important role in the formation of chromosome order in interphase nuclei. Vice versa, the majority of the centromeric alpha-satellite sequences can be removed from the nucleus independent of the activation state of the cell. There also exist, however, DNA stretches, like the ribosomal gene repeats, where an attachment to a nucleoskeleton is not only dependent on the activation state of the DNA fragment/gene in question (actively transcribed versus silent genes), but probably also on the cell type. Furthermore, our results on ribosomal genes indicate that more than one attachment mechanism has to be taken into account. Additional studies will be necessary to understand properly the nature of these attachment mechanisms and their functional significance.
Acknowledgments
This work was supported by KONTAKT/AKTION Program II.26/ME 279, by the Grant Agency of the Academy of Sciences of the Czech Republic (A5039701) and by the Grant Agency of the Czech Republic (304/98/1035).
References
- Allshire R.C., Dempster M., Hastie N.D. Human telomeres contain at least three types of G-rich repeat distributed non-randomly. Nucleic Acids Res. 1989;17:4611–4627. doi: 10.1093/nar/17.12.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allsopp R.C., Chang E., Kashefi-Aazam M., Rogaev E.I., Piatyszek M.A., Shay J.W., Harley C.B. Telomere shortening is associated with cell division in vitro and in vivo. Exp. Cell Res. 1995;220:194–200. doi: 10.1006/excr.1995.1306. [DOI] [PubMed] [Google Scholar]
- Allsopp R.C., Harley C.B. Evidence for a critical telomere length in senescent human fibroblasts. Exp. Cell Res. 1995;219:130–136. doi: 10.1006/excr.1995.1213. [DOI] [PubMed] [Google Scholar]
- Azzalin C.M., Mucciolo E., Bertoni L., Giulotto E. Fluorescence in situ hybridization with a synthetic (T2AG3)n polynucleotide detects several intrachromosomal telomere-like repeats on human chromosomes. Cytogenet. Cell Genet. 1997;78:112–115. doi: 10.1159/000134640. [DOI] [PubMed] [Google Scholar]
- Barinaga M. Cells count proteins to keep their telomeres in line. Science. 1997;275:92. doi: 10.1126/science.275.5302.928. [DOI] [PubMed] [Google Scholar]
- Berezney R., Coffey D.S. Nuclear protein matrixassociation with newly synthesized DNA. Science. 1975;189:291–293. doi: 10.1126/science.1145202. [DOI] [PubMed] [Google Scholar]
- Berezney R., Coffey D.S. Nuclear matrixisolation and characterization of a framework structure from rat liver nuclei. J. Mol. Biol. 1977;73:616–637. doi: 10.1083/jcb.73.3.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billia F., De Boni U. Localization of centromeric satellite and telomeric DNA sequences in dorsal root ganglion neurons, in vitro. J. Cell Sci. 1991;100:219–226. doi: 10.1242/jcs.100.1.219. [DOI] [PubMed] [Google Scholar]
- Blasco M.A., Lee H.W., Hande M.P., Samper E., Lansdorp P.M., DePinho R.A., Greider C.W. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- Boulikas T. Chromatin domains and prediction of MAR sequences. Int. Rev. Cytol. 1995;162:279–388. doi: 10.1016/s0074-7696(08)61234-6. [DOI] [PubMed] [Google Scholar]
- Brown K.E., Guest S.S., Smale S.T., Hahm K., Merkenschlager M., Fisher A.G. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Cockell M., Renauld H., Watt P., Gasser S.M. Sif2p interacts with Sir4p amino-terminal domain and antagonizes telomeric silencing in yeast. Curr. Biol. 1998;8:787–790. doi: 10.1016/s0960-9822(98)70304-5. [DOI] [PubMed] [Google Scholar]
- Craig J.M., Boyle S., Perry P., Bickmore W.A. Scaffold attachments within the human genome. J. Cell Sci. 1997;110:2673–2682. doi: 10.1242/jcs.110.21.2673. [DOI] [PubMed] [Google Scholar]
- Cremer T., Kurz A., Zirbel R., Dietzel S., Rinke B., Schröck E., Speicher M.R., Mathieu U., Jauch A., Emmerich P. Role of chromosome territories in the functional compartmentalization of the cell nucleus. Cold Spring Harbor Symp. Quant. Biol. 1993;53:777–792. doi: 10.1101/sqb.1993.058.01.085. [DOI] [PubMed] [Google Scholar]
