Abstract
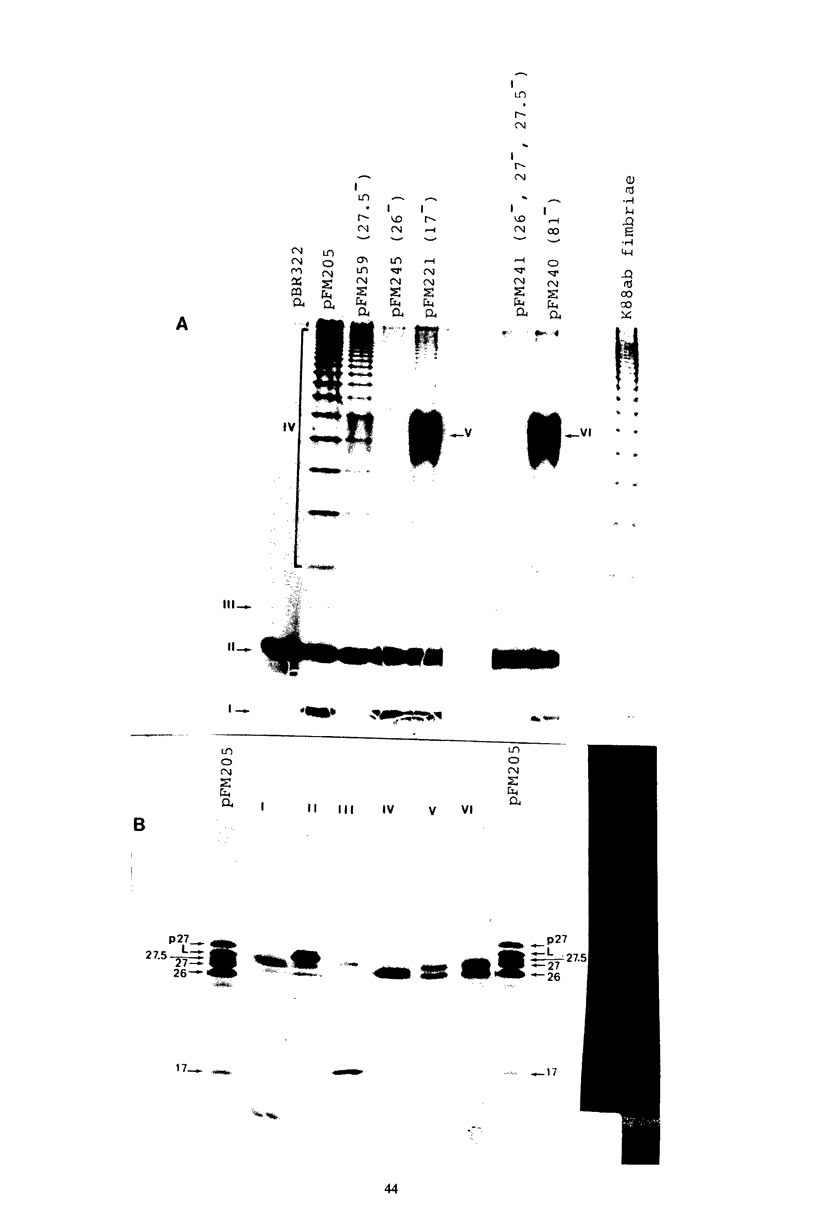
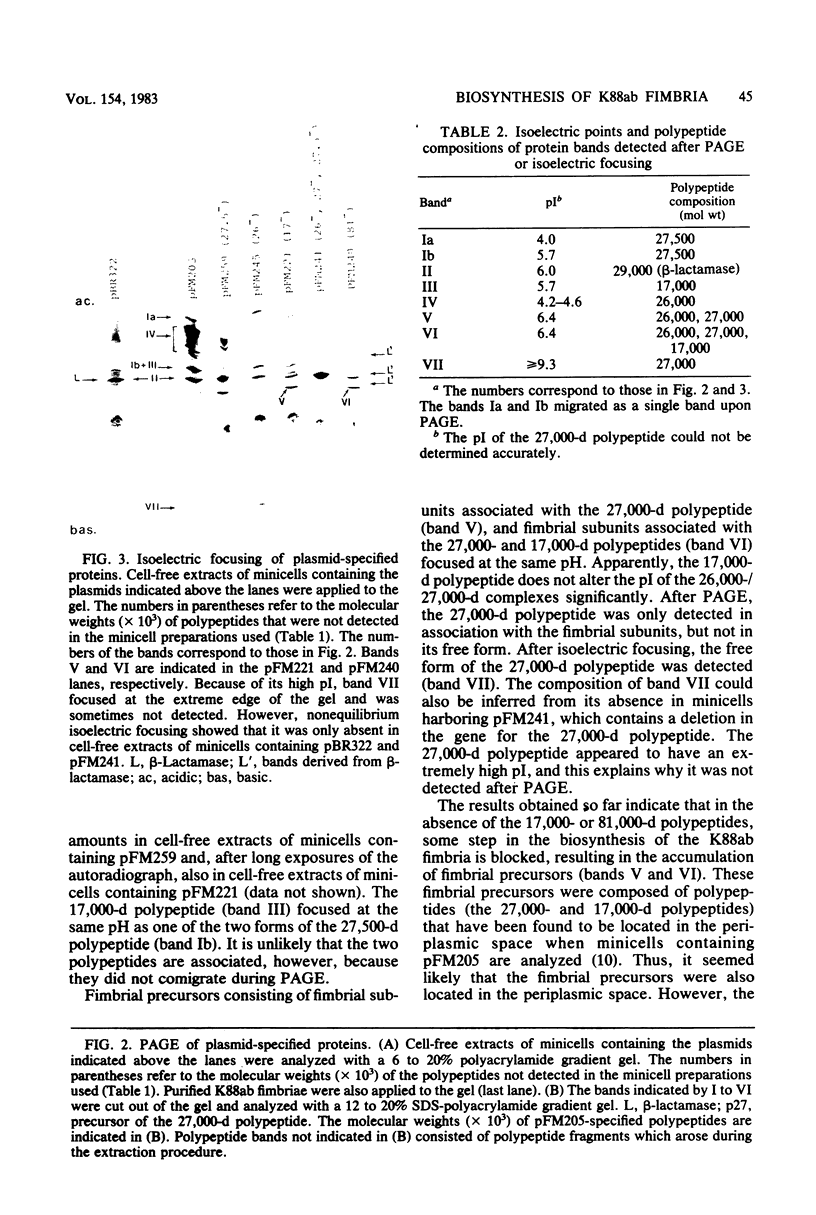
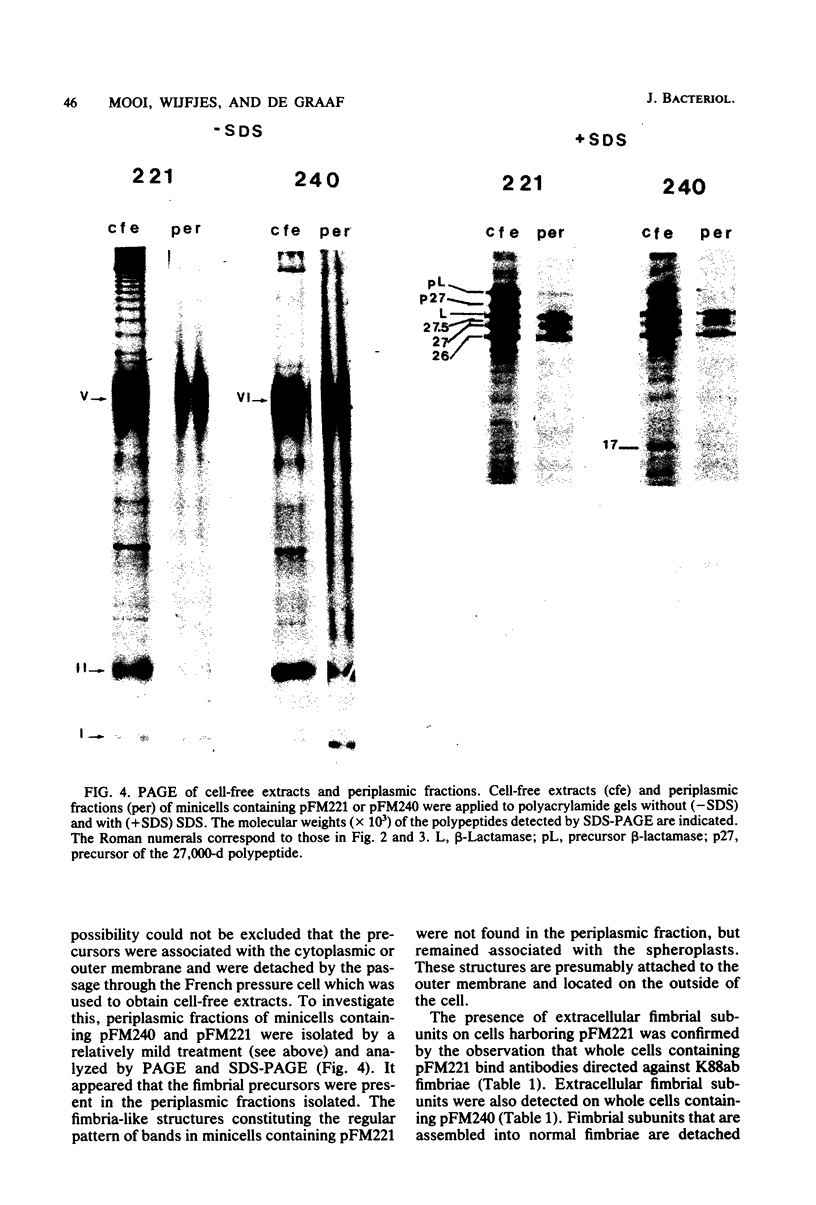
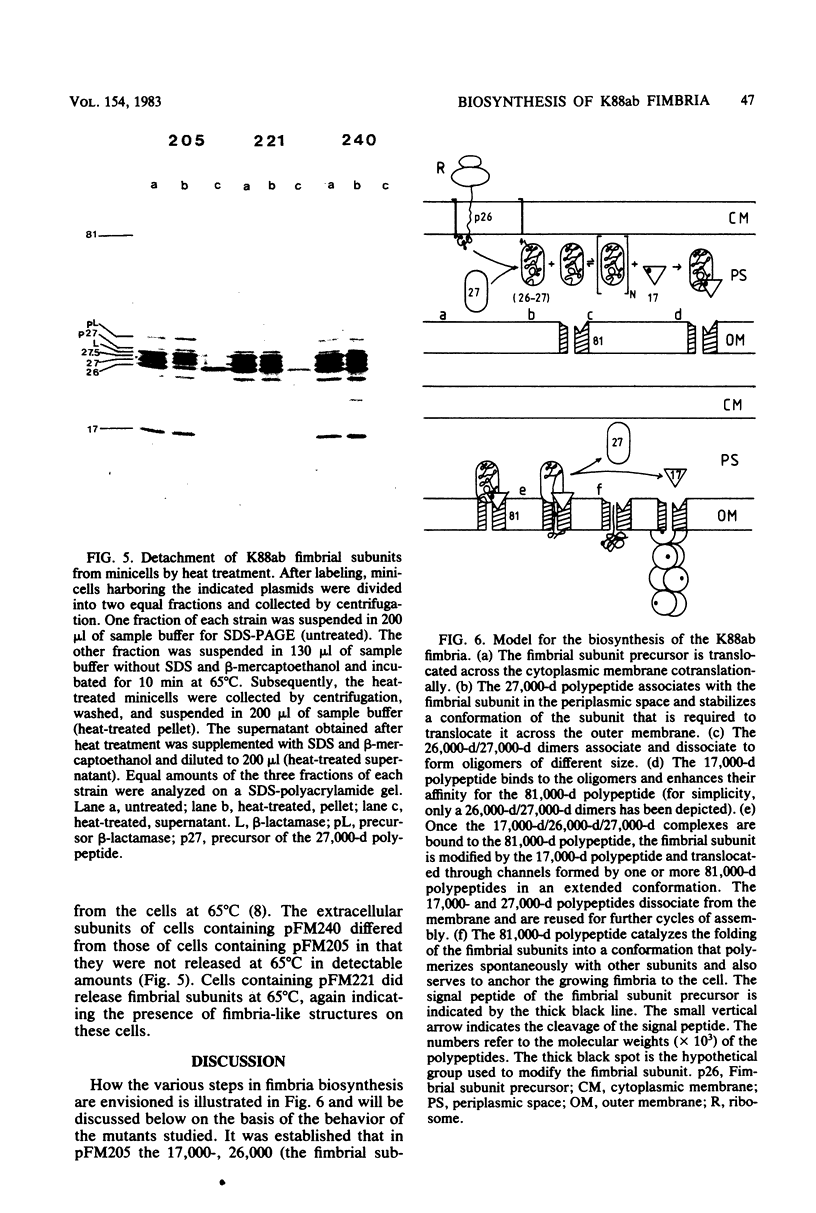
Four genes encoding for polypeptides with apparent molecular weights of 17,000, 26,000 (the fimbrial subunit), 27,000, and 81,000 have been implicated in the biosynthesis of the K88ab fimbria (Mooi et al., J. Bacteriol. 150:512-521, 1982). Escherichia coli mutants with defects in these genes were examined for the presence of fimbrial precursors. An analysis of these mutants revealed that fimbrial subunits accumulated transiently in the periplasmic space before being translocated across the outer membrane. The 81,000-dalton (d) polypeptide is probably involved in translocating fimbrial subunits across the outer membrane, because in the absence