- De Boer R.J., Noest A.J. T cell renewal rates, telomerase, and telomere length shortening. J. Immunol. 1998;160:5832–5837. [PubMed] [Google Scholar]
- de Lange T. Human telomeres are attached to the nuclear matrix. EMBO (Eur. Mol. Biol. Organ.) J. 1992;11:717–724. doi: 10.1002/j.1460-2075.1992.tb05104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson P., Cook P.R., Jackson D.A. Active RNA polymerase I is fixed within the nucleus of HeLa cells. EMBO (Eur. Mol. Biol. Organ.) J. 1990;9:2207–2214. doi: 10.1002/j.1460-2075.1990.tb07390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks R.W., Raap A.K., Van Minnen J., Vreugdenhil E., Smit A.B., Van Der Ploeg M. Detection of mRNA molecules coding for neuropeptide hormones of the pond snail Lymnaea stagnalis by radioactive and non-radioactive in situ hybridizationa model study for mRNA detection. J. Histochem. Cytochem. 1989;37:7–14. doi: 10.1177/37.1.2642295. [DOI] [PubMed] [Google Scholar]
- Gerdes M.G., Carter K.C., Moen P.T., Jr., Lawrence J.B. Dynamic changes in the higher-level chromatin organization of specific sequences revealed by in situ hybridization to nuclear halos. J. Cell Biol. 1994;126:289–304. doi: 10.1083/jcb.126.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Getzenberg R.H., Pienta K.J., Ward W.S., Coffey D.S. Nuclear structure and the three-dimensional organization of DNA. J. Cell. Biochem. 1991;47:289–299. doi: 10.1002/jcb.240470402. [DOI] [PubMed] [Google Scholar]
- Gonzales I.L., Sylvester J.E. Complete sequence of the 43-kb human ribosomal DNA repeatanalysis of the intergenic spacer. Genomics. 1995;27:320–328. doi: 10.1006/geno.1995.1049. [DOI] [PubMed] [Google Scholar]
- Gotta M., Laroche T., Formenton A., Maillet L., Scherthan H., Gasser S.M. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae . J. Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haaf T., Schmid M. Chromosome topology in mammalian interphase nuclei. Exp. Cell Res. 1991;192:325–332. doi: 10.1016/0014-4827(91)90048-y. [DOI] [PubMed] [Google Scholar]
- Haaf T., Ward D.C. Structural analysis of α-satellite DNA and centromere proteins using extended chromatin and chromosomes. Hum. Mol. Genet. 1994;3:697–709. doi: 10.1093/hmg/3.5.697. [DOI] [PubMed] [Google Scholar]
- Hawley R.S. Unresolvable endingsdefective telomeres and failed separation. Science. 1997;275:1441–1443. doi: 10.1126/science.275.5305.1441a. [DOI] [PubMed] [Google Scholar]
- He D., Brinkley B.R. Structure and dynamic organization of centromeres/prekinetochores in the nucleus of mammalian cells. J. Cell Sci. 1996;109:2693–2704. doi: 10.1242/jcs.109.11.2693. [DOI] [PubMed] [Google Scholar]
- He D., Nickerson J.A., Penman S. Core filaments of the nuclear matrix. J. Cell Biol. 1990;110:569–580. doi: 10.1083/jcb.110.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D., Zeng C., Woods K., Zhong L., Turner D., Busch R.K., Brinkley B.R., Busch H. CENP-Ga new centromeric protein that is associated with the alpha-1 satellite DNA subfamily. Chromosoma (Berl.) 1998;107:189–197. doi: 10.1007/s004120050296. [DOI] [PubMed] [Google Scholar]
- Henderson S., Allsopp R., Spector D., Wang S.S., Harley C. In situ analysis of changes in telomere size during replicative aging and cell transformation. J. Cell Biol. 1996;134:1–12. doi: 10.1083/jcb.134.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker A.J., Appels R. The arrangement of interphase chromosomesstructural and functional aspects. Exp. Cell Res. 1989;185:297–318. doi: 10.1016/0014-4827(89)90301-7. [DOI] [PubMed] [Google Scholar]
- Hozák P. The nucleoskeleton and attached activities. Exp. Cell Res. 1996;229:267–271. doi: 10.1006/excr.1996.0370. [DOI] [PubMed] [Google Scholar]
- Hozák P., Cook P.R., Schöfer C., Mosgöller W., Wachtler F. Site of transcription of ribosomal RNA and intranucleolar structure in HeLa cells. J. Cell Sci. 1994;107:639–648. doi: 10.1242/jcs.107.2.639. [DOI] [PubMed] [Google Scholar]
- Hozák P., Geraud G., Hernandez-Verdun D. Revealing nucleolar architecture by low ionic strength treatment. Exp. Cell Res. 1992;203:128–133. doi: 10.1016/0014-4827(92)90047-c. [DOI] [PubMed] [Google Scholar]
- Hozák P., Hassan A.B., Jackson D.A., Cook P.R. Visualization of replication factories attached to a nucleoskeleton. Cell. 1993;73:361–373. doi: 10.1016/0092-8674(93)90235-i. [DOI] [PubMed] [Google Scholar]
- Hozák P., Sasseville A.M.J., Raymond Y., Cook P.R. Lamin proteins form an internal nucleoskeleton as well as a peripheral lamina in human cells. J. Cell Sci. 1995;108:635–644. doi: 10.1242/jcs.108.2.635. [DOI] [PubMed] [Google Scholar]
- Hozák P., Zatsepina O., Likovksy Z. In situ separation of nucleolar components by hypotonic treatment of cells. Biol. Cell. 1990;68:167–170. doi: 10.1016/0248-4900(90)90303-k. [DOI] [PubMed] [Google Scholar]
- Hyrien O., Maric C., Lucas I. Role of nuclear architecture in the initiation of eukaryotic DNA replication. Biochimie. 1997;79:541–548. doi: 10.1016/s0300-9084(97)82001-9. [DOI] [PubMed] [Google Scholar]
- Ivanchenko M., Avramova Z. Interaction of MAR-sequences with nuclear matrix proteins. J. Cell. Biochem. 1992;50:190–200. doi: 10.1002/jcb.240500209. [DOI] [PubMed] [Google Scholar]
- Jackson D.A. Chromatin domains and nuclear compartmentsestablishing sites of gene expression in eukaryotic nuclei. Mol. Biol. Rep. 1997;24:209–220. doi: 10.1023/a:1006873614521. [DOI] [PubMed] [Google Scholar]
- Jackson D.A., Bartlett J., Cook P.R. Sequences attaching loops of nuclear and mitochondrial DNA to underlying structures in human cellsthe role of transcription units. Nucleic Acids Res. 1996;24:1212–1219. doi: 10.1093/nar/24.7.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D.A., Cook P.R. A general method for preparing chromatin containing intact DNA EMBO (Eur. Mol. Biol. Organ.) J 4 1985. 913 918a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D.A., Cook P.R. Transcription occurs at a nucleoskeleton EMBO (Eur. Mol. Biol. Organ.) J 4 1985. 919 925b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D.A., Cook P.R. Replication occurs at a nucleoskeleton. EMBO (Eur. Mol. Biol. Organ.) J. 1986;5:1403–1410. doi: 10.1002/j.1460-2075.1986.tb04374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D.A., Cook P.R. Visualization of a filamentous nucleoskeleton with a 23 nm axial repeat. EMBO (Eur. Mol. Biol. Organ.) J. 1988;7:3667–3777. doi: 10.1002/j.1460-2075.1988.tb03248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D.A., Yuan J., Cook P.R. A gentle method for preparing cyto- and nucleo-skeletons and associated chromatin. J. Cell Sci. 1988;90:365–378. doi: 10.1242/jcs.90.3.365. [DOI] [PubMed] [Google Scholar]
- Keppel F. Transcribed human ribosomal RNA genes are attached to the nuclear matrix. J. Mol. Biol. 1986;187:15–21. doi: 10.1016/0022-2836(86)90402-x. [DOI] [PubMed] [Google Scholar]
- Lamond A.I., Earnshaw W.C. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- Lansdorp P.M., Verwoerd N.P., van de Rijke F.M., Dragowska V., Little M.T., Dirks R.W., Raap A.K., Tanke H.J. Heterogeneity in telomere length of human chromosomes. Hum. Mol. Genet. 1996;5:685–691. doi: 10.1093/hmg/5.5.685. [DOI] [PubMed] [Google Scholar]