of this polypeptide the fimbrial subunits remained in the periplasmic space, where they were found to be associated with the 17,000- and 27,000-d polypeptides. In mutants with a deletion in the gene for the 27,000-d polypeptide, fimbrial precursors were not detected, because the fimbrial subunits were degraded. The 27,000-d polypeptide might be involved in stabilizing a conformation of the fimbrial subunit required to translocate it across the outer membrane. In the absence of the 17,000-d polypeptide, most fimbrial subunits were found in the periplasmic space associated with the 27,000-d polypeptide. However, small amounts of subunits were also translocated across the outer membrane. These extracellular subunits did not adhere to brushborders, suggesting that fimbrial subunits must be modified by the 17,000-d polypeptide to be assembled into functional fimbriae. A model for the biosynthesis of the K88ab fimbria is proposed.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bertschinger H. U., Moon H. W., Whipp S. C. Association of Escherichia coli with the small intestinal epithelium. I. Comparison of enteropathogenic and nonenteropathogenic porcine strains in pigs. Infect Immun. 1972 Apr;5(4):595–605. doi: 10.1128/iai.5.4.595-605.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Davis B. D., Tai P. C. The mechanism of protein secretion across membranes. Nature. 1980 Jan 31;283(5746):433–438. doi: 10.1038/283433a0. [DOI] [PubMed] [Google Scholar]
- Dougan G., Sherratt D. The transposon Tn1 as a probe for studying ColE1 structure and function. Mol Gen Genet. 1977 Mar 7;151(2):151–160. doi: 10.1007/BF00338689. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mooi F. R., Harms N., Bakker D., de Graaf F. K. Organization and expression of genes involved in the production of the K88ab antigen. Infect Immun. 1981 Jun;32(3):1155–1163. doi: 10.1128/iai.32.3.1155-1163.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooi F. R., Wouters C., Wijfjes A., de Graaf F. K. Construction and characterization of mutants impaired in the biosynthesis of the K88ab antigen. J Bacteriol. 1982 May;150(2):512–521. doi: 10.1128/jb.150.2.512-521.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooi F. R., de Graaf F. K., van Embden J. D. Cloning, mapping and expression of the genetic determinant that encodes for the K88ab antigen. Nucleic Acids Res. 1979 Mar;6(3):849–865. doi: 10.1093/nar/6.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]