- Lejnine S., Makarov V.L., Langmore J.P. Conserved nucleoprotein structure at the ends of vertebrate and invertebrate chromosomes. Proc. Natl. Acad. Sci. USA. 1995;92:2393–2397. doi: 10.1073/pnas.92.6.2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludérus M.E.E., van Steensel B., Chong L., Sibon O.C.M., Cremers F.F.M., de Lange T. Structure, subnuclear distribution and nuclear matrix association of the mammalian telomeric complex. J. Cell Biol. 1996;135:867–881. doi: 10.1083/jcb.135.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancini M.A., He D., Ouspenski I.I., Brinkley B.R. Dynamic continuity of nuclear and mitotic matrix proteins in the cell cycle. J. Cell. Biochem. 1996;62:158–164. doi: 10.1002/(SICI)1097-4644(199608)62:2%3C158::AID-JCB3%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. Different central nervous system cell types display distinct and nonrandom arrangements of satellite DNA sequences. Proc. Natl. Acad. Sci. USA. 1984;81:3123–3127. doi: 10.1073/pnas.81.10.3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maric C., Hyrien O. Remodelling of chromatin loops does not account for specification of replication origins during Xenopus development. Chromosoma. 1998;107:155–165. doi: 10.1007/s004120050292. [DOI] [PubMed] [Google Scholar]
- Mattern K.A., van Goethem R.E., de Jong L., van Driel R. Major internal nuclear matrix proteins are common to different human cell types. J. Cell. Biochem. 1997;65:42–52. doi: 10.1002/(sici)1097-4644(199704)65:1<42::aid-jcb5>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- Mirkovitch J., Mirault M.E., Laemmli U.K. Organization of the higher-order chromatin loopspecific DNA attachment sites on nuclear scaffold. Cell. 1984;39:223–232. doi: 10.1016/0092-8674(84)90208-3. [DOI] [PubMed] [Google Scholar]
- Mitchell A., Jeppesen P., Hanratty D., Gosden J. The organisation of repetitive DNA sequences on human chromosomes with respect to the kinetochore analysed using a combination of oligonucleotide primers and CREST anticentromere serum. Chromosoma. 1992;101:333–341. doi: 10.1007/BF00346012. [DOI] [PubMed] [Google Scholar]
- Mondello C., Riboni R., Casati A., Nardo T., Nuzzo F. Chromosomal instability and telomere length variations during the life span of human fibroblast clones. Exp. Cell Res. 1997;236:385–396. doi: 10.1006/excr.1997.3756. [DOI] [PubMed] [Google Scholar]
- Murphy T.D., Karpen G.H. Centromeres take flightalpha satellite and the quest for the human centromere. Cell. 1998;93:317–320. doi: 10.1016/s0092-8674(00)81158-7. [DOI] [PubMed] [Google Scholar]
- Nakayasu H., Berezney R. Nuclear matrinsidentification of the major nuclear matrix proteins. Proc. Natl. Acad. Sci. USA. 1991;88:10312–10316. doi: 10.1073/pnas.88.22.10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickerson J.A., Krockmalnic G., Wan K.M., Penman S. The nuclear matrix revealed by eluting chromatin from a cross-linked nucleus. Proc. Natl. Acad. Sci. USA. 1997;94:4446–4450. doi: 10.1073/pnas.94.9.4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notaro R., Cimmino A., Tabarini D., Rotoli B., Luzzatto L. In vivo telomere dynamics of human hematopoietic stem cells. Proc. Natl. Acad. Sci. USA. 1997;94:13782–13785. doi: 10.1073/pnas.94.25.13782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs R.L., Press R.I. Centromere autoantigens are associated with the nucleolus. Exp. Cell Res. 1992;200:339–350. doi: 10.1016/0014-4827(92)90181-7. [DOI] [PubMed] [Google Scholar]
- Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkel D., Straume T., Gray J.W. Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. Proc. Natl. Acad. Sci. USA. 1986;83:2934–2938. doi: 10.1073/pnas.83.9.2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl C. Über Zelltheilung. Gegenbaur's Morphol. Jahrb. 1885;10:214–330. [Google Scholar]
- Raska I. Nuclear ultrastructures associated with the RNA synthesis and processing. J. Cell. Biochem. 1995;59:11–26. doi: 10.1002/jcb.240590103. [DOI] [PubMed] [Google Scholar]
- Rattner J.B. The structure of the mammalian centromere. Bioessays. 1991;13:51–56. doi: 10.1002/bies.950130202. [DOI] [PubMed] [Google Scholar]
- Razin S.V. The nuclear matrix and spatial organization of chromosomal DNA domains 1997. Medical Intelligence Unit, Springer Verlag; Heidelberg, Germany: pp. 35–61 [Google Scholar]
- Schwarzacher H.G., Wachtler F. The nucleolus. Anat. Embryol. 1993;188:515–536. doi: 10.1007/BF00187008. [DOI] [PubMed] [Google Scholar]
- Shaw P.J., Jordan E.G. The nucleolus. Annu. Rev. Cell Dev. Biol. 1995;11:93–121. doi: 10.1146/annurev.cb.11.110195.000521. [DOI] [PubMed] [Google Scholar]
- Sirri V., Roussel P., Trere D., Derenzini M., Hernandez-Verdun D. Amount variability of total and individual Ag-NOR proteins in cells stimulated to proliferate. J. Histochem. Cytochem. 1995;43:887–893. doi: 10.1177/43.9.7642962. [DOI] [PubMed] [Google Scholar]
- Slijepcevic P., Hande M.P., Bouffler S.D., Lansdorp P., Bryant P.E. Telomere length, chromatin structure and chromosome fusigenic potential. Chromosoma. 1997;106:413–421. doi: 10.1007/s004120050263. [DOI] [PubMed] [Google Scholar]
- Smith H.C., Rothblum L.I. Ribosomal DNA sequences attached to the nuclear matrix. Biochem. Genet. 1987;25:863–879. doi: 10.1007/BF00502606. [DOI] [PubMed] [Google Scholar]
- Stein G.S., van Wijnen A.J., Stein J.L., Lian J.B., Pockwinse S., McNeil S. Interrelationships of nuclear structure and transcriptional controlfunctional consequences of being in the right place at the right time. J. Cell. Biochem. 1998;70:200–212. [PubMed] [Google Scholar]
- Stephanova E., Stancheva R., Avramova Z. Binding of sequences from the 5′- and 3′-nontranscribed spacers of the rat rDNA locus to the nucleolar matrix. Chromosoma. 1993;102:287–295. doi: 10.1007/BF00352403. [DOI] [PubMed] [Google Scholar]
- Strissel P.L., Espinosa R., III, Rowley J.D., Swift H. Scaffold attachment regions in centromere-associated DNA. Chromosoma. 1996;105:122–133. doi: 10.1007/BF02509522. [DOI] [PubMed] [Google Scholar]
- van Steensel B., Smogorzewska A., de Lange T. TRF2 protects human telomeres from end-to-end fusion. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- Vaziri H., Schächter F., Uchida I., Wei L., Zhu X., Effros R., Cohen D., Harley C.B. Loss of telomeric DNA during aging of normal and trisomy 21 human lymphocytes. Am. J. Hum. Genet. 1993;52:661–667. [PMC free article] [PubMed] [Google Scholar]
- Wachtler F., Hopman A.H., Wiegant J., Schwarzacher H.G. On the position of nucleolus organizer regions (NORs) in interphase nuclei. Exp. Cell Res. 1986;167:227–240. doi: 10.1016/0014-4827(86)90219-3. [DOI] [PubMed] [Google Scholar]
- Wachtler F., Schöfer C., Schedle A., Schwarzacher H.G., Hartung M., Stahl A., Gonzalez I., Sylvester J. Transcribed and nontranscribed parts of the human ribosomal gene repeat show a similar pattern of distribution in nucleoli. Cytogenet. Cell Genet. 1991;57:175–178. doi: 10.1159/000133140. [DOI] [PubMed] [Google Scholar]
- Wachtler F., Schwarzacher H.G., Ellinger A. The influence of the cell cycle on structure and number of nucleoli in cultured human lymphocytes. Cell Tissue Res. 1982;225:155–163. doi: 10.1007/BF00216225. [DOI] [PubMed] [Google Scholar]
- Weipoltshammer K., Schöfer C., Almeder M., Sylvester J., Wachtler F. Spatial distribution of sex chromosomes and ribosomal genesa study on human lymphocytes and testicular cells Cytogenet. Cell Genet 73 1996. 108 113a [DOI] [PubMed] [Google Scholar]
- Weipoltshammer K., Schöfer C., Wachtler F., Hozák P. The transcription unit of ribosomal genes is attached to the nuclear skeleton Exp. Cell Res 227 1996. 374 379b [DOI] [PubMed] [Google Scholar]
- Weng N., Palmer L.D., Levine B.L., Lane H.C., June C.H., Hodes R.J. Tales of tailsregulation of telomere length and telomerase activity during lymphocyte development, differentiation, activation, and aging. Immunol. Rev. 1997;160:43–54. doi: 10.1111/j.1600-065x.1997.tb01026.x. [DOI] [PubMed] [Google Scholar]
- Wood K.W., Sakowicz R., Goldstein L.S.B., Cleveland D.W. CENP-E is a plus end-directed kinetochore motor required for metaphase chromosome alignment. Cell. 1997;91:357–366. doi: 10.1016/s0092-8674(00)80419-5. [DOI] [PubMed] [Google Scholar]
- Zakian V.A. Telomeresbeginning to understand the end. Science. 1995;270:1601–1607. doi: 10.1126/science.270.5242.1601. [DOI] [PubMed] [Google Scholar]